Abstract
The relationships among 55 wheat accessions (47 accessions collected from Iran and eight accessions provided by the Institute of Plant Biology of the University of Zurich, Switzerland) belonging to eight species carrying A genome (Triticum monococcum L., T. boeoticum Boiss., T. urartu Tumanian ex Gandilyan, T. durum Desf., T. turgidum L., T. dicoccum Schrank ex Schübler, T. dicoccoides (Körn. ex Asch. & Graebner) Schweinf. and T. aestivum L.) were evaluated using 31 A genome specific microsatellite markers. A high level of polymorphism was observed among the accessions studied (PIC = 0.77). The highest gene diversity was revealed among T. durum genotypes, while the lowest genetic variation was found in T. dicoccoides accessions. The analysis of molecular variance (AMOVA) showed a significant genetic variance (75.56%) among these accessions, representing a high intra-specific genetic diversity within Triticum taxa in Iran. However, such a variance was not observed among their ploidy levels. Based on the genetic similarity analysis, the accessions collected from Iran were divided into two main groups: diploids and polyploids. The genetic similarity among the diploid and polyploid species was 0.85 and 0.89 respectively. There were no significant differences in A genome diversity from different geographic regions. Based on the genetic diversity analyses, we consider there is value in a greater sampling of each species in Iran to discover useful genes for breeding purposes.
Keywords: Triticum, SSRs, Iran, wheat, genetic analysis
1. Introduction
The genus Triticum L. is one of the most important genera in the tribe Triticeae and has been the focus of many biosystematic studies. Four basic genomes, A, B, D and G are involved in the genomic constitution of all Triticum species [1,2]. The ancestral diploid species of A, B and D genome have diverged from a common ancestor about three million years ago [3]. From these ancestral diploids, two species hybridized somewhere along the Fertile Crescent to form the first tetraploid Triticum species [4]. The processes of polyploidization and genomic differentiation finally resulted in the present day genus Triticum with a ploidy series of di-, tetra- and hexaploid species, all based on x = 7 [5]. The A and D genomes which are less differentiated from those of the parental diploids, are considered as pivotal genomes [6,7]. Many reports indicated that the A genome has suffered different changes in T. urarto Thum. ex Gandil. (AuAu) and T. boeoticum Boiss. (AbAb) [2,8].
Since wheat cultivation commenced, the breeding and selection of particular genotypes have resulted in enormous loss of alleles and limited the genetic diversity of modern wheat cultivars [9,10]. Therefore, the remaining variability in the cultivated wheat gene pool is insufficient to address current and future breeding efforts [11]. For that reason, there is an essential and urgent need to explore the genetic potential among natural populations of wheat species and their closely related taxa. Germplasm accessions distinct from modern wheat cultivars are predicted to contain potentially useful alleles to broaden the genetic base of wheat [12].
Since the bread wheat (T. aestivum) most probably originated from the south eastern or south western Caspian Sea in Iran [13–15], the wild species and populations growing in Iran, as one of the putative centers of origin of cultivated wheat, can be valuable from this point of view. This opinion is strengthened by the fact that the chromosomes of A genome carry important genes such as adult plant resistance genes [16], milling yield genes [17], flour color genes[18], white salted noodle quality genes [19], supernumerary spikelet (SS) genes [20], sprouting resistance genes [21], chlorophyll synthesis genes [22], total florets per spike genes [23], cold tolerance genes [24], size of stomata genes [25], forest resistance genes [26,27] and yield traits such as tiller number, heading date and plant height genes [28]. Many workers have studied the Triticum species from different points of view: morphology [29,30], isozymes [14,31,32] restriction fragment length polymorphismes (RFLPs) [33–35], and microsatellites [36–39]. A high level of polymorphism in RFLPs and microsatellites among Triticum species accessions has been detected [37,40–43].
Microsatellites or simple sequence repeats (SSRs) have become the markers of choice among a variety of different molecular markers in order to evaluate genetic diversity and phylogenetic relationships [44,45]. It has been demonstrated that microsatellites are highly informative markers in many plant species [40,41,46–61] and it is believed that microsatellites show a much higher level of polymorphism in hexaploid wheat than any other marker systems.
More than a thousand wheat mapped microsatellite markers are available that are useful tools for genetic analyses. Genomic SSRs have been used in wheat for a variety of purposes including genomic mapping [33,40,62,63], gene tagging [39,64–66] and genetic diversity [41,67,68] analyses.
This study was aimed to use SSR markers to estimate the level of A genome polymorphism and to identify the relationships among the species carrying A genome of the genus Triticum native to Iran.
2. Results and Discussion
All 31 A genome specific SSR primers yielded 410 bands (alleles) from genomic DNA of all 55 accessions of eight A genome containing Triticum species from which 316 (0.77) were polymorphic (Table 1).
Table 1.
Amplification of the homologous microsatellites in 55 accessions of the genus Triticum using 31 primer sets originally designed for the microsatellites of A genome (for the primer sequence see Röder et al. 1998 [36]).
| Marker | Chr. Loc. | Ann. Temp. | Allele Fr. | Allele No | HE | HO | PIC |
|---|---|---|---|---|---|---|---|
| gwm-601 | 4A | 60 | 0.37 | 15 | 0.7 | 0.85 | 0.66 |
| gwm-135 | 1A | 60 | 0.28 | 14 | 0.84 | 0.66 | 0.83 |
| gwm-71 | 2A | 60 | 0.22 | 18 | 0.86 | 0.86 | 0.85 |
| gwm-666 | 1A, 3A, 5A, 7A | 60 | 0.22 | 22 | 0.86 | 0.98 | 0.84 |
| gwm-311 | 2A, 2B, 6B | 60 | 0.23 | 12 | 0.86 | 0.27 | 0.85 |
| gwm-359 | 2A | 55 | 0.23 | 14 | 0.86 | 0.75 | 0.85 |
| gwm-512 | 2A | 60 | 0.29 | 5 | 0.75 | 0.12 | 0.70 |
| gwm-372 | 2A | 60 | 0.23 | 14 | 0.88 | 0.24 | 0.87 |
| gwm-391 | 3A | 55 | 0.22 | 18 | 0.79 | 0.81 | 0.77 |
| gwm-757 | 3A | 60 | 0.27 | 14 | 0.84 | 0.74 | 0.83 |
| gwm-155 | 3A | 60 | 0.34 | 8 | 0.77 | 0 | 0.74 |
| gwm-291 | 5A | 60 | 0.41 | 16 | 0.77 | 0.59 | 0.74 |
| gwm-494 | 6A, 4A, 3A, 1B | 60 | 0.34 | 12 | 0.77 | 0.87 | 0.74 |
| gwm-427 | 6A | 50 | 0.13 | 20 | 0.92 | 0.24 | 0.92 |
| gwm-635 | 7A, 7B, 7D | 60 | 0.20 | 11 | 0.86 | 0.63 | 0.85 |
| gwm-332 | 7A | 60 | 0.23 | 12 | 0.85 | 0.67 | 0.84 |
| gwm-296 | 2A, 2D, 7D | 55 | 0.14 | 18 | 0.78 | 0.49 | 0.76 |
| gwm-471 | 7A, 7B | 60 | 0.26 | 12 | 0.85 | 0.39 | 0.84 |
| gwm-260 | 7A | 55 | 0.21 | 13 | 0.87 | 0.83 | 0.86 |
| gwm-459 | 6A | 55 | 0.46 | 9 | 0.73 | 0.25 | 0.71 |
| gwm-179 | 5A | 55 | 0.31 | 5 | 0.77 | 0.62 | 0.74 |
| gwm-382 | 2A, 2B, 2D | 60 | 0.27 | 15 | 0.86 | 0.26 | 0.85 |
| gwm-205 | 5A, 5D | 60 | 0.23 | 19 | 0.9 | 0.8 | 0.89 |
| gwm-136 | 1A | 60 | 0.41 | 6 | 0.68 | 0.5 | 0.63 |
| wmc-104 | 1A, 6B | 55 | 0.44 | 10 | 0.74 | 0.22 | 0.72 |
| barc-56 | 5A | 55 | 0.32 | 15 | 0.78 | 0.5 | 0.75 |
| barc-151 | 5A, 7A | 55 | 0.19 | 13 | 0.88 | 0.14 | 0.87 |
| cfa-2086 | 2A | 60 | 0.19 | 17 | 0.86 | 0.46 | 0.85 |
| cfa-2028 | 7A | 55 | 0.33 | 9 | 0.75 | 0.72 | 0.72 |
| cfa-2262 | 3A | 55 | 0.22 | 13 | 0.77 | 0.2 | 0.74 |
| cfa-2263 | 2A | 60 | 0.17 | 11 | 0.88 | 0.25 | 0.87 |
| Mean | 0.29 | 12.8 | 0.79 | 0.49 | 0.77 | ||
| Sum | 410 | ||||||
The number of alleles per microsatellite ranged from 5 (Xgwm512 and Xgwm179) to 22 (Xgwm666) with an average of 12.8 alleles per locus (Table 1). Major allele frequency ranged from 0.13 to 0.46 averaging 0.29 (Table 1). The mean value for polymorphism information content (PIC) for all microsatellites was 0.77. The microsatellite Xgwm427 with 20 alleles had the highest (0.92) and the microsatellite Xgwm136 with 6 alleles had the lowest (0.63) PIC value (Table 1).
2.1. Genetic Similarity Analysis
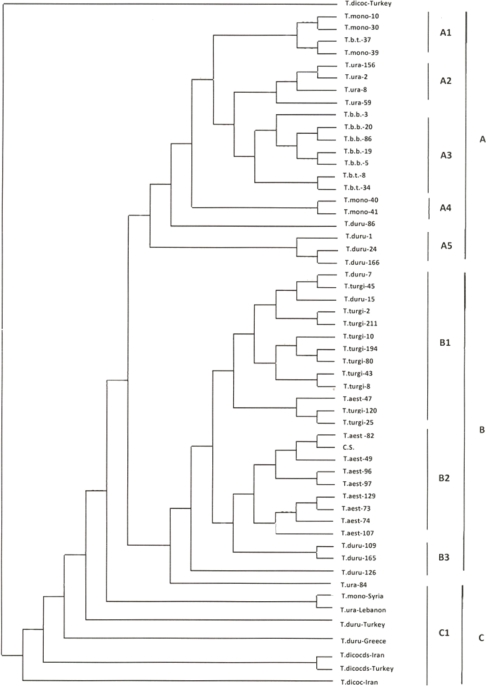
The results distinguished all the 55 accessions (Figure 1), from which 46 were divided into two major groups designated as A and B in Figure 1 with 100% bootstrap support (data not shown).
Figure 1.
A genetic similarity based dendrogram showing relationships among Triticum accessions using 31 microsatellite markers. The main groups are denoted on the right side as A, B and C and the sub-groups as A1, A2, A3, A4, A5, B1, B2, B3 and C1. (T.mono = Triticum monococcum, T.b.t. = T. boeoticum subsp. taodar, T.b.b. = T. boeoticum subsp. boeoticum, T.ura = T. urartu, T.duru = T. durum, T.turgi. = T. turgidum, T.dicoc = T. dicoccum, T.dicocds = T. dicoccoides, T.aest = T. aestivum, and C.S. = Chinese spring).
These two groups, with several subgroups, were heterogeneous. The accessions of diploid species were grouped with considerable genetic similarities (except T.ura-84). Four accessions of tetraploid cultivated wheat T. durum were grouped with diploid accessions (group A, Figure 1). The group B included 14 tetraploid, 11 hexaploid and one diploid accession. The remaining eight accessions (provided by the Institute of Plant Biology, University of Zurich, Switzerland) were not grouped with the above main groups, and were clearly separated from the Iranian ones (group C, Figure 1).
At the species level (Table 2), the highest genetic similarity (0.89) was found between T. aestivum and T. durum; although T. aestivum and T. turgidum with a genetic similarity of 0.86 appeared relatively close too. The two species T. dicoccum and T. dicoccoides with 0.64 and 0.67 genetic similarity respectively, were grouped well away from the other species, indicating that the A genome in tetraploids was distant from the genome in the diploid and polyploid species.
Table 2.
The analysis of genetic similarity between A genomes of diploid and diploid, diploid and tetraploid, diploid and hexaploid, tetraploid and tetraploid, and tetraploid and hexaploid pair species of 55 accessions belonging to eight Triticum L. species as revealed by SSR markers.
| Groups | Species | Genetic Similarity |
|---|---|---|
| diplo & diplo | T. monococcum & T.boeoticum | 0.89 |
| T. monococcum & T. urartu | 0.90 | |
| T.boeoticum & T. urartu | 0.90 | |
| tetra & tetra | T. durum & T. turgidum | 0.86 |
| T. durum & T. dicoccum | 0.79 | |
| T. durum & T. dicoccoides | 0.78 | |
| T. turgidum & T. dicoccum | 0.79 | |
| T. turgidum & T. dicoccoides | 0.78 | |
| T. dicoccum & T. dicoccoides | 0.70 | |
| diplo & tetra | T. monococcum & T. durum | 0.85 |
| T. monococcum & T. turgidum | 0.66 | |
| T. monococcum & T. dicoccum | 0.74 | |
| T.monococcum&T.dicoccoides | 0.74 | |
| T. boeoticum & T. durum | 0.82 | |
| T. boeoticum & T. turgidum | 0.65 | |
| T. boeoticum & T. dicoccum | 0.66 | |
| T. boeoticum & T. dicoccoides | 0.69 | |
| T. urartu & T. durum | 0.84 | |
| T. urartu & T. turgidum | 0.64 | |
| T. urartu & T. dicoccum | 0.75 | |
| T. urartu & T. dicoccoides | 0.76 | |
| diplo & hexa | T. monococcum & T. aestivum | 0.77 |
| T. boeoticum & T. aestivum | 0.72 | |
| T. urartu & T. aestivum | 0.72 | |
| tetra & hexa | T. durum & T. aestivum | 0.89 |
| T. turgidum & T. aestivum | 0.86 | |
| T. dicoccum & T. aestivum | 0.64 | |
| T. dicoccoides & T. aestivum | 0.67 |
In the UPGMA dendrogram (Figure 1), the eight Triticum species studied were divided into three groups: (1) three diploids (T. monococcum, T. boeoticum, T. urartu), (2) three cultivated wheats (T. aetivum, T. durum and T. turgidum), and (3) two tetraploids (T. dicoccum and T. dicoccoides).
2.2. Analysis of Molecular Variance (AMOVA)
The main portion of genetic variance (75.56%) was attributed to the variation among populations within species. A significant genetic variation (17.44%) was calculated between different species. There was no significant difference between A genome of species with different ploidy levels (Table 3).
Table 3.
The analysis of molecular variance (AMOVA) of 55 accessions of eight A genome containing species of the genus Triticum calculated at ploidy level (groups), species within each ploidy level (within groups) and accessions of each species (within species).
| Source of variation | d.f | Sum of squares | Mean of squares | Percentage of variation | Variance components | P-value |
|---|---|---|---|---|---|---|
| Among Ploidy levels (groups) | 2 | 235.865 | 117.932 | 7.00 | 2.30879 | <0.001 |
| Among species Within groups | 5 | 286.738 | 57.347 | 17.44 | 5.75630 | <0.001 |
| Among accessions | 47 | 1172.161 | 24.939 | 75.56 | 24.93959 | 0.10948 ± 0.00939 |
| Total | 54 | 1694.764 | 200.218 | 100 | 33.00468 | |
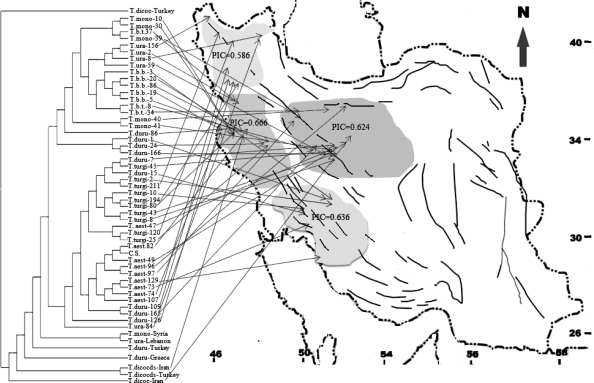
The accessions were mainly collected from Iran as it is considered to be a part of centre of origin of cultivated wheat [15]. The germplasms presented in the centre of origin of a taxon are considered to be more diverse than those growing at the margins of its geographic distribution. The higher genetic diversity observed in this study concur with previous reports [37,41,42,69], indicating Iran as a likely part of the centre of diversity of this genus. The differences can also be attributed to the number of accessions studied, their genetic background, and the number of markers used. A total of 410 polymorphic bands (alleles) detected in this study seem to be enough to assess genetic variation among accessions. Zhang et al. (2002) [70] discussed that the presence of 350–400 alleles is enough for objective assessment of genetic relationship between wheat accessions. From the geographic point of view, the A genome SSR differentiations were not correlated with geographic distribution (Figure 2); however, some groupings related to both taxa and geographic origin (e.g., of T. durum accessions collected from NW and T. boeoticum subsp. boeoticum accessions collected from the West) were evident. The data were able to group geographically closely related collections (Figure 2). There were no significant differences between diversity measures calculated for different regions.
Figure 2.
An integrated illustration of a dendrogram showing relationships among the accessions carrying A genome and the map of their geographic origin. Average PICs of all microsatellites are shown as average in each region.
The recognition of the tetraploid species T. durum as a subspecies of T. turgidum by Kihara (1994) [1] and Mc Fadden and Sears (1966) [71] has been followed up by some other botanists. In spite of high genetic similarity between the two taxa showed in this study, they were enough apart to be considered as a distinct species; this is supported by morphological studies [72]. Analyses at the accessional level (Figure 1) indicated higher genetic distances among the accessions of these two species. The origin of the A genome encountered in the hexaploid wheat has always been under discussion, and its two closely related species, i.e., T. durum and T. turgidum, have been generally known as the putative A genome donors to T. aestivum [71,73]. When one considers the very close genetic similarities between the above tetra- and the latter hexaploid Triticum species provided by this study (0.89 and 0.86 respectively, see Table 2), this notion is strengthened.
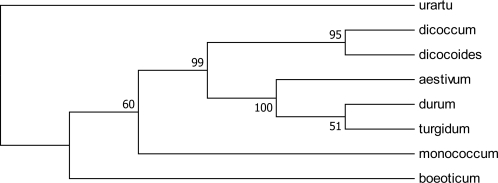
Considering the topology of the UPGMA dendrogram (Figure 3), the tetraploid species were divided into two groups: (1) T. turgidum and T. durum, and (2) T. dicoccoides and T. dicoccum. This can be interpreted either as different post hybridization A genome modifications among the tetraploid species or involving two different origins as the A genome donor to them or preferential gene flow occurring between pair species within each group.
Figure 3.
A bootstrap dendrogram based on genetic distances, constructed using UPGMA method, showing relationships between A genomes of 8 Triticum species.
The SSR analysis showed a close relationship between the diploid Triticum species. Based on the calculated genetic similarities (Table 2), the A genomes occurring among the polyploids appeared to be more similar to that of the diploid species T. monococcum than the other diploids. This observation is partly in accordance with Johnson (1975) [74] and Tsunewaki (1999) [75] who pointed out that the A genome donor to T. aestivum, T. turgidum and T. durum, is T. monococcum, and to T. dicoccoides and T. dicoccum is T. urartu.
3. Experimental Section
A total of 47 accessions particularly collected for this study and eight accessions provided by the Institute of Plant Biology, University of Zurich (Table 4) were examined.
Table 4.
The species name, collection label, genome combination and the origin of accessions used in this study.
| Species | Collection label | Genome | Locality and altitude (m) |
|---|---|---|---|
| T. monococcum | T. mono-30 | A | Kermanshah, Gardaneh Reno (1480) |
| T. mono -10 | A | Kordestan, 3 km to Saghez (1620) | |
| T. mono -41 | A | Isfahan, Semirom to yasooj (2100) | |
| T. mono -39 | A | Arak to Malayer (2020) | |
| T. mono -40 | A | Tehran, Taleghan valley (1850) | |
| T. mono Syria | A | Provided by Institute of Plant Biology of the University of Zurich | |
| T.boeoticum subsp. thaodar | T.b.t.-37 | A | Kordestan, 5 km after Jenan to Saghez (1770) |
| T.b.t.-8 | A | Chaharmahal Bakhtiari, Shahr-e-Kord, Shapoorabad to Jooneghan (2090) | |
| T.b.t.-34 | A | Arak 15 km to Malayer (1840) | |
| T.boeoticum subsp. boeoticum | T.b.b.-19 | A | Ilam to Kermanshah, Gardaneh Reno (1370) |
| T.b.b.-5 | A | Lorestan, 35 km to Khoramabad from Malavi (1100) | |
| T.b.b.-20 | A | Kermanshah 10 km to Harsin (1330) | |
| T.b.b.-86 | A | Kermanshah to Kamyaran (1340) | |
| T.b.b.-3 | A | Kohkiloye & Boyerahmad,Yasooj, Amirabad (1650) | |
| T.urartu | T.ura-156 | A | West Azarbaijan, Makoo (1580) |
| T.ura-84 | A | Ardabil (1320) | |
| T.ura-2 | A | Kordestan, 10 km Saghez from Asadabad (1440) | |
| T.ura-8 | A | Aradbil, 10 km to Kaghazkanan (1349) | |
| T.ura-59 | A | Chaharmahal Bakhtiari, between Gandoman and Lordegan (2080) | |
| T.ura-Lebanon | A | Provided by Institute of Plant Biology of the University of Zurich | |
| T.durum | T.duru-86 | AB | Kermanshah, Kamyaran (1440) |
| T.duru-24 | AB | Lorestan, Malavi toward Khoram Abad (1200) | |
| T.duru-166 | AB | Chahar mahal Bakhtiari, DoAb Samsami (2000) | |
| T.duru-1 | AB | Kohkiloye & Boyerahmad (990) | |
| T.duru-165 | AB | Chahar mahal Bakhtiari, near Chaghakhor lake (2190) | |
| T.duru-109 | AB | West Azarbaijan, Sardasht to Baneh (1050) | |
| T.duru-15 | AB | Khoosestan, Haftgel to Masjed Soleiman (550) | |
| T.duru-126 | AB | Kordestan, Alamoot 6 Km (1660) | |
| T.duru-7 | AB | Chahar mahal Bakhtiari, Borojen to Izeh (2190) | |
| T.duru-Turky | AB | Provided by Institute of Plant Biology of the University of Zurich | |
| T.duru-Greece | AB | Provided by Institute of Plant Biology of the University of Zurich | |
| T. turgidum | T.turgi-211 | AB | West Azarbaijan, Khoi (1110) |
| T.turgi-45 | AB | Chahar mahal Bakhtiari, Bazoft (2190) | |
| T.turgi-2 | AB | Kohkiloye & Boyerahmad, Yasooj (2880) | |
| T.turgi-43 | AB | Chahar mahal Bakhtiari, Bazoft, Morez valley (2000) | |
| T.turgi-8 | AB | Chahar mahal Bakhtiari, Borojen to Izeh (2190) | |
| T.turgi-10 | AB | Khoosestan, Izeh (900) | |
| T.turgi-194 | AB | Kordestan, between Sanandaj and Saghez (1595) | |
| T.turgi-80 | AB | Kermanshah, Mahi Dasht (1290) | |
| T.turgi-25 | AB | Lorestan, Malavi toward Khoram Abad (1200) | |
| T.turgi-120 | AB | East Azarbaijan, Ahar (1320) | |
| T. dicoccum | T.dicoc-Turkey | AB | Provided by Institute of Plant Biology of the University of Zurich |
| T.dicoc-Iran (Isfahan) | AB | Provided by Institute of Plant Biology of the University of Zurich | |
| T. dicoccoides | T.dicocds-Iran (Kermanshah) | AB | Provided by Institute of Plant Biology of the University of Zurich |
| T.dicocds-Turkey | AB | Provided by Institute of Plant Biology of the University of Zurich | |
| T. aestivum | T.aest-47 | ABD | Chahar mahal Bakhtiari (2000) |
| T.aest-74 | ABD | Ilam, Do Rahe (1410) | |
| T.aest-129 | ABD | Booshehr, Bandargah to Deilam (17) | |
| T.aest-73 | ABD | Khoosestan, Karkheh (13) | |
| T.aest-97 | ABD | Mlayer toward Arak, 50Km (2010) | |
| T.aest-96 | ABD | Tehran, Firooz kouh1 (700) | |
| T.aest-107 | ABD | West Azarbaijan, Boukan to Mahabad (1290) | |
| T.aest-49 | ABD | Isfahan, Daran (2190) | |
| T.aest-82 | ABD | Kermanshah, Mahi Dasht (1290) | |
| Chinese spring | C.S. | ABD | Provided by Institute of Plant Biology of the University of Zurich |
The materials were taxonomically identified based on Rahiminejad and Kharazyan [72]. DNA was isolated from fresh leaves of twenty individuals of each accession using CTAB DNA extraction method [76]. In order to assess genetic relationships and diversity of the species carrying A genome in the genus Triticum, 31 A genome specific SSR primers [36] were used. Markers name and other details regarding the SSR markers are presented in Table 1.
Polymerase chain reactions (PCRs) were performed based on Jakson and Matthews (2000) [77]. Briefly, the PCR was carried out in 10 μL containing 2.5 μL of the 13 ng/μL genomic DNA sample, 1 μL of 10 × reaction buffer, 0.3 μL of 30 mM MgCl2, 1 U Taq polymerase, 0.5 μL of 2.5 mM dNTPs and 0.3 pmol each of IRD 800 dye and IRD 700 dye labeled arbitrary primers. The amplification program consisted of the following cycles: 95 °C for 4 min, 95 °C for 30 sec, 50 °C to 60 °C (depending on the primer set) for 30 sec, 72 °C for 1.5 min and a final extension at 72 °C for 10 min. After 4 min at 95 °C, 35 cycles were performed for 30 sec at 95 °C, 30 sec at annealing temperature (50–60 °C, see Table 1), 1.5 min at 72 °C, followed by a final extension step of 10 min at 72 °C. Upon completing the PCR cycles, 0.8 μL of PCR products of each sample was loaded onto a 6% polyacrylamide sequencing gel in a Li-COR Global DNA Sequencer, and electrophoresed at 2000 V for 1.5 h.
Statistical Analyses
In order to perform a simple phenetic analysis, the presence (1) and absence (0) of each band with particular mobility was scored. Genetic similarities were calculated using Powermarker [78] and NTSYS-PC softwares from the data tables. A genetic similarity based dendrogram (based on Jaccard’s similarity coefficient) [56] was constructed to show relationships between populations and species using the UPGMA clustering method implemented in NTSYS-PC [79].
For the genetic diversity analysis, allele number per locus, major allele frequencies, polymorphism information content ( , where pi and pj are the frequencies of the ith and jth alleles of a given marker, respectively [80]) and gene diversity (Di) were calculated as genetic parameters of polymorphism for each marker in all accessions, and for all markers in the accessions collected from the same geographical regions (see Figure 2). For all the parameters, the overall estimates were calculated as the averages across all loci. The analysis of molecular variance (AMOVA; implemented in ARLEQUIN software [81]) was carried out to estimate the variance components of fingerprinting patterns, and to partition the variation among ploidy levels (groups), among the species with a same ploidy level (within groups) and among the accessions within the species.
4. Conclusion
Based on the results of this study, and those previously reported [82,83], it can be concluded that a hypothetical process of hybridization, polyploidization and genomic differentiation can result in the A genome of bread wheat as below:
Huang et al. (2002) [29], and Wicker et al. (2003) [84] suggested that the genome of T. urartu (Au) diverged about 0.5–3 Mya from the genome of T. monococcum (Am). Therefore, it can be proposed that T. monococcum is the parental A genome species in the genus Triticum and the A genome of di-, tetra- and hexaploid species carrying A genome are directly or indirectly originated from this species. The populations of T. monococcum grow in restricted areas and the data (not shown) indicated low seed germination ability (about 10%) of this species. Therefore, this can be interpreted that T. monococcum is facing the risk of extinction in this area and we suggest that carrying out a wider sampling with more depth across its geographic range of distribution would be vital for gene pool conservation proposes.
As Figure 1 shows the materials provided by the Institute of Plant Biology, University of Zurich are clearly separated from the Iranian accessions. These originated from Turkey, Syria, Lebanon, Greece and Iran. In dendrogram, these accessions were mainly divided according to their country of origin. The T. dicoccoides accessions originating from Iran and Turkey were grouped together; however, because of less materials being studied no interpretation can be made about their genetic relationship. Since there were no materials belonging to T. dicoccum and T. dicoccoides among the 47 accessions specifically collected from Iran for this study, no comparison was made with their Iranian genepool (Table 4).
AMOVA analyses showed that the main portion of diversity is attributed to the variation among the accessions within species, suggesting high genetic diversity within the Triticum species in Iran. We would expect that a great sampling of each species in Iran and analysis of genetic diversity would be worthwhile to reveal the genetic structure of their gene pools and to discover new useful alleles for breeding proposes.
Acknowledgments
The authors wish to thank the Office of Graduate Studies of the University of Isfahan and the Institute of Plant Biology, University of Zurich for their support.
References
- 1.Kihara H. Cytologische und genetische Studien bei wichtigen Getreidearten mit besonderer Rücksicht auf das Verhalten der Chromosomen und die Sterilität in den Bastarden. Mem. Coll. Sci. Kyoto Imp. Univ. 1924;1:1–200. [Google Scholar]
- 2.Lilienfeld F, Kihara H. Genomanalyse bei Triticum und Aegilops. V. Triticum timopheevi Zhuk. Cytologia. 1934;6:87–122. [Google Scholar]
- 3.Gill BS, Appels R, Botha-Oberholster A-M, Buell CR, Bennetzen JL, Chalhoub B, Chumley F, Dvorak J, Iwanaga M, Keller B, et al. A workshop report on whaet genome sequencing: International genome research on wheat consortium. Genetics. 2004;168:1087–1096. doi: 10.1534/genetics.104.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harlan JR. American Soc. of Agronomy. 2nd ed. CSSA; Madison, WI, USA: 1992. Crops and Man. [Google Scholar]
- 5.Sakamura T. Kurze Mitteilung über die Chromosomenzahlen und die Verwandtschaftsverhältnisse der Triticum-Arten. Bot. Mag. Tokyo. 1918;32:151–154. [Google Scholar]
- 6.Zohary D, Feldman M. Hybridization between amphiploids and the evolution of polyploids in the wheat (Aegilops-Triticum) group. Evolution. 1962;16:44–61. [Google Scholar]
- 7.Kimber G, Feldman M. Wild Wheat, an Introduction. College of Agriculture, University of Missouri; Columbia, MO, USA: 1987. [Google Scholar]
- 8.Mandy G. New concept of the origin of Triticum aestivum. Acta Agron. Hung. 1970;19:413–417. [Google Scholar]
- 9.Allard RW. Genetic basis of the evolution of adaptedness in plants. Euphytica. 1996;92:1–11. [Google Scholar]
- 10.Hoisington D, Khairallah M, Ribaut JM, Skovmand B, Taba S, Warburton M. Plant genetic resources: What can they contribute toward increased crop productivity? Proc. Natl. Acad. Sci.USA. 1999;99:8133–8138. doi: 10.1073/pnas.96.11.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rejesus RM, van Ginkel M, Smale M. CYMMIT; Texcoco, Mexico: 1996. Wheat breeder’s perspectives of genetic diversity and germplasm use; Wheat special report 40. [Google Scholar]
- 12.Vavilov NI. The theory of the origin of cultivated plants after Darwin. Navka (Science) 1940;2:55–75. [Google Scholar]
- 13.Tsunewaki K. Comparative gene analysis of common wheat and its ancestral species. II. Waxiness, growth habit and awnedness. Jpn. J. Bot. 1966;19:175–229. doi: 10.1093/genetics/53.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakai Y. Isosyme variation in Aegilops and Triticum, IV The origin of the common wheats revealed from the study on esterase isozymes in synthesized wheats. Jpn. J. Genet. 1979;54:175–189. [Google Scholar]
- 15.Nishikawa K, Furuta Y, Wada T. Genetic studies on alpha-amylase isozymes in wheat. III. Intraspecific variation in Aegilops squarrosa and the birthplace of hexaploid wheat. Jpn. J. Genet. 1980;55:325–336. [Google Scholar]
- 16.Helguera M, Khan IA, Dubcovsky J. Development of PCR markers for wheat leaf rust resistance gene Lr47. Theor. Appl. Genet. 2000;101:625–631. [Google Scholar]
- 17.Parker GD, Chalmers KJ, Rathjen AJ, Langridge P. Mapping loci associated with milling yield in wheat (Triticum aestivum L.) Mol. Breed. 1999;5:561–568. [Google Scholar]
- 18.Parker GD, Langridge P. Development of a STS marker linked to a major locus controlling flour colour in wheat (Triticum aestivum L.) Mol. Breed. 2000;6:169–174. [Google Scholar]
- 19.Zhao ZD, Liu JJ, Dong JY, Zhang L, Li Q. Association between TOM and cooking quality of Chinese noodle. Acta Agronomica Sin. 1998;24:738–742. [Google Scholar]
- 20.Peng ZS, Yen C, Yang JL. Chromosomal Location of genes for supernumerary spikelet in bread wheat. Euphytica. 1998;103:109–114. [Google Scholar]
- 21.Zanetti S, Winzeler M, Keller M, Keller B, Messmer M. Genetic Analysis of Pre-Harvest Sprouting Resistance in a Wheat 3 Spelt Cross. Crop Sci. 2000;40:1406–1417. [Google Scholar]
- 22.Ramana JV, Singh D. Major genes involved in chlorophyll synthesis in bread wheat (Triticum aestivum L.) Cultivars. Indian J. Genet. Plant Breed. 2002;62:67–80. [Google Scholar]
- 23.Zheng YL, Yen C, Yang JL. Monosomic analysis for total and sterile floret number per spike in the multispikelet line 10-A of common wheat. Wheat Inf. Service. 1993;77:25–28. [Google Scholar]
- 24.Vagujfalvi A, Crosatti C, Galiba G, Dubcovsky J, Cattivelli L. Two loci on wheat chromosome 5A regulate the differential cold-dependent expression of the cor14b gene in frosttolerant and frost-sensitive genotypes. Mol. Gen. Genet. 2000;263:194–200. doi: 10.1007/s004380051160. [DOI] [PubMed] [Google Scholar]
- 25.Davydov VA. Characterization of the stomatal apparatus in monosomic lines of chinese spring wheat. Genetika (Moskva) 1999;35:546–550. [Google Scholar]
- 26.Sutka J, Galiba G, Snape JW. Inheritance of frost resistance in wheat (Triticum aestivum L.) Acta Agron. Hung. 1997;45:257–263. [Google Scholar]
- 27.Veisz O, Sutka J. Role of the chromosome 4B in suppressing frost resistance in winter wheat (Triticum aestivum L.). Cereal Res. Commun. 1998;26:47–52. [Google Scholar]
- 28.Yuanwu Z, Yujun Y. Study on the γ radiosensitivity of monosomic lines of Sumai 3 to radiation from 60Co. Acta Agron. Sin. 1995;21:429–433. [Google Scholar]
- 29.Hammer K, Filatenko AA, Alkhanjari S, Al-Maskri A, Buerkert A. Emmer (Triticum dicoccon Schrank) in Oman. Genet. Resour. Crop Evol. 2004;51:111–113. [Google Scholar]
- 30.Filatenko AA, Hammer K. A new gross morphological variation in the genus Triticum L. Genet Resour. Crop Evol. 2007;54:231–232. [Google Scholar]
- 31.Tsegaye S, Tesemma T. The genetics of some isozyme variants in tetraploid wheats (Triticum turgidum L.) Hereditas. 1995;123:147–154. [Google Scholar]
- 32.Cheniany M, Ebrahimzadeh H, Salimi A, Niknam V. Isozyme variation in some populations of wild diploid wheats in Iran. Biochem. Syst. Ecol. 2007;35:363–371. [Google Scholar]
- 33.Mesfin A, Frohberg RC. RFLP Markers Associated with High Grain Protein from Triticum turgidum L. var. dicoccoides Introgressed into Hard Red Spring Wheat. Crop Sci. 1999;39:508–513. [Google Scholar]
- 34.D’ovidio R, Tanzarella OA, Masci S, Lafiandra D, Porceddu E. RFLP and PCR analyses at Gli-1, Gli-2, Glu-1 and Glu-3 loci in cultivated and wild wheats. Hereditas. 1992;116:79–85. [Google Scholar]
- 35.Figliuolo G, Perrino P. Genetic diversity and intra-specific phylogeny of Triticum turgidum L. subsp. dicoccon (Schrank) Thell. revealed by RFLPs and SSRs. Genet. Resour. Crop Evol. 2004;51:519–527. [Google Scholar]
- 36.Röder MS, Korzun V, Wendehake K, Plashke J, Tixier MH, Leory P, Ganal MW. A microsatellite map of wheat. Genetics. 1998;149:2007–2023. doi: 10.1093/genetics/149.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohammadi SA, Khodarahmi M, Jamalirad S, Jalal Kamali MR. Genetic diversity in a collection of old and new bread wheat cultivars from Iran as revealed by simple sequence repeat-based analysis. Ann. Appl. Biol. 2009;154:67–76. [Google Scholar]
- 38.Gupta PK, Balyan HS, Edvards KJ, Isaac P, Korzun V, Roder M, Gautier M-F, Joudrier P, Schlatter AR, Dubcovskly J, et al. Genetic mapping of 66 new microsatellite (SSR) loci in bread wheat. Theor. Appl. Genet. 2002;105:413–422. doi: 10.1007/s00122-002-0865-9. [DOI] [PubMed] [Google Scholar]
- 39.Bossolini E, Krattinger SG, Keller B. Development of simple sequence repeat markers specific for the Lr34 resistance region of wheat using sequence information from rice and Aegilops tauschii. Theor. Appl. Genet. 2006;113:1049–1062. doi: 10.1007/s00122-006-0364-5. [DOI] [PubMed] [Google Scholar]
- 40.Bryan GJ, Collins AJ, Stephenson P, Orry A, Smith JB, Gale MD. Isolation and characterization of microsatellites from hexaploid bread wheat. Theor. Appl. Genet. 1997;94:557–563. [Google Scholar]
- 41.Plaschke J, Ganal MW, Röder MS. Detection of genetic diversity in closely related bread wheat using microsatellite markers. Theor. Appl. Genet. 1995;91:1001–1010. doi: 10.1007/BF00223912. [DOI] [PubMed] [Google Scholar]
- 42.Stachel M, Lelley T, Grausgruber H, Vollmann J. Application of microsatellites in wheat (Triticum aestivum L.) for studying genetic differentiation caused by selection for adaptation and use. Theor. Appl. Genet. 2000;100:242–248. [Google Scholar]
- 43.Huang SX, Sirikhachornkit A, Su XJ, Faris J, Gill B, Haselkorn R, Gornicki P. Genes encoding plastid acetyl-CoA carboxylase and 3-Phosphoglycerate kinase of the Triticum/Aegilops complex and evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA. 2002;99:8133–8138. doi: 10.1073/pnas.072223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eujayl I, Sorrells M, Baum M, Wolters P, Powell W. Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor. Appl. Genet. 2002;104:339–407. doi: 10.1007/s001220100738. [DOI] [PubMed] [Google Scholar]
- 45.Buchanan FC, Adams LJ, Littlejohn RP, Maddox JF, Crawford AM. Determination of evolutionary relationships among sheep breeds using microsatellites. Genomics. 1994;22:397–403. doi: 10.1006/geno.1994.1401. [DOI] [PubMed] [Google Scholar]
- 46.Condit R, Hubbell S. Abundance and DNA sequence of two-base repeat regions in tropical tree genomes. Genomics. 1991;34:66–71. doi: 10.1139/g91-011. [DOI] [PubMed] [Google Scholar]
- 47.Kkaya MS, Bhagwat AA, Cregan PB. Length polymorphisms of simple sequences repeat DNA in soybean. Genetics. 1992;132:1131–1139. doi: 10.1093/genetics/132.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagercrantz U, Ellegren H, Andersson L. The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res. 1993;21:1111–1115. doi: 10.1093/nar/21.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Senior ML, Heun M. Mapping maize microsatellites and polymerase chain reaction confirmation of the targeted repeats using a CT primer. Genome. 1993;36:884–889. doi: 10.1139/g93-116. [DOI] [PubMed] [Google Scholar]
- 50.Wu KS, Tanksley SD. Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol. Gen. Genet. 1993;241:225–235. doi: 10.1007/BF00280220. [DOI] [PubMed] [Google Scholar]
- 51.Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 52.Saghai-Maroof MA, Soliman K, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosome location and population dynamics. Proc. Natl. Acad. Sci. USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rongwen J, Akkaya MS, Bhagwat AA, Lavi U, Crecan PB. The use of microsatellite DNA markers for soybean genotype identification. Theor. Appl. Genet. 1995;90:43–48. doi: 10.1007/BF00220994. [DOI] [PubMed] [Google Scholar]
- 54.Liu ZW, Biyashev RM, Saghai-Maroof MA. Development of simple sequence repeat DNA markers and their integration into a barley linkage map. Theor. Appl. Genet. 1996;93:869–876. doi: 10.1007/BF00224088. [DOI] [PubMed] [Google Scholar]
- 55.Mörchen M, Cuguen J, Michaelis G, Hänni C, Saumitou-Laprade P. Abundance and length polymorphism of microsatellite repeats in Beta vulgaris L. Theor. Appl. Genet. 1996;92:326–333. doi: 10.1007/BF00223675. [DOI] [PubMed] [Google Scholar]
- 56.Proven J, Powell W, Waugh R. Microsatellite analysis of relationships within cultivated potato (Solanum tuberosum) Theor. Appl. Genet. 1996;92:1078–1084. doi: 10.1007/BF00224052. [DOI] [PubMed] [Google Scholar]
- 57.Szewc-McFadden AK, Kresovich S, Bliek SM, Mrrchell SE, McFerson JR. Identification of polymorphic, conserved simple sequence repeats (SSRs) in cultivated Brassica species. Theor. Appl. Genet. 1996;93:534–538. doi: 10.1007/BF00417944. [DOI] [PubMed] [Google Scholar]
- 58.Taramino G, Tingey S. Simple sequence repeats for germplasm analysis and mapping in maize. Genome. 1996;39:277–287. doi: 10.1139/g96-038. [DOI] [PubMed] [Google Scholar]
- 59.Smulders MJM, Bredemeijer G, Rus-Kortekaas W, Arens P, Vosman B. Use of short microsatellites from database sequences to generate polymorphisms among Lycopersicum esculentum cultivars and accessions of other Lycopersicum species. Theor. Appl. Genet. 1997;91:681–690. [Google Scholar]
- 60.Röder MS, Plaschke J, König SU, Börner A, Sorrells ME, Tanksley SD, Ganal MW. Abundance, variability and chromosomal location of microsatellites in wheat. Mol. Gen. Genet. 1995;246:327–333. doi: 10.1007/BF00288605. [DOI] [PubMed] [Google Scholar]
- 61.Ma ZQ, Röder M, Sorrells ME. Frequencies and sequence characteristics of di-, tetra-nucleotide microsatellites in wheat. Genome. 1996;39:123–130. doi: 10.1139/g96-017. [DOI] [PubMed] [Google Scholar]
- 62.Stephenson P, Bryan G, Kirby J, Collins A, Devos K, Busso C, Gale M. Fifty new microsatellite loci for the wheat genetic map. Theor. Appl. Genet. 1998;97:946–949. [Google Scholar]
- 63.Varshney RK, Prasad M, Roy JK, Röder MS, Balyan HS, Gupta PK. Intregated physical maps of 2DL, 6BS and 7DL carrying loci for grain protein content and preharvest sprouling tolerance in bread wheat. Creal Res. Commun. 2001;29:33–40. [Google Scholar]
- 64.Korzun V, Röder MS, Ganal MW, Worland AJ, Law CN. Genetic analysis of the dwarfing gene Rht8 in wheat Part 1. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.) Theor. Appl. Genet. 1998;96:1104–1109. [Google Scholar]
- 65.Prasad M, Varshney RK, Kumar A, Balyan HS, Sharma PC, Edwards KJ, Singh H, Dhaliwal HS, Roy JK, Gupta PK. A microsatellite marker associated with a QTL for grain protein content on chromosome arm 2DL of bread wheat. Theor. Appl. Genet. 1999;99:341–345. [Google Scholar]
- 66.Roy JK, Prasad M, Varshney RK, Balyan HS, Blake TK, Dhaliwal HS, Singh H, Edwards KJ, Gupta PK. Identification of a microsatellite on chromosomes 6B and a STS on 7D of bread wheat showing an association with preharvest sprouting tolerance. Theor. Appl. Genet. 1999;99:336–340. [Google Scholar]
- 67.Prasad M, Varshney RK, Roy JK, Balyan HS, Gupta PK. The use of microsatellites for detecting DNA polymorphism, genotype identification and genetic diversity in wheat. Theor. Appl. Genet. 2000;100:584–592. [Google Scholar]
- 68.McLauchlan A, Henry RJ, Issac PG, Edwards KJ. Microsatellite analysis in cultivated hexaploid wheat and wild wheat relatives. In: Henry RJ, editor. Plant Genotyping: The DNA Fingerprinting of Plants. CABI Publishing, CAB International; Wallingford, UK: 2001. pp. 147–159. [Google Scholar]
- 69.Huang XQ, Börner A, Röder MS, Ganal MW. Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor. Appl. Genet. 2002;105:699–707. doi: 10.1007/s00122-002-0959-4. [DOI] [PubMed] [Google Scholar]
- 70.Zhang XY, Li GW, Wang LF, Wang HM, You GX, Dong YS. An estimation of the minimum number of SSR alleles needed to reveal genetic relationships in wheat varieties. I. Information from large-scale planted varieties and cornerstone breeding parents in Chinese wheat improvement and production. Theor. Appl. Genet. 2002;106:112–117. doi: 10.1007/s00122-002-1016-z. [DOI] [PubMed] [Google Scholar]
- 71.McFadden ES, Sears ER. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 1966;37:81–89. doi: 10.1093/oxfordjournals.jhered.a105590. [DOI] [PubMed] [Google Scholar]
- 72.Rahiminegad MR, Kharazian N. Illustration of and a key to the genus Triticum L. in Iran. Wheat Inf. Service. 2005;99:29–34. [Google Scholar]
- 73.Kihara H. The diversity of DD analyzer, one of the ancestors of common wheat (preliminary reports) Aric. Hort. 1944;19:889–890. [Google Scholar]
- 74.Johnson BL. Identification of the apparent B-genome donor of wheat. Can. J. Genet. Cytol. 1975;17:21–39. [Google Scholar]
- 75.Tsunekawi K, Ebona K. Production of near-isogenic lines of common wheat for glaucousness and genetic basis of this trait clarified by their use. Genes Genet. Syst. 1999;74:33–41. [Google Scholar]
- 76.Rogers SO, Bendich AJ. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 1985;5:69–76. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- 77.Jackson JA, Matthews D. Modified inter-simple sequence repeat PCR protocol for use in conjunction with the Li-Cor gene ImagIR(2) DNA analyzer. Biotechniques. 2000;28:914–917. [PubMed] [Google Scholar]
- 78.Liu K, Muse SV. PowerMarker: Integrated analysis enviornment for genetic marker data. Bioinformatic. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 79.Jaccard P. Novelles rescherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 1908;44:223–270. [Google Scholar]
- 80.Rohlf FJ. NTSYS-PC numerical taxonomy and multivariate analysis system, version 2.02. Exter Publications; Setauket, NY, USA: 1998. [Google Scholar]
- 81.Nei M. Genetic distance between populations. Am. Nat. 1972;106:283–293. [Google Scholar]
- 82.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–791. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gill BS, Kimber G. Giemsa C-banding and the evolution of wheat. Proc. Natl. Acad. Sci. USA. 1974;71:4086–4090. doi: 10.1073/pnas.71.10.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dvořák J. The relationships between the genomes of Triticum urartu and the A and B genomes of Triticum aestivum. Can. J. Genet. Cytol. 1976;18:371–377. [Google Scholar]
- 85.Wicker T, Yahiaui N, Guyot R, Schlagenhauf E, Liu Z, Dubcovsky J, Keller B. Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell. 2003;15:1186–1197. doi: 10.1105/tpc.011023. [DOI] [PMC free article] [PubMed] [Google Scholar]