Abstract
The retrotransposons HeT-A, TART, and TAHRE, which maintain Drosophila telomeres, transpose specifically onto chromosome ends to form long arrays that extend the chromosome and compensate for terminal loss. Because they transpose by target-primed reverse transcription, each element is oriented so that its 5′ end serves as the extreme end of the chromosome until another element transposes to occupy the terminal position. Thus 5′ sequences are at risk for terminal erosion while the element is at the chromosome end. Here we report that TART elements in Drosophila melanogaster and Drosophila virilis show species-specific innovations in promoter architecture that buffer loss of sequence exposed at chromosome ends. The two elements have evolved different ways to effect this protection. The D. virilis TART (TARTvir) promoter is found in the 3′ UTR of the element directly upstream of the element transcribed. Transcription starts within the upstream element so that a “Tag” of extra sequence is added to the 5′ end of the newly transcribed RNA. This Tag provides expendable sequence to buffer end erosion of essential 5′ sequence after the RNA is reverse transcribed onto the chromosome. In contrast, the D. melanogaster TART (TARTmel) promoter initiates transcription deep within the 5′ UTR, but the element is able to replace and extend the 5′ UTR sequence by copying sequence from its 3′ UTR, we believe while being reverse transcribed onto the chromosome end. Astonishingly, end-protection in TARTvir and HeT-Amel are essentially identical (using Tags), whereas HeT-Avir is clearly protected from end erosion by an as-yet-unspecified program.
Keywords: chromosome end protection, promoter evolution, pseudo-LTR promoter, perfect nonterminal repeat, antisense RNA
The retrotransposable elements that maintain telomeres in Drosophila are in many respects typical non-LTR retrotransposons but also have characteristics not found in their nontelomeric relatives (1). These telomere-specific characteristics illuminate the biology of retrotransposon telomeres.
The telomere-specific retrotransposons HeT-A, TART, and TAHRE were originally characterized in Drosophila melanogaster but have been found in many other Drosophila. We have identified HeT-A and TART in Drosophila virilis, showing that these elements have maintained telomeres since before separation of the extant Drosophila species (40–60 Mya). Like their homologs in other species, the D. virilis elements transpose specifically to chromosome ends, where they form the mixed head-to-tail arrays that make up Drosophila telomeres (2, 3).
Non-LTR retrotransposons are reverse transcribed onto the chromosome by target-primed reverse transcription (4). The 3′ end of the RNA associates with a nick in chromosomal DNA. Reverse transcription of the RNA, primed by the 3′ hydroxyl of the nick, links the new copy of the element to the chromosome. The Drosophila telomere-specific elements are exceptional because they associate with the ends of chromosomes, rather than internal nicks, and transpose as terminal additions (1).
The 5′ end of each newly transposed telomere-specific element serves as the extreme end of the chromosome until another element transposes to cap it and take over the terminal position. During the time that an element forms the end of the chromosome, its 5′ end is at risk for terminal erosion. Analyses of D. melanogaster telomere arrays suggest there are at least two modes of loss from chromosome ends: relatively slow, gradual erosion and sporadic, more drastic, loss by terminal deletion. Even relatively slow loss would incapacitate the promoter of a typical non-LTR retrotransposon because these elements have their promoter sequences within the 5′ UTR and initiate transcription immediately upstream of the promoter (5). The promoter is included in the RNA and moved to the new site, ensuring that the retrotransposon can be transcribed in its new location.
Analysis of the D. melanogaster HeT-A promoter showed that this element has an unusual solution to the problem of protecting its 5′ end. Its promoter is reminiscent of the promoters of LTR retrotransposons and retroviruses (6, 7). Sequences in the 3′ UTR of HeT-A drive transcription of another immediately downstream HeT-A element; the upstream 3′ UTR acts as a “pseudo-LTR” for the downstream neighbor. Transcription starts within this pseudo-LTR so that terminal nucleotides plus the 3′ oligoA from the upstream element are added to the 5′ end of the RNA copy of the downstream element. (We define this added sequence as a “Tag.”) In the new element, Tags become de facto extensions of the 5′ UTR sequence that can be eroded to protect essential sequence of the element. Full-length elements in telomere arrays have several partially eroded Tags on their 5′ ends, showing that Tags do provide protection of the end (8).
Tags do not contain the complete promoter sequence (6). Thus the pseudo-LTR promoter has a cost for HeT-A: an element can be transcribed only if it is capped by another HeT-A to provide a promoter. This cost is largely mitigated because most elements are capped by other HeT-As, and even severely 5′-truncated HeT-As usually retain enough 3′ sequence to serve as promoters for a downstream neighbor.
D. virilis HeT-A shares many of the characteristics of D. melanogaster HeT-A (9, 10). Surprisingly, it does not have a pseudo-LTR promoter. Instead it has a typical non-LTR promoter located in the 5′ UTR and this promoter adds no 5′ protective sequence (7). Nevertheless, the ratio of complete to truncated HeT-As seems to be as high in D. virilis telomere arrays as in D. melanogaster arrays. Apparently D. virilis has some other mechanism for protecting end sequence. The mechanism is not known but may be related to a notable difference between the UTR sequences of HeT-As of these two species. D. melanogaster elements differ by numerous indels and nucleotide changes in the UTRs. D. virilis UTRs do not show this variability (7). We suggest that these conserved sequences in D. virilis UTRs may be necessary for precise associations with proteins and/or RNAs that process the extreme end of the telomere, prevent its erosion, and align reverse transcription of the next element.
Here we report characterization of the promoters and 5′ end protection of D. virilis TART and D. melanogaster TART. These two elements differ significantly. TARTvir has a pseudo-LTR promoter similar to that of HeT-Amel. TARTmel has a more typical promoter in its 5′ UTR but this produces an RNA transposition intermediate with a 5′ UTR shorter than those found in almost all identified elements. We suggest a model for extending an element's 5′ UTR when the element transposes; evidence for this model is presented in Results and the model delineated in Discussion.
Results
D. virilis TART Promoter Resembles the D. melanogaster HeT-A Pseudo-LTR Promoter.
The first indication that D. virilis TART has a pseudo-LTR promoter came from the λ phage we used to identify D. virilis telomere retrotransposons (2). Two elements in these clones were full length, and each had multiple Tags of 3′ UTR sequence at its 5′ end (evidence for multiple transpositions onto the telomere). The Tags were variably eroded, as expected if they had been eroded while protecting essential sequence at the 5′ end (Fig. 1A).
Fig. 1.
D. virilis TART transcripts have 5′ Tags. (A) Genomic DNA with two complete and one 5′-truncated element. Transcription of the central element from the −55 start site in the upstream element produces a transposition intermediate RNA with a new Tag on the 5′ end. Light gray: 5′ and 3′ UTRs. Dark gray triangle: 3′ end of element from −55 through oligoA tail (sequence at bottom of figure). This is added to the new transcript as a Tag and forms the end of the chromosome after reverse transcription. Thin black triangles: partially eroded Tags from earlier transpositions (sequences shown in box). Tag strings are found on 5′ ends of complete, but not 5′-truncated, elements. (B) Organization of partially eroded Tags into strings found on individual elements. “R” denotes Tag strings sequenced from RACE products. “G” denotes strings from genomic DNA. For each element, Tags are ordered with most distal on the left. Each element has several Tags showing multiple transpositions. Tag lengths show variable erosion in successive transpositions.
Further evidence that transcription starts within the 3′ UTR of an adjacent upstream TART comes from 5′ rapid amplification of cDNA ends (RACE) analysis of RNA from larval brains. Seven clones were sequenced, and four transcription starts were identified, 55, 109, 134, and 153 nt upstream of the oligoA of the upstream neighbor. Small sequence differences in these clones indicate that the RNA is transcribed from several different elements and that elements might use different start sites.
Like elements in the λ phage clones, TART transcripts from the brain have multiple variably truncated Tags, evidence that they were transcribed from elements that had transposed multiple times (Fig. 1). Truncated TARTvir Tags tend to be shorter than those of D. melanogaster HeT-A, suggesting that TARTvir elements erode more rapidly or remain on the extreme end for a longer time than HeT-Amel.
As a further test of TARTvir promoter location, we studied the ability of segments of TARTvir 3′ and/or 5′ UTR sequence to drive expression of a bacterial lacZ reporter gene (6, 7). Constructs containing these sequences were transiently transfected into cultured D. virilis cells, and promoter activity was measured by β-galactocidase activity (Fig. 2). Like HeT-Amel in D. melanogaster cells, the 3′ UTR of the downstream element has significant promoter activity. Addition of 5′ UTR sequence from the transcribed element enhances promoter strength (Fig. 2), as was found for D. melanogaster HeT-A (6). This enhancement may have the effect of increasing transcription from promoters upstream of complete elements relative to promoters upstream of truncated HeT-As.
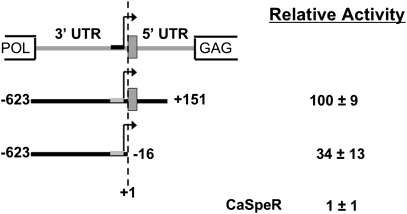
Fig. 2.
Relative sense strand promoter activity of sequences from D. virilis TART elements tested in D. virilis cells. Diagram shows junction between two elements with the 3′ UTR of the upstream element and the 5′ UTR of the downstream element. Arrow: transcription start at −55, with the region containing other starts indicated in black. Gray box: string of eroded tags. Constructs are shown below with starting and ending nucleotide numbered from the 3′ end of the upstream element. The first nucleotide of the downstream element (eroded Tags included) is +1. Negative numbers run into the 3′ UTR, and positive numbers run into the 5′ UTR. Activity (±SD) is relative to a very active construct, set at 100% and used in each experiment to allow comparison between experiments. CaSpeR, empty expression vector.
D. melanogaster TART Has Perfect Nonterminal Repeats.
D. melanogaster TART does not have “pseudo-LTR” promoters, but it has another feature resembling retroviruses and LTR retrotransposons: each TART contains a pair of long repeated sequences that are clearly evolving together (11), as are the LTRs of those other retroelements (12). We propose that, as in LTR elements, the two TART repeats coevolve because both are reverse transcribed from one of two repeats in the RNA. However, in TART (a non-LTR element), the details of the coevolution mechanism differ radically from that in LTR elements: most notably, the 3′ TART repeat does not extend to the 3′ end of the element. Thus the TART repeats are not strictly terminal, and we refer to them as perfect nonterminal repeats (PNTRs). To understand the structure and proposed function of the 5′ end of D. melanogaster TART, it is important to understand the structure of the PNTR (Fig. 3).
Fig. 3.
Comparison of typical members of the subfamilies of D. melanogaster TART. White: ORF1 (Gag) and ORF2 (Pol). Gray: 5′ and 3′ UTRs. Bent arrows: sense strand transcription starts, determined here and by Maxwell et al. (17). Arrows in 5′ and 3′ UTRs: 5′ and 3′ PNTRs. Length of the 3′ PNTR is determined by identity with 5′ PNTR, so the 5′ limit of the 3′ arrow is determined by the 5′ extent of the 5′ UTR. Note that different elements in these same subfamilies can have longer or shorter 5′ UTRs.
D. melanogaster has three TART subfamilies: A, B, and C (11). They seem to be functionally interchangeable but differ in sequence. The UTRs show little or no sequence similarity between subfamilies at high stringency, but coding regions have at least 85% nucleotide identity. Nevertheless, all TARTmel subfamilies share unusual UTR structures: all have PNTRs, and furthermore, the 5′ PNTRs not only fill the 5′ UTRs but, depending on subfamily, extend 84–126 nt into the gag coding region. PNTR DNA differences in the ORFs lead to subfamily-specific Gag differences in 10 of the first 32 aa, showing that nucleotide, rather than amino acid, sequence is dominant here. The 3′ end of the 3′ PNTR ends 308–439 nt upstream of the terminal oligoA, depending on subfamily. We refer to this terminal region of the 3′ UTR as the post-PNTR.
The 5′ ends of the PNTRs are the more complicated. The 5′ end of TART seems to be highly variable; the few available 5′ UTR sequences range from 33 nt (13) to 3,934 nt (14), and each element sequenced has a different end. Lengths do not correlate with subfamily; both the longest and the shortest 5′ UTRs are TART-A. In every case the entire 5′ UTR is essentially identical to sequence in the 3′ UTR. Because PNTRs are defined by the identity of 5′ and 3′ sequences, elements with longer 5′ UTRs perforce have longer 3′ PNTRs that extend further 5′ to match their 5′ UTR. Different lengths of 5′ UTR do not affect the length of the post-PNTR at the 3′ end of the element.
Comparison of the Two Longest Available TART Sequences Provides Strong Support for the Conclusion that 5′ and 3′ PNTRs are Evolving Together.
The paucity of complete TART sequences precludes extensive analysis of PNTR sequence divergence; nonetheless, in database searches we found two TART elements that provide clear evidence that changes in one PNTR are copied in the partner. These elements belong to the same subfamily and are from D. melanogaster stocks separated long enough to have undergone some sequence change. The canonical TART-A, an Oregon-R element (AY561850), is 13,424 nt; the other, a Canton-S TART-A (AJ566116) has >99.9% sequence identity throughout this 13,424 nt but has an additional 2,175 nt of 5′ UTR (and hence PNTR) sequence.
These two elements overlap for the entire 1,850 nt of the shorter element's PNTR; most of the sequence differences between elements lies in this overlap. The two elements differ by one 35-nt indel, one 5-nt indel, and eight single nucleotides lost, gained, or replaced (see Fig. S1 for details of this comparison). Significantly, almost all differences in the two 5′ PNTRs are also in the two 3′ PNTRs. The two indel differences and five of the eight single-nucleotide differences are found in both 5′ and 3′ PNTRs of each element, a clear demonstration that sequence changes in one PNTR have been transferred to its mate. Interestingly, the three single-nucleotide differences that are not found in both PNTRs are found in only one PNTR of the Oregon-R element, one in the 5′ PNTR and two in the 3′ PNTR. If coevolution of the two PNTRs, like that of LTRs, occurs during reverse transcription when the element transposes, the three changes found only in one PNTR of the Oregon-R element may well have occurred after that element was reverse transcribed onto the telomere. We assume that differences occurring after transposition will be erased when a transcript of this element is reverse transcribed to yield a new copy.
Promoter Studies of D. melanogaster TART.
To investigate the 5′ end of TART we searched the 5′ PNTR sequence for promoter activity with reporter constructs made from 3′ UTR sequence because the entire 5′ PNTR is contained within the 3′ UTR (Fig. 3). Thus we could use 3′ PNTR sequences to make constructs to simulate longer 5′ PNTRs. (The 5′ limit of the 5′ PNTR is not yet defined because the lengths of known elements are so varied that there is no assurance that the longest element in any subfamily has been found. Thus the 5′ limit of any 3′ PNTR, determined perforce by identity to its 5′ PNTR, is not known.) Therefore, we began our largest reporter construct within the 3′ end of ORF2 so that we could test sequence upstream of the longest 5′ end now known. The last 84 nt of these PNTRs contains the first codons of ORF1 in the 5′ PNTR. These we replaced with our reporter gene to make the construct we consider the “putative full-length” 5′ PNTR. This construct and its modifications were used to search for promoter activity that would yield sense strand TART transcripts.
5′ Sense Strand Promoter Initiates Transcription Slightly Upstream of the First ORF.
The longest of the putative 5′ PNTR constructs gave strong promoter activity (Fig. 4). The activity was not significantly diminished as large segments of 5′ sequence were deleted. In contrast, removal of 70 nt from the 3′ end almost completely eliminated the activity from both the longest and shortest constructs. This 70-nt deletion removed the downstream promoter element but left the initiator of a very good match to the initiator motif of 5′ UTR promoters typical of many non-LTR retrotransposons (5, 15). Deletion of 212 additional 3′ nucleotides removed the rest of the match to the promoter and yielded essentially no promoter activity. We conclude that the only transcription start site driven by the promoter activity in the “putative full-length” 5′ PNTR is 75 nt 5′ of the ATG of ORF 1. This seems to be the only start that can produce the transposition intermediate RNA for TART.
Fig. 4.
Relative sense strand promoter activity of sequences from D. melanogaster TART elements tested in D. melanogaster cells. Gray bar diagram at top shows the putative 5′ PNTR of TART, ending in the first few nucleotides of the Gag ORF. This ORF was replaced with a reporter gene to make constructs testing promoter activities in the 5′ PNTR (see text). Those constructs are shown below the diagram. The lower gray bar diagram shows the same sequence with the ORF replaced by the post-PNTR sequence plus terminal oligoA (thin black line). The reporter gene follows the oligoA to test the effect of termination sequences on the promoter in the 3′ PNTR. Shown below each PNTR diagram are sequences tested, identified by nucleotide number at either end. Nucleotides are numbered from the transcription start site (arrow) identified here and by Maxwell et al. (17). Positive numbers run downstream, and negative numbers run upstream. Activity (±SD) was determined as in Fig. 2.
Major Sense Strand Promoter in the 3′ UTR Does Not Direct Transcription of the Downstream Element.
Because the 5′ and 3′ PNTRs are identical, the promoter activity found at the 5′ end should also be active at the 3′ end. We do see small transcripts on Northern blots that might be the products of this promoter. We also considered the possibility that the 3′ promoter might be a pseudo-LTR promoter directing transcription of the downstream element. However, if the 3′ promoter drives transcription of its downstream neighbor, the transcript must continue through the termination signals at the 3′ end of the promoting element. To test this, we removed the reporter gene from the 5′ PNTR construct used above and added back the post-PNTR sequence and terminal oligoA to reconstruct the actual 3′ PNTR and its environment. The reporter gene was then added 3′ of the element's oligoA. We found no activity with these constructs. Because the constructs had strong activity when the 3′ end of the element was not included, the 3′ sequence must have caused transcription to terminate before reaching the reporter (Fig. 4). We conclude that the 3′ sense strand promoter is responsible for the small 3′ UTR transcripts but rarely, if ever, reads through into the downstream element.
Both TARTmel and TARTvir Have Strong Antisense Promoters.
TART elements from each species are the only two Drosophila retrotransposons known to produce a nearly full-length antisense RNA. In each, this antisense RNA is significantly more abundant than sense strand RNA (3, 13).
The function of the large antisense transcript is not understood. Many antisense RNAs are regulatory. In the D. melanogaster germ line, TART transposition has been shown to be under RNAi control by 24–30 nt piwi-interacting RNAs, rather than by the large antisense transcript (16). The large RNA might code for protein, but it has only a few short ORFs; nevertheless, its abundance and ubiquitous presence in different tissues and developmental stages strongly suggest it is important to TART biology. Because sense strand promoters have dramatically different architecture in D. virilis and D. melanogaster, we wondered whether the antisense promoters were equally divergent. By reporter mapping, we found that TART antisense promoters in the two species are very similar; each has a single or small cluster of start sites in the 3′ UTR several hundred nucleotides upstream of the 3′ terminus (Figs. S2 and S3). Conservation of this feature suggests this long antisense RNA plays a significant role in TART biology, but, given the notable differences between TARTmel and TARTvir sense strand promoter architecture and 5′ end protection, it seems unlikely that antisense has a role in either of these activities.
Discussion
TART and HeT-A Homologs in D. melanogaster and D. virilis Have Evolved Different Methods of 5′ End-Protection.
The study of TARTmel and TARTvir reported here, together with our earlier study of HeT-A from these two species (7), show that promoters of these telomere-specific retrotransposons have been reconfigured in unusual ways: ways that protect against loss of essential 5′ sequence whenever elements are exposed on the chromosome end. That protection increases the probability that some elements in the telomere always will be capable of transposition to maintain those chromosome ends. The pseudo-LTR mechanism seen for TART in D. virilis resembles that of HeT-A in D. melanogaster: in that each adds extra sequence by starting transcription outside the element (7). The mechanism used by TART in D. melanogaster is more complicated. As discussed below, our promoter assays and sequence analyses lead us to suggest that the TARTmel 5′ PNTR is renewed at each retrotransposition and is itself the sacrificial sequence that protects from erosion and small-scale terminal deletions.
TARTmel End-Protection Mechanism Differs from That of Other Telomere Elements.
TARTmel is the only telomere-specific retrotransposon with PNTRs and also the only element for which the exact 5′ end has not been determined. Each of the available TARTmel sequences has a different length 5′ UTR, and the lengths differ significantly. This is a dramatic contrast to the 5′ ends of the other three telomeric elements we have studied. HeT-A, from both D. melanogaster and D. virilis, and TARTvir each have a single promoter for the full-length sense strand transcript, although that promoter drives two to four closely spaced transcription start sites.
For D. melanogaster TART we have also found a single promoter for full-length sense strand transcripts. As described above, it directs transcription from a start site 75–77 nt upstream of the first codon of ORF1. All but one known TARTmel have 5′ UTRs too long to have been transcribed from this promoter. Thus the promoter architecture seems to be at odds with the longer 5′ UTRs found on most elements in telomeric arrays.
Although, by itself, our promoter study explains the length of only the shortest 5′ UTR, our results are entirely consistent with the analysis of TART initiation, polyadenylation, and splice sites reported by Maxwell et al. (17). These authors used 5′ and 3′ RACE in an extensive study of TART RNA in larvae, adults, and cultured cells, using primers specific for each of the three TART subfamilies. As in our experiments, the 3′ UTR served as proxy for the 5′ UTR because primers designed for 3′ PNTR sequence also identify sequence in the 5′ PNTR. Surprisingly, almost all sense strand RNAs from the three subfamilies started at one or the other of two major sites, a site 75–77 nt upstream from the start of ORF1, and its duplicate sequence in the 3′ PNTR. These are the same two transcription starts identified by our promoter studies. Maxwell et al. (17) found only one polyadenylation site at the 3′ end of TART, suggesting that both the 5′ and 3′ start sites use the same termination point. This suggestion is supported by earlier studies showing that RNAs of the appropriate sizes are abundant on Northern blots (13). No function is known for the short RNA initiated at the 3′ start site.
Only two other possible sense strand starts were found by Maxwell et al. (17). One, seen only in cultured cells, would start transcription near the 3′ end of ORF1 in TART-B. A likely product of this start site has been reported in cultured cells (13). Its function is not known. Their fourth start site was found only once and may be an artifact.
Thus, two different approaches, our study and that of Maxwell et al. (17), find a single highly conserved start of transcription for the transposition intermediate RNA. Maxwell et al. (17) found this site in all three subfamilies, and our TART-B reporter constructs also identify the same site. The unexpected finding was that this start site is in the PNTR. The three TART subfamilies differ greatly in their PNTR sequences, yet the start site is conserved. Comparing subfamilies by the least stringent BLAST (blastn), we find small islands of nucleotide similarity surrounding the sense strand start sites, suggesting that these sequences have been conserved while surrounding UTR sequences have diverged.
Structure of the PNTRs and Their Coevolution Provide a Mechanism for Replacing Sequence Upstream of the Transcription Start Site.
The results discussed above make a strong case that TARTmel transposition intermediate RNA does not encode large segments of the 5′ UTR found in transposed elements. We believe that the mechanism by which D. melanogaster TART maintains its 5′ end also involves its PNTRs. It is notable that the short 5′ UTR of the putative transposition intermediate is identical to the 3′ end of the 3′ PNTR. This, with the evidence discussed above, that sequence changes in one PNTR are copied into the other PNTR, suggests a model to derive a transposed element with a long 5′ UTR from RNA with a short 5′ UTR (Fig. 5).
Fig. 5.
Proposed mechanism for extending the 5′ end of D. melanogaster TART. Transcription starts (bent arrow) near the ATG of ORF 1 (Gag), producing a transposition intermediate RNA (dashed line) lacking most of the 5′ UTR. This RNA has a small piece of 5′ PNTR (short arrow) and complete 3′ PNTR (long arrow). Steps 1, 2, and 3 show the RNA as it is reverse transcribed onto the chromosome end. 1: The polyA tail associates with the chromosome, and reverse transcriptase begins to copy the RNA. The gray oval represents proteins proposed to hold the RNA in a conformation that brings the 5′ PNTR sequence into proximity to the 3′ end of the 3′ PNTR. It is omitted for clarity in later steps. 2: When reverse transcriptase reaches the 5′ end of the transcript, it makes a template jump back to the 3′ end of the 3′ PNTR. 3: Reverse transcriptase dissociates the RNA–DNA complex and recopies some or all of the 3′ PNTR. As a result the transposed element will have more 5′ UTR sequence than the RNA did, and possibly more sequence than the element from which it was derived.
We suggest that a 5′ extension is added during transposition by repeating the reverse transcription of the 3′ PNTR. Specifically, we postulate that when reverse transcriptase reaches the 5′ end of RNA that is being reverse transcribed onto the chromosome, the enzyme makes a template jump to the identical sequence in the 3′ end of the 3′ PNTR (Fig. 5). It then continues to extend the 5′ UTR of the new element by making a second copy of the 3′ UTR, thus elongating the 5′ UTR of the transposed element and incorporating any changes that have occurred in the 3′ PNTR.
This repetitious transcription adds sacrificial sequence that, like the Tags of TARTvir and HeT-Amel, can be lost to terminal erosion without affecting the transposition competence of the transposed element. The extreme variability in the length of the 5′ UTR in genomic TARTmel elements can be explained by variable termination of reverse transcription, by terminal erosion, by terminal deletions, or some combination thereof. D. melanogaster TART apparently has evolved a mechanism for maintaining its 5′ end that differs, in all details, but not in principle, from the one found for D. virilis TART.
Our proposed model provides one mechanism by which TART RNA with the short 5′ UTR found in our study could produce transposed elements with long 5′ UTRs. Alternatively, elements with long 5′ UTRs could be formed if RNA initiated in an upstream HeT-A or TART and read through a TART with a long 5′ UTR. Several read-through sequences have been found in EST libraries (17). Whether or not these transcripts successfully transpose onto chromosome ends, the striking conservation of only one full-length sense strand promoter in the divergent UTRs of the three TARTmel subfamilies argues that this promoter is the major contributor to the population of RNA transposition intermediates. In addition, coevolution of sequences in the two PNTRs is nicely explained if both are reverse transcribed from the same segment of RNA. Read-through transcription from an upstream element would not provide the correction of one sequence against the other.
For TART reverse transcriptase to make a second copy of the 3′ PNTR, it must make a template jump to the 3′ end of the 3′ PNTR after its first reverse transcription of the template RNA. Reverse transcriptases from some other non-LTR retrotransposons can make such template jumps. The reverse transcriptase enzyme of Bombyx mori element R2Bm makes template jumps to the 3′ end of a new RNA after reaching the end of its template (18, 19); template jumps have also been reported for mouse L1 (20).
TART reverse transcriptase also must dissociate its RNA template from the cDNA made on its first pass over the 3′ PNTR so that it can make a second copy of the RNA. Retroviral reverse transcriptases have an RNase H domain that degrades its RNA template to free the cDNA for second strand synthesis. This RNase H domain is not found in non-LTR elements of the jockey clade to which TART belongs (21, 22). Studies of the non-LTR element, R2Bm, show that its enzyme dissociates the cDNA–RNA complex without degrading the RNA before synthesizing the second DNA strand (19, 23). If the TART enzyme can also extract the RNA template from the cDNA–RNA complex, it should be able to use the RNA for a second reverse transcription. Although the TART enzyme remains to be tested, we expect it will be able to do the job.
Conclusion.
Analyses of Drosophila telomere arrays show that the retrotransposons making up these arrays are subject to terminal erosion. Although truncated elements can form the necessary structural extension of the telomere, these arrays must also contain some transposition-competent elements as breeding stock for continued extension of the telomeres. This study of TARTmel and TARTvir suggests that these two homologs have evolved different but effective ways of adding nonessential sequence to buffer loss and assure a supply of nontruncated elements in each array. Our earlier study of HeT-Amel and HeT-Avir showed that these elements also have evolved ways to maintain a similar supply of complete elements. Remarkably, these four telomere elements use three different mechanisms to protect 5′ end sequence.
Both HeT-A and TART have unusual characteristics that have been conserved over the 40–60 million years separating D. melanogaster and D. virilis. For example, HeT-As lack a gene for reverse transcriptase, and their Gag proteins are targeted to chromosome ends; TARTs produce abundant nearly full-length antisense RNA. It is intriguing that mechanisms of 5′ end protection have not been similarly conserved. Only HeT-Amel and TARTvir use the same mechanism. The shared mechanism could reflect inheritance from a common ancestor, convergent evolution, or even horizontal transfer. (That three of these four telomere elements use different mechanisms of end protection suggests that these mechanisms are relatively labile on an evolutionary time scale.) In any case, only transposition-competent elements can give rise to lineages of new elements, which provides a very strong drive for evolving efficient 5′ end protection. The diversity of mechanisms seen in these studies seems to be a result of this drive.
Materials and Methods
Cell Lines.
The D. virilis cell line WR Dv-1 and the D. melanogaster S3 cell line were maintained at room temperature in Schneider's Drosophila media (Gibco) with 10% heat-inactivated FBS (HyClone).
Sequence Anlyses.
D. melanogaster TART-A, AY561850 (13,424 nt, from Oregon R stock) and AJ566116 (15,576 nt, from Canton S stock); TART-B, U14101 (nt 1–10515, omitting the repeat at the 3′ end); TART-C, AY600955, were used for dot plot comparisons. Additional sequences analyzed used only BAC, P1, or λ phage clones to avoid possible misassembly artifacts associated with whole genome sequencing.
Constructs for Promoter Activity.
Constructs were in the pCaSpeR-AUG-β-gal vector. TART sequences in the polylinker drove expression of the lacZ reporter gene. Constructs are named by the nucleotide at each end of the construct. The numbering scheme for each set of constructs is explained in the figure legends. All D. virilis constructs were PCR amplified from the 3′ UTR of element TARTa phage V2 nt 3973 joined to the 5′ UTR of element TARTb phage V2 nt 4832 (AY219709). All D. melanogaster constructs were made from the 3′ end of the canonical TART-B (U14101). Constructs were verified by sequencing.
Transient Transfection.
Promoter strength was measured by the activity of β-galactosidase expressed from each construct. To normalize transfection, pCMV-Luc (from N. Kamoshita, Jichi Medical University, Tochigi, Japan), which constitutively expresses luciferase under the control of the CMV promoter, was cotransfected with each experimental construct as previously described (7). D. virilis cells were transfected with a calcium phosphate-DNA coprecipitation method, as previously described (7). D. melanogaster cells were transfected with effectene as previously described (24).
Expression Assays.
Cells were harvested 48 h after transfection. Luciferase activity was measured by the Luciferase Assay System (Promega), and β-galactosidase activity was measured by the Beta-Glo Assay System (Promega). β-Galactosidase activity was normalized to luciferase activity. Data for analysis came from at least three independent experiments. Data from each experiment were normalized by the activity of one construct used for this purpose in every experiment.
Supplementary Material
Acknowledgments
We thank E. Casacuberta (CSIC-Pompeu Fabra University, Barcelona, Spain) and N. Kamoshita for clones; T. RajBhandary for use of the luminometer; C. Koehrer for instruction in its use; and K. Lowenhaupt, E. Casacuberta, and members of the Pardue laboratory for helpful discussions and critical reading of the manuscript. The D. virilis cell line WR Dv-1 was obtained from the Drosophila Genomics Resource Center (Bloomington, IN). Flybase has provided important information. This work was supported by National Institutes of Health Grant GM50315 (to M.-L.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015926107/-/DCSupplemental.
References
- 1.Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu Rev Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- 2.Casacuberta E, Pardue ML. Transposon telomeres are widely distributed in the Drosophila genus: TART elements in the virilis group. Proc Natl Acad Sci USA. 2003;100:3363–3368. doi: 10.1073/pnas.0230353100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casacuberta E, Pardue ML. HeT-A elements in Drosophila virilis: Retrotransposon telomeres are conserved across the Drosophila genus. Proc Natl Acad Sci USA. 2003;100:14091–14096. doi: 10.1073/pnas.1936193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 5.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 6.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: The promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 7.Traverse KL, George JA, DeBaryshe PG, Pardue ML. Evolution of species-specific promoter-associated mechanisms for protecting chromosome ends by Drosophila Het-A telomeric transposons. Proc Natl Acad Sci USA. 2010;107:5064–5069. doi: 10.1073/pnas.1000612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George JA, DeBaryshe PG, Traverse KL, Celniker SE, Pardue ML. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 2006;16:1231–1240. doi: 10.1101/gr.5348806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casacuberta E, Pardue ML. HeT-A and TART, two Drosophila retrotransposons with a bona fide role in chromosome structure for more than 60 million years. Cytogenet Genome Res. 2005;110:152–159. doi: 10.1159/000084947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casacuberta E, Marín FA, Pardue ML. Intracellular targeting of telomeric retrotransposon Gag proteins of distantly related Drosophila species. Proc Natl Acad Sci USA. 2007;104:8391–8396. doi: 10.1073/pnas.0702566104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheen FM, Levis RW. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc Natl Acad Sci USA. 1994;91:12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voytas DF, Boeke JD. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: American Society for Microbiology; 2002. pp. 631–662. [Google Scholar]
- 13.Danilevskaya ON, Traverse KL, Hogan NC, DeBaryshe PG, Pardue ML. The two Drosophila telomeric transposable elements have very different patterns of transcription. Mol Cell Biol. 1999;19:873–881. doi: 10.1128/mcb.19.1.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abad JP, et al. Genomic analysis of Drosophila melanogaster telomeres: Full-length copies of HeT-A and TART elements at telomeres. Mol Biol Evol. 2004;21:1613–1619. doi: 10.1093/molbev/msh174. [DOI] [PubMed] [Google Scholar]
- 15.Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maxwell PH, Belote JM, Levis RW. Identification of multiple transcription initiation, polyadenylation, and splice sites in the Drosophila melanogaster TART family of telomeric retrotransposons. Nucleic Acids Res. 2006;34:5498–5507. doi: 10.1093/nar/gkl709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bibiłło A, Eickbush TH. The reverse transcriptase of the R2 non-LTR retrotransposon: Continuous synthesis of cDNA on non-continuous RNA templates. J Mol Biol. 2002;316:459–473. doi: 10.1006/jmbi.2001.5369. [DOI] [PubMed] [Google Scholar]
- 19.Bibillo A, Eickbush TH. High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J Biol Chem. 2002;277:34836–34845. doi: 10.1074/jbc.M204345200. [DOI] [PubMed] [Google Scholar]
- 20.Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 22.Malik HS, Eickbush TH. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 23.Kurzynska-Kokorniak A, Jamburuthugoda VK, Bibillo A, Eickbush TH. DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon. J Mol Biol. 2007;374:322–333. doi: 10.1016/j.jmb.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuller AM, Cook EG, Kelley KJ, Pardue M-L. Gag proteins of Drosophila telomeric retrotransposons: Collaborative targeting to chromosome ends. Genetics. 2010;184:629–636. doi: 10.1534/genetics.109.109744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.