Abstract
Centrosomes are primary microtubule (MT)-organizing centers (MTOCs). During mitosis, they dramatically increase their size and MT-nucleating activity and participate in spindle assembly from spindle poles. These events require the serine/threonine kinase, Aurora A (AurA), and the centrosomal protein of 192 kDa (Cep192)/spindle defective 2 (Spd-2), but the underlying mechanism remains unclear. We have found that Cep192, unlike targeting protein for Xklp2 (TPX2), a known MT-localizing AurA activator, is an AurA cofactor in centrosome-driven spindle assembly. Cep192, through a direct interaction, targets AurA to mitotic centrosomes where the locally accumulating AurA forms homodimers or oligomers. The dimerization of endogenous AurA, in the presence of bound Cep192, triggers potent kinase activation that, in turn, drives MT assembly. Depletion of Cep192 or specific interference with AurA-Cep192 binding did not prevent AurA oligomerization on MTs but abrogated AurA recruitment to centrosomes and its activation by either sperm nuclei or anti-AurA antibody (αAurA)-induced dimerization. In these settings, MT assembly by both centrosomes and αAurA-coated beads was also abolished or severely compromised. Hence, Cep192 activates AurA by a mechanism different from that previously described for TPX2. The Cep192-mediated mechanism maximizes AurA activity at centrosomes and appears essential for the function of these organelles as MTOCs.
Keywords: aurora kinase, microtubule-organizing center, protein recruitment
Centrosomes consist of a pair of centrioles surrounded by pericentriolar material (PCM). They function as microtubule-organizing centers (MTOCs) and control vital cellular processes including cell polarity and division (1, 2). Centrioles duplicate only once per cell cycle, and do so during interphase, via a process that relies on the sequential recruitment of several specific centrosomal proteins (reviewed in refs. 1 and 2). At the onset of mitosis, the size and microtubule (MT)-nucleating capacity of centrosomes increase dramatically as a result of PCM recruitment (1). The molecular details of this process, termed centrosome maturation, are incompletely understood.
Protein recruitment during both centrosome duplication and maturation is controlled by centrosomal protein of 192 kDa (Cep192)/spindle defective 2 (Spd-2), a protein that operates by an unknown mechanism (3–6). Among the proteins that localize to centrosomes in a Cep192-dependent manner is aurora A (AurA) (4, 6), a serine/threonine kinase involved in mitotic entry, centrosome maturation, bipolar spindle assembly, and cell polarity (7, 8). AurA activation depends on the phosphorylation of threonine (T) 288/295 (in human/Xenopus AurA, respectively) in its kinase activation loop (9, 10) and, in one specific mitotic setting, on the binding of targeting protein for Xklp2 (TPX2), a MT-nucleating protein (10–12). TPX2, when released from importin by RanGTP, activates AurA (both allosterically and by protecting T288/T295 from dephosphorylation) (10–12) and targets AurA to spindle MTs (13, 14). The AurA–TPX2 complex participates in spindle assembly promoted by chromatin/RanGTP but not by centrosomes and in setting a proper spindle length (11, 15, 16). Although several factors have been implicated in AurA regulation at centrosomes (7, 8, 17–19), the mechanism of AurA recruitment to and activation at these organelles has been unclear. Hence, the existence of a centrosome-specific AurA activator distinct from TPX2 and other known AurA cofactors has been proposed (20). Here, we identify Cep192 as an AurA centrosome-targeting and -activating cofactor.
Results
Detectable T-Loop Phosphorylation of Endogenous AurA Depends on the Presence of Centrosomes.
To study AurA regulation during centrosome-mediated spindle assembly, we used cell-free metaphase-arrested Xenopus egg extract (extract) (21, 22). When extract is supplemented with demembranated sperm nuclei, which contain a pair of centrioles, the latter recruit PCM, giving rise to a functional centrosome that acts as a MTOC (22). Centrosomal MT assembly begins 2–3 min after sperm addition to extract, peaking ≈7–8 min later (22) (Fig. 1A, Upper). By contrast, chromatin-mediated MT assembly does not commence until ≈20 min (22, 23). AurA and Cep192, although absent in naïve sperm nuclei/centrioles, were present in extract (Fig. 1B, lanes 1 and 2). Both were rapidly recruited to centrioles and colocalized during recruitment (Fig. 1A).
Fig. 1.
AurA activation and MT assembly promoted in egg extract by sperm nuclei and αAurA beads. (A) Localization of AurA and Cep192 during MT assembly promoted by sperm nuclei-associated centrosomes (time course) and RanGTP. (Scale bars: 5 μm.) (B) W-blots showing the time course of AurA activation in extract by sperm nuclei (4 × 104/μL). (C) Extract was driven into interphase by the addition of calcium and supplemented with demembranated sperm nuclei (2 × 103/μL). Extract aliquots were withdrawn at the indicated times after sperm addition and analyzed by W-blotting using the indicated antibodies. (D) W-blots of extracts supplemented with XB buffer, nocodazole (50 μM), RanT24N (30 μM), Ran(Q69L)GTP (30 μM), or DNA beads (0.1 μL/μL). Where indicated, reaction mixtures contained sperm nuclei (4 × 104/μL). (E and F) Beads coated with nonimmune IgG (N/Imm), αAurA, or αAurA-Fab were incubated in rhodamine-tubulin+Ran(Q69L)GTP-supplemented extract and analyzed by fluorescence microscopy (E) and W-blotting (F). (Scale bar: 10 μm.) (G) W-blots of extract incubated with 1.7 μM goat anti-rabbit IgG, IgG fraction (αIgG) and increasing concentrations of αAurA-Fab [detected as IgG light chain (IgG L.C.)]. The concentration of AurA in egg extract is ≈0.35 μM.
To assay endogenous AurA activity, we monitored phosphorylation of T295, which reflects the active state of the kinase (9, 10). Supplementation of metaphase-arrested extract with sperm nuclei led to the accumulation of T295-phosphorylated AurA (P-AurA), as assessed by Western blotting (W-blotting) (Fig. S1A). Similarly, in cycling extract, replicated sperm nuclei promoted AurA T295 phosphorylation at the onset of mitosis (Fig. 1C). AurA activation occurred independently of MTs and RanGTP, because P-AurA abundance was unaffected by nocodazole or by a dominant-negative Ran mutant (RanT24N) (Fig. 1D). Moreover, P-AurA was undetectable in extract supplemented with DNA-coated beads, which mimic chromatin (24), or with a constitutively active Ran mutant, Ran(Q69L)GTP, which facilitates AurA interaction with and activation by TPX2 and promotes MT assembly (10–12, 25) (Fig. 1D and Fig. S1B). It therefore appears that in egg extract, AurA activation, as manifested by T295 phosphorylation, depends on the presence of centrosomes, and occurs via a mechanism distinct from and more potent than the allosteric effect of TPX2. Consistently, and in agreement with a previous report (26), in human (HeLa) cells, P-AurA colocalized with a fraction of AurA associated with centrosomes and not MTs (Fig. S1C).
AurA Activation by Dimerization Is Essential for AurA-Mediated MT Assembly.
AurA and Cep192 recruitment to and MT nucleation by centrosomes coincided with AurA activation (Fig. 1 A and B). To test for a link between these processes, we exploited the prior observation that in extract, anti-AurA antibody (αAurA)-coated beads, which recruit endogenous AurA complexes, act as MTOCs (27) (Fig. 1E, ii). Interestingly, αAurA beads, like sperm nuclei/centrosomes, promoted AurA T295-phosphorylation; by contrast, beads coated with an αAurA monovalent fragment (αAurA-Fab) (Fig. S1D, Left) did not (Fig. 1F, quantified in Fig. S1E). Moreover, soluble αAurA also activated AurA, whereas soluble αAurA-Fab fragments did not unless they were bridged with a bivalent anti-IgG (IgG) antibody (αIgG) (Fig. S1F). Thus, AurA was activated by aggregation/cross-linking, a mechanism that has also been reported for Aurora B (AurB) (28).
To clarify this activation mechanism, P-AurA levels were measured in extract supplemented with a constant amount of bivalent αIgG and increasing concentrations of αAurA-Fab. As the αAurA-Fab concentration rose, P-AurA abundance first increased, peaking at an approximately equimolar ratio of αAurA-Fab/endogenous AurA, and then declined (Fig. 1G). This behavior is characteristic of kinase activation by dimerization (29), because the probability of bivalent bridging of the αAurA-Fab–bound AurA by αIgG is predicted to first increase and then decrease as the concentration of free αAurA-Fab rises. Of note, the αIgG used in the aforementioned experiments contained only bivalent IgG molecules, because it was affinity purified from the relevant immune IgG fraction. Moreover, nonreducing SDS/PAGE revealed the presence of only monomeric (i.e., bivalent) Ig molecules both in αIgG, and in αAurA (Fig. S1D). Thus, in naive extract, Ab-driven dimerization activates AurA. Moreover, the αAurA-Fab beads, unlike the αAurA beads, although recruiting a similar amount of AurA, failed to act as MTOCs (Fig. 1E, iii, quantified in Fig. S1G). These observations suggest that the function of αAurA beads as MTOCs is a product of αAurA/dimerization-mediated AurA activation occurring on the bead surface.
Cep192 Is an AurA-Binding Partner in Centrosome/MTOC-Driven Spindle Assembly.
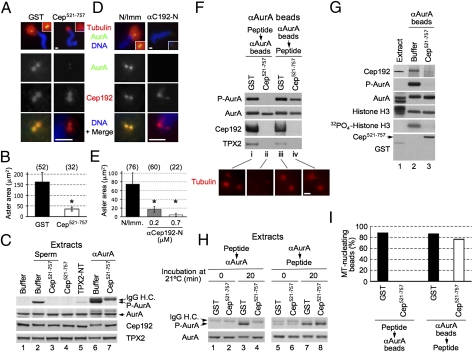
Mass spectrometry of αAurA bead-associated proteins revealed multiple peptides derived from the Xenopus ortholog of Cep192/SPD-2 (Joukov et al., unpublished data). This finding, along with the concurrence of AurA/Cep192 colocalization and AurA activation at centrosomes, as well as the central role of Cep192/SPD-2 in centrosome biogenesis (3–6), suggested that a common, Cep192-driven process underlies the development of MTOC activity by both centrosomes and αAurA beads. To explore this notion, we isolated a Xenopus laevis Cep192 cDNA encoding a 2,638 amino acid (aa), 289-kDa protein and raised Cep192-specific N- and C-terminally directed antibodies (αCep192-N and αCep192-C, respectively) (Fig. 2A, Fig. S2, and Fig. S3A). Immunoprecipitation (IP) of AurA from naïve extract resulted in co-IP of both Cep192 and TPX2 (Fig. S3B). Cep192 was not detected in TPX2 IPs, and TPX2 was absent from Cep192 IPs, whereas AurA was present in both (Fig. 2B). Thus, extract contains distinct complexes of AurA bound to TPX2 or to Cep192. AurA interacted with Cep192 directly (Fig. 2C and Fig. S3 C–E), likely through a segment also involved in TPX2 binding, because endogenous AurA was displaced from both proteins by an AurA-binding fragment of either Cep192 (Cep521–757) or TPX2 (TPX2-NT) (Fig. 2D).
Fig. 2.
Cep192 is a cofactor of AurA at centrosomes. (A) Schematic diagram of Xenopus Cep192. The AurA-BD and the three ASH (ASPM, SPD-2, Hydin) domains (40) are shown. The numbers denote aa. The underlying gray lines indicate the polypeptides used for antibody production. (B) W-blot of Cep192 and TPX2 IPs from extract. IgG H.C., IgG heavy chain. Note that the nonimmune IgG, even when used in approximately a twofold excess over αCep192-C and αTPX2 (as assessed by the intensity of IgG H.C.), brings down only a small, background amount of AurA. (C) Binding of recombinant Xenopus AurA to glutathione Sepharose-immobilized glutathione S-transferase (GST), GST-Cep521–757, and GST-TPX2-NT. (D) W-blot of αAurA beads retrieved from extracts incubated with 3 μM of the indicated GST polypeptides. (E) W-blot of mock-treated, Cep192-, and TPX2-depleted extracts incubated with 4 × 104/μL sperm nuclei (30 min) or with αAurA (0.3 μM) for the indicated times. (F) MT structures assembled in extracts supplemented with sperm nuclei, αAurA beads, or Ran(Q69L)GTP. (Scale bars: 10 μm.) (G) P-AurA and AurA detected by W-blot of αAurA beads retrieved from extracts in Fii. (H) W-blot showing partial rescue of the AurA activation defect by rCep192 in Cep192-depleted, fivefold diluted, extract. (I) Visualization of AurA oligomers by BiFC in extracts and transfected HeLa cells. BiFC complexes are shown in green; MTs (columns 1–3) and CFP (which denotes transfected cells; column 4, pseudocolor) in red; and DNA (DAPI; row 1) in blue. (Scale bars: 10 μm.)
To assess the function of AurA–Cep192 complexes, extract was immunodepleted of Cep192 (TPX2 and mock depletions served as controls). Efficient (>95%) depletion of Cep192 or TPX2 did not significantly affect the abundance of the other protein or AurA (Fig. 2E), confirming that a minor fraction of endogenous AurA is bound to Cep192 and TPX2 in distinct complexes. Cep192 depletion did not affect the assembly of RanGTP-induced MT asters or AurA targeting to MTs, which are TPX2-dependent processes (13, 14, 25) (Fig. 2F, iii, quantified in Fig. S4A). However, it abrogated AurA centrosomal localization and MT assembly by both centrosomes and αAurA beads (Fig. 2F, i and ii and Fig. S4 B–D). This finding suggests that Cep192 directly targets AurA to centrosomes. Depleting the extract of Cep192 abrogated the earliest, centrosome-driven, step of spindle assembly and inhibited, but did not prevent, bipolar spindle formation (Fig. S4E). This result is consistent with the earlier observations that, both in egg extract and in cells, bipolar spindle formation can proceed without centrosomes, presumably via the chromatin/RanGTP/TPX2-mediated pathway (24, 30, 31). By contrast, and as expected (13, 14, 25, 27, 32), TPX2-depleted extract was inactive in RanGTP-driven MT assembly, while supporting AurA recruitment to and MT nucleation by centrosomes (Fig. 2F, i and iii and Fig. S4 A–C). Nevertheless, TPX2-depleted extract also failed to promote MT assembly by αAurA beads (Fig. 2F, ii and Fig. S4D), a process shown to require both RanGTP and TPX2 (27). This discrepancy might be due to inherent differences between natural (centrosomes) and artificial (αAurA beads) MTOCs. Nevertheless, the differences are likely not absolute, because centrosomal MT assembly, although it does not, per se, require RanGTP and TPX2, is significantly enhanced by excess RanGTP (16, 33, 34). Of note, the TPX2–AurA interaction is not required for either the stimulating effect of RanGTP on centrosomal MTs or bipolar spindle formation (15, 16). Conceivably, MT assembly in an artificial setting by αAurA beads, unlike that by centrosomes, may require not only Cep192 but also TPX2. Taken together, these data reveal that Cep192 and TPX2 control AurA localization and MT assembly promoted by centrosomes and chromatin/RanGTP, respectively.
Cep192 Is Required for AurA Activation by Sperm Nuclei and Forced Dimerization.
Because, as shown above, both centrosomes and αAurA beads activated AurA and required Cep192 to act as MTOCs, we asked whether Cep192 and/or AurA dimerization are involved in AurA activation at centrosomes. AurA T295-phosphorylation promoted by both sperm nuclei and αAurA was abolished in Cep192-depleted extract and nearly unaffected in TPX2-depleted extract (Fig. 2 E and G). Recombinant Cep192 (rCep192) (Fig. S4G) was added to Cep192-depleted extract to test the specificity of this depletion effect. It resulted only in minimal reconstitution of Cep192 relative to mock-treated extract and insignificant rescue of AurA activation (data not shown). Reasoning that the ratio of Cep192 to other extract components could be important in achieving AurA activation, rCep192 was tested in Cep192-depleted extract diluted fivefold. In this setting, partial rescue of the AurA activation defect was reproducibly observed (Fig. 2H), indicating that this defect is, at least in part, a specific outcome of Cep192 depletion. Thus, in extract, Cep192, and not TPX2, is required for dimerization-driven AurA activation. Accordingly, density gradient sedimentation analysis revealed that AurA activated by αAurA was complexed with Cep192 and not with TPX2 (Fig. S5 A and B, rows 11–14 vs. 6–9).
If the dimerization-driven mechanism operates in cells, endogenous AurA would be expected to form homodimers at spindle structures. Indeed, using Bimolecular Fluorescence Complementation (BiFC) (35), AurA homodimers and/or higher order oligomers were detected at both centrosomes and spindle MTs in egg extract and in mitotic cells (Fig. 2I and Fig. S6 A and B). Moreover, AurA oligomerized on MTs of RanGTP-induced asters, even in Cep192-depleted extract (Fig. 2I). As shown above, P-AurA was undetectable in this setting (Fig. 1D, lane 5, and Fig. S1B). Thus, AurA oligomerization on spindle structures can occur in the absence of Cep192 and is, by itself, insufficient to promote readily measurable AurA T295 phosphorylation. We also detected oligomers of AurB on mitotic chromosomes and in the midbody, where this kinase normally resides (8) (Fig. S6 C and D), suggesting that oligomerization might be a common property of the Aurora family kinases.
AurA–Cep192 Interaction Is Essential for AurA Activity and MTOC Function.
To further explore the function of AurA–Cep192 complexes, dominant-negative and antibody antagonists were used. We found that the AurA-binding domain (AurA-BD)-containing fragment of Cep192, Cep521–757, when added to extract before αAurA, did not affect AurA di/oligomerization promoted by αAurA. However, it disrupted endogenous AurA–Cep192 complexes and prevented activation of the di/oligomerized kinase (Fig. S5B, rows 21–23 vs. 11–13). Together with the results of Cep192 depletion, these data demonstrate that only when bound to endogenous Cep192 is AurA capable of activation upon dimerization. Cep521–757 also inhibited the targeting of AurA to centrosomes, as well as MT assembly (Fig. 3 A, B, and F, ii) and AurA activation (Fig. 3C, lanes 3 vs. 2 and 7 vs. 6) promoted by either sperm nuclei/centrosomes or αAurA-induced dimerization. Supplementing extract with an affinity-purified αCep192-N antibody directed against the AurA-BD led to similar inhibitory defects (Fig. 3 D and E, and Fig. S5C), which were observed in Cep192-depleted extract, as shown above (Table S1). By contrast, αCep192-C antibody, which did not react with the AurA-BD, lacked these effects. Instead, it activated AurA, presumably by dimerizing preexisting AurA–Cep192 complexes (Fig. S5D and data not shown). These results confirm that Cep192 targets AurA to centrosomes and promotes activation of the dimerized enzyme.
Fig. 3.
Cep192-mediated AurA activation is essential for MTOC function. (A and D) Localization of AurA and Cep192 on centrosomal asters assembled in extracts supplemented with 3 μM GST or Cep521–757 (A) or with 0.7 μM nonimmune IgG or αCep192-N (D). (Scale bars: 2.5 μm.) (B and E) Quantitation of MT aster surface area in A and D, respectively. The results obtained in the presence of 0.2 μM αCep192-N are also shown for comparison (E). Error bars, SD. *P < 0.0001 compared with the corresponding control extract, as determined by a two-tailed Student's t test. The number of asters analyzed is shown in parentheses. (C) W-blot of extracts incubated with XB buffer, Cep521–757 (3 μM), or TPX2-NT (6 μM) in the absence or presence of sperm nuclei (4 × 104/μL) or αAurA (0.6 μM). (F) W-blots and images of αAurA beads that were added to extract before (iii and iv) or after (i and ii) its incubation with 3 μM GST or Cep521–757 on ice. (Scale bar: 10 μm.) (G) In vitro kinase assay of endogenous AurA complexes isolated from egg extract incubated with XB buffer or Cep521–757 (3 μM). The reaction products were resolved by SDS/PAGE and transferred onto a nitrocellulose membrane, which was subjected to autoradiography (32PO4-histone H3 row) followed by W-blotting (the remaining rows). (H) W-blots of extracts incubated with soluble αAurA (0.5 μM) added either before (lanes 5–8) or after (lanes 1–4) incubation of extracts with 3 μM GST or Cep521–757 on ice. (I) Quantitation of MT-nucleating αAurA beads in F, Lower.
Interfering with the AurA–Cep192 interaction abrogated the MTOC function of both centrosomes and αAurA beads, but it did not affect Cep192 targeting to centrosomes (Fig. 3 A and D) or AurA binding to the beads (Fig. 3F, lane ii). Thus, neither Cep192 nor AurA alone is sufficient for MTOC function, which appears to require intact AurA–Cep192 complexes. Collectively, our findings imply that Cep192, through a direct interaction, targets AurA to centrosomes, where AurA–Cep192 complexes undergo di/oligomerization (likely as a consequence of their proximity and not by a direct effect of Cep192). The latter event, in turn, triggers AurA activation, which drives MT assembly.
Cep192 Maximizes AurA Activity at Centrosomes.
The notion that Cep192 promotes activation of dimerized AurA was tested further. Unlike binding to TPX2 (10–12), binding to Cep192 did not activate AurA directly, because endogenous AurA complexed to either full-length Cep192 (as is the case in naïve extract) or its AurA-BD (Cep521–757) lacked T295 phosphorylation (Fig. 3C, lane 4 vs. 1 and 5 and Fig. 3G, lane 3 vs. 2). AurA activation ensued when endogenous AurA–Cep192 complexes, but not AurA free of full length Cep192, were dimerized by bivalent αAurA or αCep192-C. To test whether the AurA–Cep192 interaction is required for kinase activation after dimerization has occurred, extract was preincubated with αAurA on ice to dimerize AurA–Cep192 complexes. This step was followed by the addition of Cep521–757 and incubation at 21 °C. Dimerization of endogenous AurA by αAurA, even when performed on ice, resulted in a slight activation of the kinase (Fig. 3H, lanes 5 and 6 vs. 1 and 2). Displacement of endogenous Cep192 from dimerized AurA in this setting and subsequent incubation at 21 °C did not prevent further AurA activation (Fig. 3F, lane iv vs. iii; and Fig. 3H, lanes 5–8). By contrast, and as expected, kinase activation was abolished when Cep192 was displaced from AurA before its dimerization (Fig. 3F, lane ii vs. i and Fig. 3H, lanes 1–4). Thus, the endogenous AurA–Cep192 interaction is essential for steps that link AurA dimerization to its activation but not for the sustained activity of the dimerized enzyme.
Moreover, when Cep192 was displaced from AurA–Cep192 complexes predimerized on αAurA beads, not only AurA activation but also MT assembly proceeded (Fig. 3F, lane iv, quantitated in Fig. 3I). Thus, after AurA–Cep192 complexes have dimerized, AurA activity and not Cep192 drives MT assembly. In support of this view, centrosomes that contained Cep192 but lacked AurA failed to promote AurA activation and to function as MTOCs (Fig. 3 A–E and Fig. S5C).
As a test of the possible biological rationale underlying the different mechanisms of AurA activation at MTs and centrosomes, we compared the maximum extent of endogenous AurA activation that can be achieved in extract in the presence of TPX2-NT and αAurA. TPX2-NT activates AurA by mimicking endogenous TPX2 (12, 16), whereas αAurA does so by dimerizing endogenous AurA–Cep192 complexes (see above). Of note, both treatments promoted AurA activation independent of MTs and RanGTP (refs. 12 and 16) and data not shown). Moreover, throughout a titration of αAurA, endogenous AurA activation was abrogated by Cep521–757, indicating dependence on an endogenous AurA–Cep192 interaction (Fig. S7A). Consistent with the above-described results, the Cep192/dimerization-mediated mechanism resulted in significantly higher AurA T295 phosphorylation (as assessed by W-blotting) and in measurably greater kinase activity (as assessed by an in vitro kinase assay) than did saturated TPX2 binding (Fig. 4A and Fig. S7 B–D). These results provide a possible explanation for the more prominent AurA T288/T295 phosphorylation at centrosomes than at MTs.
Fig. 4.
Comparison of AurA regulation by Cep192 and TPX2. (A) Quantitative comparison of AurA T295 phosphorylation induced by TPX2-NT and αAurA (see Fig. S7B for the relevant data). (B and C) Intact AurA–Cep192 complexes are essential for the efficient phosphorylation of endogenous maskin in sperm nuclei- or αAurA-supplemented extracts. Mock-treated and Cep192-depleted (B) or naïve (C) egg extracts were incubated with the indicated components and [γ-32PO4]ATP. Extracts were diluted with radio-immunoprecipitation assay (RIPA) buffer and subjected to IP with anti-maskin Ab. IPs were analyzed by autoradiography and the results quantified as described in SI Materials and Methods (see Fig. S7 G and H, Upper for the relevant data). The calculated values for the corresponding control extracts were set at 100%. (D) Model of AurA regulation by TPX2 (10–16, 25) and Cep192.
Such potent AurA activation might be critical for the efficient phosphorylation of AurA substrates at centrosomes. One of them is TACC3/maskin. Phosphorylation of TACC3/maskin by AurA at three sites is critical for centrosomes to function as potent MTOCs (reviewed in ref. 7). Our analysis (Fig. S7 E–I) revealed that, in crude extract, maskin undergoes constitutive phosphorylation, approximately half of which is attributed to AurA (Fig. S7G, Upper, lane 3 vs. 1). This constitutive phosphorylation was Cep192-independent (Fig. 4B, Buffer; and Fig. S6G, Upper, lane 2 vs. 1). By contrast, when extracts were supplemented with either sperm nuclei or αAurA, AurA-mediated maskin phosphorylation became Cep192-dependent. For example, by comparison with corresponding mock-treated extracts, this phosphorylation was decreased by ≈60–70% in Cep192-depleted or Cep521–757-supplemented extracts (Fig. 4 B and C, and Fig. S7 G–I). These data reinforce the conclusion that the AurA–Cep192 complex is specifically required for the centrosome/dimerization-mediated AurA activation.
Discussion
This study has revealed the mechanism of AurA targeting to and activation at mitotic centrosomes that underlies their function as MTOCs (Fig. 4D). To our knowledge, it also offers the first molecular insight into the function of Cep192, a key protein in centrosome biogenesis.
Cep192 binds AurA and targets it to centrosomes, where AurA forms dimers or higher order oligomers (as detected by BiFC) and undergoes activation (as evidenced by T288/T295 phosphorylation). Cep192 does not seem to be essential for AurA oligomerization in vivo, because AurA formed oligomers both at centrosomes and at MTs, likely as a result of its high intraorganelle concentration. Although the BiFC assay does not allow one to distinguish between the formation of true dimers and higher order oligomers of AurA, our experiments using αAurA demonstrate that the dimerization of endogenous AurA, in the presence of bound Cep192, is sufficient to initiate T295 phosphorylation and kinase activation. This process might occur via transphosphorylation, i.e., in a manner described for certain other protein kinases (36). Hence, Cep192 activates AurA at centrosomes via a mechanism fundamentally different from that used by TPX2 at spindle MTs (10–12). The former mechanism is inherently associated with more-prominent kinase T-loop phosphorylation and results in an apparently higher AurA activity. Nonetheless, and consistent with previous reports (10–12), TPX2-NT also promoted AurA activation, as assessed by an in vitro kinase assay (Fig. S7C) and by the stimulating effect of TPX2-NT on RanGTP-induced MT aster formation (refs. 11, 15, and 16) and data not shown). These observations, along with the higher abundance of the MT-bound versus centrosome-localized AurA, strongly suggest that TPX2 plays a significant overall role in AurA activation in the vicinity of chromatin and on MTs.
Of note, phosphorylation of AurA at T295 increased its avidity toward Cep192 (Fig. 1F and Fig. S1E) and was accompanied by the formation of high-molecular weight AurA complexes (Fig. S5B). Thus, activated AurA may well facilitate centrosomal recruitment of Cep192, further enhancing accumulation and activation of AurA at centrosomes in a positive feedback loop. High local concentration of AurA–Cep192 complexes may also favor their multimerization. Hence, by targeting AurA to centrosomes, Cep192 appears to initiate a process inherently capable of maximizing AurA activity at these organelles. Such a mechanism might be essential to ensure sufficient AurA phosphorylation-mediated recruitment and/or activation of spindle assembly factors, such as TACC3/maskin, responsible for centrosomes to operate as dominant MTOCs. Indeed, we found Cep192/oligomerization-mediated AurA activation to be critical for the function of MTOCs, both natural (centrosomes) and artificial (αAurA-coated beads). Parenthetically, the AurA–Cep192 function, reported here, is independent of centriole duplication/assembly (3, 5, 6), because exogenous, sperm nuclei-derived centrioles did not duplicate under the experimental conditions used (metaphase-arrested extract) (1, 2) and remained intact through the experiment (Fig. 3 A and D).
The formation of AurA oligomers on MTs, by itself, did not translate into additional AurA activation, as assessed by T295 phosphorylation, because AurA was not bound to Cep192 in this setting. Moreover, in the absence of an intact AurA–Cep192 interaction, even forced, bivalent antibody-induced dimerization failed to promote AurA activation. Similarly, in human cells measurable levels of T288-phosphorylated AurA were detected at centrosomes and not at MTs (ref. 26 and Fig. S1C), whereas AurA oligomerization occurred in both compartments (Fig. 2I, column 4). These data, along with the finding that Xenopus and human AurA each interacted in vitro with a highly conserved domain shared by both Xenopus and human Cep192 (Fig. S3E), suggest that AurA–Cep192 complexes operate in a similar fashion in amphibian and mammalian cells.
The precise mechanism of Cep192-mediated AurA activation at centrosomes will require further investigation. One wonders whether additional factors are involved in the formation of AurA di/oligomers and whether the latter dissociate into monomers before their participation in the phosphorylation of centrosomal substrates. In this regard, it appears that AurA–Cep192 binding alone is insufficient for AurA activation upon dimerization. Because displacement of endogenous AurA from Cep192 by an excess of either Cep521–757 or Cep1–1000, the 1,000-aa N-terminal fragment of Cep192 (Fig. S3C), prevented AurA activation by αAurA or sperm nuclei (see above and data not shown), Cep192 binding plus another function(s) of bound Cep192 are likely needed for AurA activation. Thus, in addition to the AurA-binding domain, another region(s) of Cep192 and/or a Cep192-interacting protein(s) likely contributes to the activation process (e.g., by scaffolding, allostery, or posttranslational modification(s)). It is also conceivable that part of the marked difference in T-loop phosphorylation of AurA bound to Cep192 and to TPX2 is a product of preferential phosphatase action directed at AurA bound to the latter (10–12). Of note, a release from the inhibitory effect of protein phosphatases was proposed to underlie local AurB activation by chromatin (28). In light of this observation and the detection of AurB oligomers in mitotic cells (Fig. S6C), one wonders whether AurB activation occurs via a mechanism analogous to that described above for AurA–Cep192.
In conclusion, the role of AurA in the two, independent pathways of spindle assembly (mediated by centrosomes and chromosomes) relies on spatial and quantitative control of its kinase activity by two distinct cofactors (Cep192 and TPX2, respectively) that use different organelle-targeting and AurA-regulation strategies.
Materials and Methods
Plasmid cDNA Constructs, Recombinant Proteins, and Antibodies.
Full-length X. laevis Cep192 cDNA was generated by RT-PCR using RNA from unfertilized eggs. All other Xenopus cDNA constructs were generated by PCR from a Xenopus stage 18 cDNA library (37). Human Cep192 and AurA cDNAs were PCR-amplified from a random primed, human IMR-90 cDNA library prepared as described (37). The recombinant proteins were produced in Escherichia coli, using cDNAs cloned into the vectors pET29a(+) (Novagen) and pGEX-6P-1 (GE Healthcare), followed by affinity purification by using Ni-NTA Agarose (Qiagen) and Glutathione Sepharose (GE Healthcare), respectively. All GST fusion proteins were eluted from beads with reduced glutathione. In addition, Cep521–757 lacking the GST moiety was generated by the cleavage of bead-bound GST-Cep521–757 with PreScission protease (GE Healthcare). The inhibitory effect of Cep521–757 on AurA activation and MTOC function was independent of the presence of the GST moiety (data not shown). Polyclonal antibodies were produced by immunizing rabbits with relevant protein fragments followed by affinity purification from immune sera using the cognate antigens immobilized on agarose beads. Further experimental details are provided in SI Materials and Methods.
Experiments in Xenopus Egg Extracts.
Crude metaphase-arrested egg extracts were prepared as described (21). For MT assembly, extracts were supplemented with 50 μg/mL rhodamine-labeled tubulin (Cytoskeleton). All assays in extracts were carried out at 21 °C, unless otherwise specified. Immunodepletions were performed by using specific antibodies or nonimmune IgG (control) bound to protein A-Sepharose (GE Healthcare).
Analysis of MT assembly promoted by sperm nuclei, RanGTP, or αAurA beads was carried out as described (27, 38, 39) with some modifications. The detailed experimental procedures are described in SI Materials and Methods.
Fluorescence Microscopy and BiFC Assay.
MTs were visualized with rhodamine tubulin and DNA with Hoechst 33258 or DAPI. Localization of AurA and Cep192 on MT structures was analyzed by direct immunofluorescence. Quantitative analysis of MT structures was performed by using the program ImageJ, version 10.2 (http://rsb.info.nih.gov/ij). The BiFC assay (35) was carried out by using recombinant, complementary, full-length human AurA BiFC proteins. A detailed outline of the experiments can be found in SI Materials and Methods.
Protein Interaction and Phosphorylation Assays.
The analysis of AurA–Cep192 complexes was carried out by using standard techniques of IP, GST pull-down, in vitro kinase activity assay, and sucrose density gradient fractionation. These procedures, as well as the analysis of maskin phosphorylation in egg extract, are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank C. D. Hu (Purdue University, West Lafayette, IN), M. Dasso (National Institutes of Health, Bethesda), A. C. Groen (Harvard Medical School, Boston), and R. L. Davis (Harvard Medical School, Boston) for generous gifts of reagents; S. Gygi and R. Tomaino for mass spectrometry analysis; L. Cameron for the assistance with microscopy; M. A. Cohn for helpful advice; and R. Bayliss for comments on the manuscript. This work was supported by the Department of Defense Breast Cancer Research Program Award W81XWH-04-1-0524 (to V.J.); Susan G. Komen for the Cure Award PDF0601163 (to A.D.N.); National Institute of Health Grants GM62267 and GM80676 and a Leukemia Lymphoma Scholar Award (to J.C.W.); and grants from the National Cancer Institute (to D.M.L.). D.M.L. is a grantee of and consultant to the Novartis Institute for Biomedical Research.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. HQ446227).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014664107/-/DCSupplemental.
References
- 1.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: Centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 2.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 3.Zhu F, et al. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol. 2008;18:136–141. doi: 10.1016/j.cub.2007.12.055. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Ferreria MA, et al. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol. 2007;17:1960–1966. doi: 10.1016/j.cub.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Pelletier L, et al. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- 7.Barr AR, Gergely F. Aurora-A: The maker and breaker of spindle poles. J Cell Sci. 2007;120:2987–2996. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- 8.Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: Regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Littlepage LE, et al. Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc Natl Acad Sci USA. 2002;99:15440–15445. doi: 10.1073/pnas.202606599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eyers PA, Erikson E, Chen LG, Maller JL. A novel mechanism for activation of the protein kinase Aurora A. Curr Biol. 2003;13:691–697. doi: 10.1016/s0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 11.Tsai MY, et al. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat Cell Biol. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- 12.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 13.Kufer TA, et al. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozlü N, et al. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev Cell. 2005;9:237–248. doi: 10.1016/j.devcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Bird AW, Hyman AA. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J Cell Biol. 2008;182:289–300. doi: 10.1083/jcb.200802005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sardon T, Peset I, Petrova B, Vernos I. Dissecting the role of Aurora A during spindle assembly. EMBO J. 2008;27:2567–2579. doi: 10.1038/emboj.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirota T, et al. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 18.Zhao ZS, Lim JP, Ng YW, Lim L, Manser E. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol Cell. 2005;20:237–249. doi: 10.1016/j.molcel.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 19.Molli PR, et al. Arpc1b, a centrosomal protein, is both an activator and substrate of Aurora A. J Cell Biol. 2010;190:101–114. doi: 10.1083/jcb.200908050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Portier N, et al. A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev Cell. 2007;12:515–529. doi: 10.1016/j.devcel.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 22.Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat Protoc. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- 23.Carazo-Salas RE, Karsenti E. Long-range communication between chromatin and microtubules in Xenopus egg extracts. Curr Biol. 2003;13:1728–1733. doi: 10.1016/j.cub.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Heald R, et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 25.Gruss OJ, et al. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 26.Dutertre S, et al. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J Cell Sci. 2004;117:2523–2531. doi: 10.1242/jcs.01108. [DOI] [PubMed] [Google Scholar]
- 27.Tsai MY, Zheng Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Curr Biol. 2005;15:2156–2163. doi: 10.1016/j.cub.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 28.Kelly AE, et al. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuh G, et al. Rational design of potent antagonists to the human growth hormone receptor. Science. 1992;256:1677–1680. doi: 10.1126/science.256.5064.1677. [DOI] [PubMed] [Google Scholar]
- 30.Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 31.Basto R, et al. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 32.Gruss OJ, et al. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat Cell Biol. 2002;4:871–879. doi: 10.1038/ncb870. [DOI] [PubMed] [Google Scholar]
- 33.Carazo-Salas RE, Gruss OJ, Mattaj IW, Karsenti E. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- 34.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 35.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 36.Pellicena P, Kuriyan J. Protein-protein interactions in the allosteric regulation of protein kinases. Curr Opin Struct Biol. 2006;16:702–709. doi: 10.1016/j.sbi.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Joukov V, Chen J, Fox EA, Green JB, Livingston DM. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc Natl Acad Sci USA. 2001;98:12078–12083. doi: 10.1073/pnas.211427098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- 39.Joukov V, et al. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 40.Ponting CP. A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics. 2006;22:1031–1035. doi: 10.1093/bioinformatics/btl022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.