Abstract
The structure and integrity of DNA is of considerable biological and biomedical importance, and it is therefore critical to identify and to characterize enzymes that alter DNA structure. DNA helicases are ATP-driven motor proteins that unwind DNA. Conversely, HepA-related protein (HARP) protein (also known as SMARCAL1 and DNA-dependent ATPase A) is an annealing helicase that rewinds DNA in an ATP-dependent manner. To date, HARP is the only known annealing helicase. Here we report the identification of a second annealing helicase, which we term AH2, for annealing helicase 2. Like HARP, AH2 catalyzes the ATP-dependent rewinding of replication protein A (RPA)-bound complementary single-stranded DNA, but does not exhibit any detectable helicase activity. Unlike HARP, however, AH2 lacks a conserved RPA-binding domain and does not interact with RPA. In addition, AH2 contains an HNH motif, which is commonly found in bacteria and fungi and is often associated with nuclease activity. AH2 appears to be the only vertebrate protein with an HNH motif. Contrary to expectations, purified AH2 does not exhibit nuclease activity, but it remains possible that AH2 contains a latent nuclease that is activated under specific conditions. These structural and functional differences between AH2 and HARP suggest that different annealing helicases have distinct functions in the cell.
Keywords: SNF2 family protein, chromatin, DNA repair, genome integrity
The unwinding of dsDNA occurs frequently during normal cellular processes such as replication, repair, and transcription (for reviews, see refs. 1 and 2). In addition, ssDNA bubbles can arise spontaneously. Because unwound DNA can be stabilized by ssDNA-binding proteins such as replication protein A (RPA), there is considerable potential for the formation of stable RPA-bound ssDNA bubbles.
HepA-related protein (HARP, also known as SMARCAL1) is an ATP-driven motor protein that rewinds RPA-bound complementary ssDNA strands (3). Because HARP anneals DNA, it is termed an annealing helicase. HARP binds preferentially to fork DNA relative to ssDNA or dsDNA, and the binding of HARP to fork DNA activates its ATPase activity. In humans, mutations in the HARP gene are responsible for a rare autosomal recessive pleiotropic disorder known as Schimke immunoosseous dysplasia, which typically leads to death in early childhood (4). It thus appears that the annealing helicase activity of HARP is essential for normal growth and development.
In addition to its ability to function as an annealing helicase, HARP binds directly to RPA via a conserved N-terminal motif (5–9). This interaction between HARP and RPA is not required for annealing helicase activity, but rather serves to recruit HARP to sites of RPA-bound ssDNA that are formed during DNA damage or replicative stress (5–8). Cells with reduced levels of HARP also exhibit increased levels of RPA-bound ssDNA during S phase relative to that seen with wild-type cells (5, 7). These findings suggest an important role for HARP in DNA structure and dynamics.
To date, HARP is the only known annealing helicase. In these studies, we found that an uncharacterized protein, which possesses a motor domain similar to that of HARP, is also an annealing helicase. The identification of a second annealing helicase broadens our understanding of annealing helicases and further suggests that annealing helicases, like helicases, have specialized roles in our cells.
Results and Discussion
The ATPase Activity of AH2 Is Stimulated by Its Preferential Binding to Fork DNA.
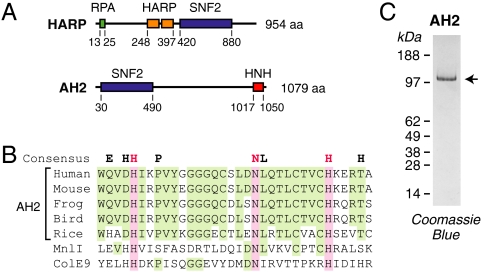
Based on the similarity between the sucrose nonfermenting 2 (SNF2) ATPase regions of HARP and a previously uncharacterized protein termed ZRanB3 (for zinc finger, Ran-binding domain containing protein 3) (10), we undertook the biochemical analysis of this protein. Aside from the SNF2 ATPase region, however, ZRanB3 does not possess any apparent relation to HARP (Fig. 1A). In addition, ZRanB3 is not structurally related to ZRanB1 or ZRanB2, neither of which is a SNF2 ATPase, and its zinc finger domain does not appear to be conserved across species. Therefore, on the basis of its biochemical activities (see below) and its status as a previously uncharacterized protein, we chose to rename ZRanB3 as annealing helicase 2 (AH2).
Fig. 1.
Purification of human AH2. (A) Schematic diagrams of human HARP and AH2. Unique features of HARP include the N-terminal RPA-binding motif as well as two conserved HARP domains, which are found in HARP proteins. AH2 has an HNH motif in its C-terminal region. The positions of the motifs in HARP and AH2 are indicated. (B) Comparison of the HNH motif in AH2 with those in the MnlI and colicin E9 (ColE9) nucleases. The sequences of the HNH motifs in AH2 from humans (Homo sapiens), mouse (Mus musculus), western clawed frog (Xenopus tropicalis), zebra finch (Taeniopygia guttata), and Asian rice (Oryza sativa) are shown. This figure shows that the key H-N-H residues are conserved among AH2 proteins. The HNH consensus is from ref. 26. (C) Polyacrylamide-SDS gel electrophoresis of purified FLAG-tagged human AH2.
AH2 contains the conserved SNF2 helicase domain as well as a putative HNH motif at its C terminus (Fig. 1B). Unlike HARP, AH2 appears to lack an RPA-binding domain. HNH motifs are common in prokaryotes. “HNH” refers to the three most conserved His and Asn amino acid residues in the motif (for reviews, see refs. 11–13). HNH proteins include a range of nucleases such as some homing endonucleases (e.g., I-HmuI), colicins (e.g., ColE7, ColE9), and restriction endonucleases (e.g., KpnI, MnlI, and HphI) (14–19). HNH motifs are less common in eukaryotes. AH2 appears to be the only vertebrate protein with an HNH motif.
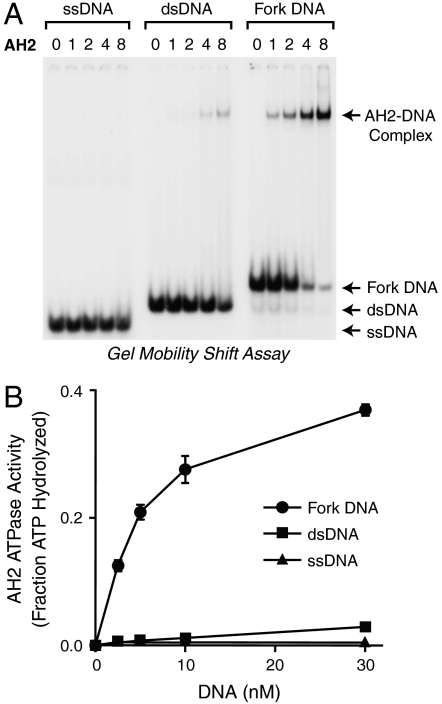
To investigate the function of AH2, we synthesized and purified full-length, FLAG-tagged human AH2 protein (Fig. 1C). We initially examined its DNA binding properties by using the gel mobility shift assay. In the experiment shown in Fig. 2A, we tested the binding of AH2 to 30 nt ssDNA [d(CT)15, which does not form hairpin or duplex structures], 30 bp dsDNA, and 30 nt fork DNA that is identical to the dsDNA except for a 9-nt mismatch at one end. We found that AH2 binds with the highest affinity to fork DNA.
Fig. 2.
AH2 protein binds selectively to fork DNA. (A) AH2 binds with higher affinity to fork DNA than to ssDNA or dsDNA. Gel mobility shift experiments were performed with 30-nt ssDNA, 30-bp dsDNA, or 30-nt fork DNA that is identical to the dsDNA except for a 9-nt mismatch at one end. The relative concentrations of AH2 are shown above the lanes; the actual concentrations of AH2 are 0, 0.05, 0.1, 0.2, and 0.4 nM. (B) The ATPase activity of AH2 is stimulated to a greater extent by fork DNA than by ssDNA or dsDNA. The DNA substrates used in the ATPase assays are identical to those used in A, except that the DNA samples were not radiolabeled. Error bars represent SD (N = 3).
We then determined the effect of each of the different DNA species on the ATPase activity of AH2 and found that the ATPase activity is stimulated to a much greater extent by fork DNA than by ssDNA or dsDNA (Fig. 2B). The specific stimulation of AH2 ATPase activity by fork DNA relative to ssDNA or dsDNA is similar to that seen with HARP (3).
AH2 Is an Annealing Helicase.
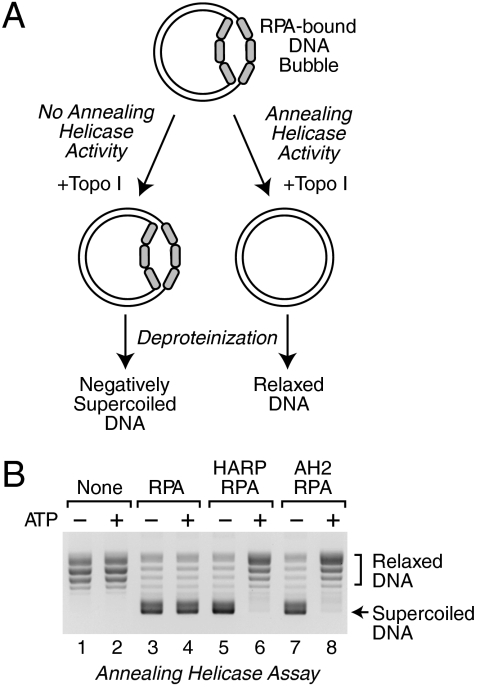
To test whether AH2 is an annealing helicase, we used the DNA-rewinding assay that we had previously developed for the characterization of HARP (Fig. 3A) (3). We generated a stable, partially unwound DNA substrate by adding RPA to plasmid DNA in the presence of topoisomerase I. This ability of RPA to form stable ssDNA bubbles in plasmids has been demonstrated previously (20, 21). Under these conditions, RPA binds to small transient bubbles and then wedges the DNA strands apart to form stable ssDNA bubbles in which RPA is bound to the unwound DNA (22). Upon addition of SDS (to inactivate enzymes such as topoisomerase I) and subsequent deproteinization, the RPA-unwound DNA yields negatively supercoiled DNA (Fig. 3B, lanes 3 and 4). As a control, when plasmid DNA is treated in an identical manner in the absence of RPA, the resulting DNA is relaxed, as expected (Fig. 3B, lanes 1 and 2). As depicted in Fig. 3A, the addition of an annealing helicase to the partially unwound DNA substrate would yield circular DNA that is relaxed by the topoisomerase I. We found that AH2 catalyzes the ATP-dependent relaxation of the RPA-unwound DNA, as observed with HARP (Fig. 3B, lanes 5–8). Therefore, like HARP, AH2 eliminates the stable single-stranded regions within the dsDNA.
Fig. 3.
AH2 is an ATP-dependent annealing helicase. (A) Schematic diagram of the annealing helicase assay. (B) AH2 catalyzes the rewinding of DNA in an ATP-dependent manner. Annealing helicase assays were carried out in the presence or absence of the indicated factors, and the resulting DNA species were resolved by agarose gel electrophoresis. An equimolar concentration of UTP was used as a control for the absence of ATP.
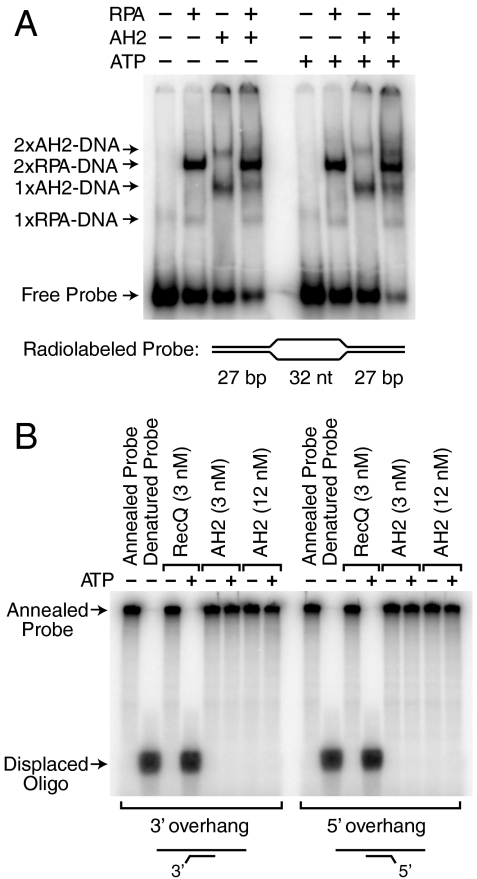
The ATP-dependent reannealing of RPA-bound ssDNA bubbles could, in principle, be accomplished either by the action of an annealing helicase or by direct displacement of RPA from the DNA. To test this latter possibility, we carried out gel mobility shift analyses of RPA binding to a DNA bubble consisting of a 32-nt loop of noncomplementary DNA flanked by two 27-bp regions of complementary dsDNA (Fig. 4A). When RPA alone is added to this substrate, we observed the binding of one or two molecules of RPA to the DNA probe. When AH2 is added to the RPA-bound substrate, there is little or no (≤ 5%) decrease in the binding of RPA to the DNA in the absence or presence of ATP (Fig. 4A). These experiments show that AH2-mediated removal of RPA from ssDNA requires the annealing of complementary ssDNA strands. Hence, AH2 does not remove RPA from ssDNA via a direct interaction, but rather is an annealing helicase.
Fig. 4.
AH2 is neither an RPA-removing enzyme nor a conventional helicase. (A) AH2 does not mediate the displacement of RPA from DNA. Gel mobility shift experiments were performed with radiolabeled bubble DNA that contains two high-affinity sites for RPA (the two 32-nt ssDNA segments) and for AH2 (the two DNA forks). A 10-fold molar excess of unlabeled d(CT)30 was added in the binding reactions subsequent to the binding of RPA to the probe DNA. AH2 (3 nM), RPA (3 nM), and ATP (1.5 mM) were included, as indicated. The apparent compositions of the shifted complexes are specified. (B) AH2 does not exhibit helicase activity. Helicase assays were performed with partial duplex DNA substrates, as indicated, containing a radiolabeled oligonucleotide annealed to single-stranded phiX174 DNA. Purified AH2 or RecQ (as a positive control for a helicase) were incubated with the DNA substrates in the absence or presence or ATP. The products were resolved by electrophoresis in a 10% polyacrylamide gel.
We additionally examined whether AH2 exhibits traditional helicase activity. We tested the ability of AH2 to unwind DNA in a standard helicase assay with partial duplex DNA substrates. As shown in Fig. 4B, we were unable to detect helicase activity for AH2. Thus, AH2 does not appear to be a helicase. Therefore, like HARP, AH2 possesses annealing helicase activity but lacks any detectable DNA unwinding activity.
AH2 Is Functionally Distinct from HARP.
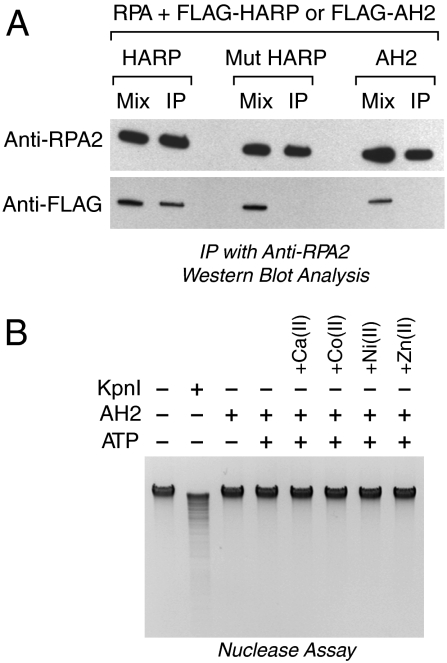
We next sought to investigate functional differences between AH2 and HARP. One notable feature of HARP is its conserved N-terminal RPA-interaction motif, which is necessary and sufficient for the binding of HARP to RPA (5–9). AH2 does not appear to contain an analogous RPA-binding motif. However, because the binding of HARP to RPA is important for the function of HARP in cells, we tested whether AH2 binds to RPA. In these experiments, purified FLAG-AH2 was incubated with purified RPA, and the resulting samples were subjected to immunoprecipitation with anti-RPA antibodies (Fig. 5A). The presence of AH2 and RPA in the immunoprecipitates was then detected by Western blot analysis with anti-FLAG or anti-RPA2 antibodies. The results revealed that, unlike wild-type HARP, AH2 does not exhibit a strong and direct association with RPA. It remains possible, however, that there is a weak transient interaction between AH2 and RPA.
Fig. 5.
Examination of differences between AH2 and HARP. (A) Unlike HARP, AH2 does not associate with RPA. Purified FLAG-tagged versions of wild-type HARP, mutant HARP lacking the conserved N-terminal RPA-binding motif, and wild-type AH2 were incubated with purified recombinant RPA, and then immunoprecipitated with anti-RPA2 resin. After the beads were washed, the bound proteins were eluted and detected by Western blot analysis with either anti-RPA2 or anti-FLAG. (B) AH2 does not exhibit nuclease activity with E. coli genomic DNA. AH2 (150 nM) or KpnI (15 u), an HNH nuclease, was incubated with E. coli genomic DNA (500 ng) for 2 h, and the digested DNA was resolved by agarose gel electrophoresis. ATP (1.5 mM final concentration) was included in the reactions where indicated. The reaction medium contained 10 mM Mg(II) and, where indicated, the following divalent metal cations: Ca(II) (1 mM), Co(II) (20 μM), Ni(II) (20 μM), and Zn(II) (20 μM).
AH2 and HARP also differ by the presence of HNH motif in AH2 but not HARP (Fig. 1 A and B). Because many HNH proteins are nucleases, we examined whether AH2 exhibits nuclease activity. To this end, we carried out a wide range of experiments with different DNA substrates and buffer conditions, but did not detect nuclease activity in purified AH2. For example, Fig. 5B shows the results of incubation of purified AH2 with Escherichia coli genomic DNA in the presence or absence of several different divalent metal cations including Mg2+, which is typically used with HNH nucleases. In those experiments, we included the HNH nuclease Kpn I as a positive control. Thus, AH2 does not exhibit nuclease activity with different DNA substrates (including both eukaryotic and prokaryotic DNA sequences) and under different experimental conditions. The presence of the HNH motif in AH2 is intriguing, and it remains possible that specific conditions may exist, such as the presence of a cofactor, in which AH2 functions as a nuclease.
Summary and Perspectives.
The identification of a second annealing helicase highlights the importance of these newly discovered enzymes. Both HARP and AH2 are distant members of the SNF2 family of ATP-dependent molecular motor proteins (3, 10, 23). Members of this family participate in a variety of nuclear processes such as chromatin assembly and remodeling, transcription, DNA repair, and recombination (10, 24, 25). Annealing helicase activity is not, however, a general property of SNF2 family proteins. For instance, Rad54, imitation switch (ISWI) [in the ATP-utilizing chromatin assembly and remodeling factor (ACF) complex], and Brg1 are SNF2 family ATPases that do not exhibit annealing helicase activity (3). In addition, there may be annealing helicases that are not members of the SNF2 protein family. Thus, the range of proteins that function as annealing helicases remains to be determined.
Comparison of the structure and activities of AH2 and HARP suggests that they have specialized functions in the cell. Both proteins are annealing helicases that lack any detectable DNA unwinding activity. However, HARP binds specifically to RPA via a conserved N-terminal motif, whereas AH2 does not interact with RPA. In addition, AH2 has a C-terminal HNH motif that is not present in HARP; hence, the HNH domain is not required for annealing helicase activity. The HNH motif is common in bacteria and fungi yet almost completely absent from metazoans (for example, see refs. 19 and 26). Notably, AH2 appears to be the only protein in vertebrates that contains an HNH motif. Although HNH proteins are often found to be nucleases, purified AH2 does not exhibit nuclease activity under a range of standard reaction conditions with a variety of DNA substrates. It is therefore possible that the HNH motif in AH2 has a non-nuclease-related function, or that the HNH motif possesses a latent nuclease activity that is activated under specific conditions. Moreover, HARP and AH2 may be members of a larger family of ATP-driven DNA-rewinding motor proteins. It will be interesting and important to investigate the full range of annealing helicases and their activities in the cell.
Materials and Methods
Protein Purification.
Human AH2 (also known as ZRanB3) cDNA was purchased from OpenBiosystems (clone ID 3077975) and sequenced to verify its integrity. The cDNA encoding N-terminally FLAG-tagged AH2 was then subcloned into pFastBac1 (Invitrogen). Baculovirus stocks were generated and amplified as recommended by the manufacturers (Life Technologies). Recombinant FLAG-AH2 and FLAG-HARP proteins were purified as described (3). Recombinant human RPA was synthesized in E. coli and purified as previously described (3, 27).
Gel Mobility Shift Assays.
The binding of AH2 (0, 0.05, 0.1, 0.2, or 0.4 nM final concentrations) to ssDNA, dsDNA, and fork DNA were carried out with oligonucleotides as described (3). The gel shift assay with the bubble DNA was performed as described (3) with the following minor modifications. Where indicated, RPA (3 nM) was incubated with the radiolabeled substrate for 10 min at 22 °C in Binding Buffer [20 mM Hepes (K+), pH 7.6, 0.1 M KCl, 5 mM MgCl2, 3% (vol/vol) glycerol, 0.25 mg/mL BSA, 0.01 mM EDTA, 0.5 mM DTT, 0.05% (vol/vol) NP-40]. A 10-fold molar excess (relative to the radiolabeled probe) of unlabeled 60 nt d(CT)30 competitor ssDNA was added along with AH2 (3 nM), where noted, and the reactions were incubated for another 20 min at 22 °C. Reactions also contained either 1.5 mM ATP or 1.5 mM UTP (as the −ATP control). The resulting products were resolved in a 5% polyacrylamide gel with 0.5× Tris-borate EDTA buffer.
ATPase Assays.
ATPase assays were carried out with the same oligonucleotides that were used in the DNA binding assays. Increasing concentrations of oligonucleotides (0, 2.5, 5, 10, and 30 nM final concentrations) were combined with purified AH2 (18 nM final concentration) in Binding Buffer containing 40 nM of [γ-32P]ATP and 100 nM of cold ATP in a final volume of 10 μL, and the reactions were incubated for 1 h at 30 °C. An aliquot (1 μL) of each reaction was applied to a polyethyleneimine-cellulose plate (Selecto Scientific), and the samples were subjected to thin-layer chromatography with 1 M formic acid, 0.5 M LiCl for 40 min. The plates were dried and quantitated by using a PhosphorImager (GE Healthcare Typhoon). Each experiment was performed in triplicate, and the mean and standard deviation for each condition were determined and displayed in a graph by using GraphPad Prism software.
Helicase and Annealing Helicase Assays.
The helicase and annealing helicase assays were performed as described (3). In the annealing helicase assays, AH2 and HARP were used at a final concentration of 300 nM.
Nuclease Assay.
E. coli genomic DNA (500 ng) was incubated with KpnI (15 units, New England Biolabs (NEB)] or AH2 (150 nM final concentration) in a 20 μL reaction containing 1x NEB buffer 1 (10 mM 1,3-bis(tris(hydroxymethyl)methylamino)propane-HCl, pH 7.0, 10 mM MgCl2, 1 mM DTT) for 2 h at 30 °C. ATP (1.5 mM final concentration) and the following metal cofactors were included where indicated: calcium chloride (1 mM), cobalt acetate (20 μM), nickel acetate (20 μM), zinc acetate (20 μM). The reactions were terminated by the addition of 125 μL of Stop Buffer [20 mM EDTA (Na+), pH 8, 0.2 M NaCl, 1% (wt/vol) SDS, 0.25 mg/mL glycogen]. The resulting DNA was deproteinized by phenol extraction and precipitated with ethanol. The reaction products were resolved by electrophoresis in 1% (wt/vol) agarose in 0.5× Tris-acetate EDTA buffer, and then visualized by staining with ethidium bromide.
Analysis of Interactions Between RPA and HARP or AH2.
Purified recombinant RPA (1 μg) and HARP or AH2 (1 μg), as indicated, were combined in IP Buffer [Binding Buffer containing 0.35 mg/mL BSA and 0.05% (vol/vol) NP-40] in a 20 μL reaction, and incubated for 30 min at room temperature. To each mixture was added IP Buffer (150 μL) and anti-RPA resin (10 μL). Anti-RPA resin was prepared by cross-linking anti-RPA2 antibodies (generous gift of Bruce Stillman, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) to Affigel-10 resin (Bio-Rad) according to the protocol from the manufacturer. The samples were incubated on a rotating platform for 2 h at 4 °C. The resin was then pelleted and washed with 3 × 500 μL of IP buffer. The bound proteins were eluted in SDS sample buffer and identified by Western blot analysis.
Acknowledgments.
We thank Sharon Torigoe, Joshua Theisen, Trevor Parry, Yuan-Liang Wang, and James Gucwa for critical reading of the manuscript. This work was supported by National Institutes of Health Grant R01 GM058272 (to J.T.K.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: Mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 2.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 3.Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boerkoel CF, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30:215–220. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- 5.Bansbach CE, et al. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciccia A, et al. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 2009;23:2400–2404. doi: 10.1101/gad.1831509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Postow L, Woo EM, Chait BT, Funabiki H. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem. 2009;284:35951–35961. doi: 10.1074/jbc.M109.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galburt EA, Stoddard BL. Catalytic mechanisms of restriction and homing endonucleases. Biochemistry. 2002;41:13851–13860. doi: 10.1021/bi020467h. [DOI] [PubMed] [Google Scholar]
- 12.Mehta P, Katta K, Krishnaswamy S. HNH family subclassification leads to commonality in the His-Me endonuclease superfamily. Protein Sci. 2004;13:295–300. doi: 10.1110/ps.03115604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2006;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 14.Pommer AJ, et al. Homing in on the role of transition metals in the HNH motif of colicin endonucleases. J Biol Chem. 1999;274:27153–27160. doi: 10.1074/jbc.274.38.27153. [DOI] [PubMed] [Google Scholar]
- 15.Ku WY, et al. The zinc ion in the HNH motif of the endonuclease domain of colicin E7 is not required for DNA binding but is essential for DNA hydrolysis. Nucleic Acids Res. 2002;30:1670–1678. doi: 10.1093/nar/30.7.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsia KC, et al. DNA binding and degradation by the HNH protein ColE7. Structure. 2004;12:205–214. doi: 10.1016/j.str.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Saravanan M, et al. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–6135. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J Mol Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 19.Cymerman IA, et al. Identification of a new subfamily of HNH nucleases and experimental characterization of a representative member, HphI restriction endonuclease. Proteins. 2006;65:867–876. doi: 10.1002/prot.21156. [DOI] [PubMed] [Google Scholar]
- 20.Georgaki A, Strack B, Podust V, Hübscher U. DNA unwinding activity of replication protein A. FEBS Lett. 1992;308:240–244. doi: 10.1016/0014-5793(92)81283-r. [DOI] [PubMed] [Google Scholar]
- 21.Treuner K, Ramsperger U, Knippers R. Replication protein A induces the unwinding of long double-stranded DNA regions. J Mol Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 22.Lao Y, Lee CG, Wold MS. Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry. 1999;38:3974–3984. doi: 10.1021/bi982371m. [DOI] [PubMed] [Google Scholar]
- 23.Coleman MA, Eisen JA, Mohrenweiser HW. Cloning and characterization of HARP/SMARCAL1: A prokaryotic HepA-related SNF2 helicase protein from human and mouse. Genomics. 2000;65:274–282. doi: 10.1006/geno.2000.6174. [DOI] [PubMed] [Google Scholar]
- 24.Gorbalenya AE, Koonin EV. Helicases: Amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 25.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H, Yuan HS. The conserved asparagine in the HNH motif serves an important structural role in metal finger endonucleases. J Mol Biol. 2007;368:812–821. doi: 10.1016/j.jmb.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 27.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: Expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]