Abstract
Whether Vibrio mimicus is a variant of Vibrio cholerae or a separate species has been the subject of taxonomic controversy. A genomic analysis was undertaken to resolve the issue. The genomes of V. mimicus MB451, a clinical isolate, and VM223, an environmental isolate, comprise ca. 4,347,971 and 4,313,453 bp and encode 3,802 and 3,290 ORFs, respectively. As in other vibrios, chromosome I (C-I) predominantly contains genes necessary for growth and viability, whereas chromosome II (C-II) bears genes for adaptation to environmental change. C-I harbors many virulence genes, including some not previously reported in V. mimicus, such as mannose-sensitive hemagglutinin (MSHA), and enterotoxigenic hemolysin (HlyA); C-II encodes a variant of Vibrio pathogenicity island 2 (VPI-2), and Vibrio seventh pandemic island II (VSP-II) cluster of genes. Extensive genomic rearrangement in C-II indicates it is a hot spot for evolution and genesis of speciation for the genus Vibrio. The number of virulence regions discovered in this study (VSP-II, MSHA, HlyA, type IV pilin, PilE, and integron integrase, IntI4) with no notable difference in potential virulence genes between clinical and environmental strains suggests these genes also may play a role in the environment and that pathogenic strains may arise in the environment. Significant genome synteny with prototypic pre-seventh pandemic strains of V. cholerae was observed, and the results of phylogenetic analysis support the hypothesis that, in the course of evolution, V. mimicus and V. cholerae diverged from a common ancestor with a prototypic sixth pandemic genomic backbone.
A Gram-negative gamma Proteobacterium, Vibrio mimicus is closely related to Vibrio cholerae. It was first described as a biochemically atypical Vibrio cholerae (1). However, it is phenotypically and genotypically distinct from V. cholerae and can be differentiated from V. cholerae by 12 specific biochemical reactions, including sucrose fermentation, Voges–Proskauer reaction (acetoin production from glucose), lipase production, sodium tartrate fermentation, and polymyxin sensitivity. showed mean pairwise divergence from V. cholerae to be ≈10%, equivalent to the divergence of Salmonella enterica LT2 from Escherichia coli K-12 (2, 3).
The natural habitat of V. mimicus is similar to that of V. cholerae, i.e., the aquatic ecosystem, including seawater, freshwater, and brackish water, where it has been found both as a free-living bacterium and in association with zooplankton, crustaceans, filter-feeding mollusks, turtle eggs, and fish. Infections in humans occur from consumption or exposure to these sources (4–9). V. mimicus human gastroenteritis is characterized by diarrhea, nausea, vomiting, abdominal cramps, and fever. However, unlike V. cholerae, V. mimicus has not been associated with epidemics of cholera-like diarrhea, probably because most isolates of V. mimicus do not produce cholera toxin (CT). In fact, Chowdhury et al. (5) reported that fewer than 10% of clinical isolates and fewer than 1% of environmental strains produce CT enterotoxin. Although a number of hypothesized virulence factors have been identified in V. mimicus, including cholera-like enterotoxin (10), heat-stable enterotoxin (11), heat-labile enterotoxin (12), hemolysin (13), protease (14), phospholipase (15), arylesterase (16), siderophore (aerobactin) (17), and hemagglutinin (18), the mechanism of its pathogenesis remains unclear.
The objective of this study was to determine the genetic basis of V. mimicus physiology, pathogenicity, and evolution and to clarify its relationship with V. cholerae. To this end, we sequenced two strains of V. mimicus, VM223, an environmental isolate collected from a bivalve in Saõ Paulo, Brazil, and MB451, a clinical isolate from a patient with diarrhea in Matlab, Bangladesh. Genome sequence comparison of these strains with each other and with V. cholerae demonstrates clear species delineation for V. mimicus but also provides evidence of probable evolution of V. mimicus and V. cholerae from a common ancestor.
Results and Discussion
Genome Features.
The combination of traditional Sanger sequencing and pyrosequencing yielded high-quality assemblies of the genomes of V. mimicus MB451 (GenBank accession number ADAF00000000) and VM223 (GenBank accession number ADAJ01000000). The genome of V. mimicus MB451 consists of two circular chromosomes of 2,971,217 and 1,304,309 bp with an average guanine-plus-cytosine (G+C) content of 46% and 45%, respectively (Fig. S1 and Table 1) and a plasmid of 37,927 bp. Chromosome II (C-II) of V. mimicus MB451 was closed, whereas chromosome I (C-I) was obtained in a single contig but was not closed because of a gap of ca. 3.5–4.0 kb, most likely a part of an rDNA repeat. RAST subsystem-based annotation (15) identified 3,802 predicted coding sequences (CDSs) and 119 RNAs in the genome of V. mimicus MB451. The genome of V. mimicus VM223 comprises eight contigs encoded on the two chromosomes, contigs 53–51-47–50-46–49, in that order, on C-1 and contigs52 and 48 on C-II. The predicted size of the genome, ca. 4,347,971 bp, is similar to that of V. mimicus MB451, with 3,290 CDSs and 111 RNAs (Table 1). Approximately 19% and 17% of the CDSs in V. mimicus MB451 and VM223, respectively, were annotated as hypothetical proteins, including proteins that are conserved in other bacteria. Like V. cholerae, most of the genes required for growth and viability are located on C-I. C-II contains relatively more hypothetical proteins. However, several genes believed to be important for normal cell function (e.g., genes encoding ribosomal proteins L20 and L35, the alkylphosphonate-utilization operon protein PhnA, and the operon transcriptional regulator encoded by uxuR, Uux) also are encoded on C-II. The overall subsystem category distributions of V. mimicus MB451 and VM223 genomes were similar to each other and to V. cholerae (Fig. S2).
Table 1.
General features of the Vibrio mimicus genome compared with other Vibrio spp.
| Feature | V. mimicus (MB-451) | V. mimicus (VM223) | V. cholerae (N16961) | V. parahaemolyticus (RIMD 2210633) | V. harveyii (BAA-1116) | V. vulnificus (CMCP6) |
| Genome size (Mb)* | 2.97/1.3 | 4.347 | 2.96/1.07 | 3.29/1.88 | 3.77/2.2 | 3.28/1.84 |
| G+C content (mol%) | 46/45 | 46 | 47/46 | 45/45 | 45/45 | 46/46 |
| Protein coding sequences | 2,321/1,125 | 3290 | 2,742/1,093 | 3,080/1,752 | 3,546/2,374 | 2,926/1,562 |
| Average CDS length (bp) | 984/955 | 1032 | 948/832 | 952/946 | 923/890 | 887/977 |
| Percent of coding region | 87/82 | 78 | 87/84 | 86/86 | 85/86 | 83/86 |
| Ribosomal RNA operons | 7/0 | 7 | 8/0 | 10/1 | 10/1 | 8/1 |
| No. of tRNAs | 97/0 | 90 | 94/4 | 112/14 | 105/16 | 98/13 |
| No. of integrons | 0/SI | SI† | 0/SI | 1/0 | NK | SI/0 |
NK, not known; SI, super integron.
*Results are shown as values for chromosome I/chromosome II.
*Results are shown as values for chromosome I/chromosome II.
†Found at small C- (C-II).
V. mimicus Plasmid.
V. mimicus MB451 contains a plasmid with high G+C content (50%), and encodes 56 CDSs with an average length of 534 bp (Fig. S1 and Table 1). Approximately 79% of the CDSs were annotated as hypothetical proteins. Most of the coding sequences with functional assignment are phage-related proteins, including phage integrase, phage portal protein, bacteriophage tail assembly protein, and others. No noticeable similarity with sequences in the database was observed, except for a few ORFs at the beginning of the sequence with significant similarity with genes of Prophage 1 of Salmonella typhimurium strain LT2. Moreover, similarity of the plasmid G+C content along with the prophage G+C (51%), the presence of multiple phage proteins, and the similarity of the few ORFs to those of a prophage in Salmonella indicate that this plasmid may, in fact, be an extrachromosomal temperate phage.
V. mimicus Super Integron.
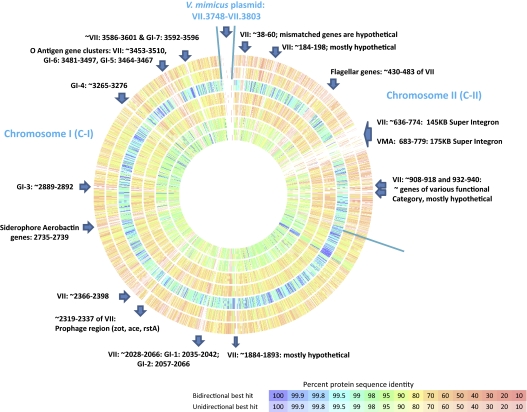
Super integrons (SI) are prevalent among Vibrio species and can be highly variable even within the same species (19). Both V. mimicus strains possess SIs on C-II that are 20–50 kb larger than V. cholerae SI (20). Significant variation was observed when SIs were compared with each other and with V. cholerae SI (Fig. 1). However, the integron integrase IntI4 of V. cholerae N16961 shows 82.5 and 83.7% nucleotide similarity with those of V. mimicus MB451 and VM223, respectively. In V. mimicus MB451 the SI encompasses VII_000636–774 (145 kb), encoding 139 ORFs, whereas in VM223 the SI encompasses VMA_000683–779 (175 kb) on contig 52 and encodes 96 ORFs. Most of the genes (55% in MB451 and 57% in VM223) are hypothetical in nature, and the coding sequences of the functional ORFs are related to homologous sequences of a wide range of microorganisms. This finding is in accordance with the postulate that the SI of the genus Vibrio serves as a capture system for acquiring DNA from the surrounding environment (21, 22).
Fig. 1.
Genome comparison of V. mimicus MB451 and reference genomes. From the outer ring to the inner ring: Vibrio fischeri MJ11, Vibrio harveyi ATCC BAA-1116, Vibrio mimicus VM223, Vibrio parahaemolyticus RIMD 2210633, Vibrio vulnificus CMCP6, Vibrio cholerae O1 El Tor N16961, and Vibrio cholerae O395. Horizontal lines are drawn to separate chromosomes and plasmid. Arrow indicates area of rearrangement or insertion.
Comparative Genomics.
The genomes of V. mimicus MB451 and V. mimicus VM223 were compared with each other and with six other completed Vibrio genomes by pairwise reciprocal BLASTP analysis (Fig. 1), and by MUMmer (Fig. S3). Overall, these genomes are most similar to the genomes of V. cholerae but are ca. 7–8% larger than V. cholerae N16961 (4.03Mb), mainly because of the significant size difference in C-II (∼23 kb), and are ca.15% smaller than both V. parahaemolyticus RIMD 2210633 and V. vulnificus CMCP6 (Table 1). A total of 1,727 nonduplicated ORFs (45.4% of MB451 and 52% of VM223 protein-coding genome) are shared by all strains, representing a “core” genome for the genus Vibrio and providing clear evidence of significant conservation among the genomes of Vibrio species. In at least six large regions on C-II and nine large regions and C-I, significant mismatch was detected (Fig. 1), explained by insertion of different genomic islands (GIs) and/or by acquisition of other mobile genetic elements, resulting in generation of strain-specific CDSs. V. mimicus MB451 encoded 215 strain-specific ORFs (5.6% a of the genome), and VM223 encoded 108 strain-specific ORFs (3.4% of the genome), most (83–85%) of which were annotated as hypothetical or proteins of unknown function. Approximately 1.1–1.3% of the genomes are conserved, having no reciprocal match with any of the reference genomes, and therefore are considered to be V. mimicus-specific ORFs. Among these, 57% are annotated as hypothetical proteins, the remaining ORFs having functional assignments in diverse categories, including metabolism, signal transduction, antibiotic resistance, virulence, pathogenicity, and other functions. Interestingly, 36% of the species-specific ORFs are encoded on C-II of V. mimicus.
Comparative ortholog cluster analysis (Table S1) revealed that V. mimicus MB451shares 2,428 nonduplicated ORFs (64% of the MB451 genome) and VM223 shares 2,428 (nonduplicated ORFs (69% of the VM223 genome) with V. cholerae (Table S1); 2,254 of these ORFs are conserved among all four strains examined. Chun et al. (23) determined that the V. cholerae core genome contains 2,613 ORFs. Taken together, these data support a close genomic relationship between V. mimicus and V. cholerae. V. mimicus MB451 and VM223 share 66 ORFs with V. cholerae O1 El Tor N16961 (Table S1) and331 ORFs with V. cholerae O1 classical O395, indicating that V. mimicus is more closely related to the classical biotype of V. cholerae than to El Tor; however, the average nucleotide identity (ANI) of conserved genes (>70% similarity) revealed no significant difference between V. cholerae classical (85.22%) and El Tor (85.25%) biotypes (Table S2) but demonstrated clear species delineation for V. mimicus.
The genomes of clinical (MB451) and environmental (VM223) V. mimicus were compared, and a set of 2,990 core ORFs was identified in the V. mimicus genome. This number may be an underestimation, because the genome of V. mimicus VM223 is incomplete. Nonetheless, this level of core gene content for V. mimicus is higher than reported for either V. cholerae or E. coli, where 2,613 and 2,200 core genes were found, respectively (23, 24). The core gene ratios of the large (C-I) and the small chromosome (C-II) are 84.7% and 68.4%, respectively. The greater core gene content of C-I reiterates the functional importance of those genes compared with those of C-II. A total of 238 nonduplicated ORFs were identified exclusively in the genome of environmental V. mimicus VM223, and 68% of these ORFs are either hypothetical or of unknown function. Most of the ORFs with functional assignments are encoded on mobile genetic elements or are proteins of nonessential function and therefore most likely were acquired by lateral gene transfer and have a role in environmental fitness, such as the Rhs elements (VMA_000863–861). The clinical V. mimicus strain MB451 has 607 nonduplicated ORFs, of which 53 are plasmid-borne. The majority (56%) of these ORFs are hypothetical or proteins of unknown function, and the ORFs with functional assignment were notably pre-cholera toxin phage (CTX-Φ) encoded genes, VPI-2 encoded genes, and multiple transcriptional regulators (TetR, Cro/Cl, ArsR, MarR family, and others).
Genome Plasticity in V. mimicus.
GIs.
GIs are non–self-mobilizing integrative and excisive elements that encode diverse functional characteristics, and their acquisition is central to bacterial evolution, serving as a mechanism of diversification and adaptation. The Island Viewer application (25) identified seven GIs in C-I of V. mimicus MB451, predicted by at least one method (Fig. S4); C-II and the plasmid have no predicted GI. Although the majority of GI-encoded ORFs are hypothetical proteins or proteins of unknown function, the ORFs with functional assignment were revealed: GIs of V. mimicus carry important functions for metabolism and adaptive traits that might be beneficial for the bacteria under certain growth or environmental conditions (SI Text).
Repeat sequences.
DNA repeats are both causes and consequences of genome plasticity, inducing deletions, amplifications, rearrangements, and gene conversion. V. mimicus MB451 genomes contain many perfect and approximate tandem repeats, 29 in C-I and 15 in C-II, with period lengths ranging from 6–387 and from 6–429, respectively (Table S3). Because tandem repeats are generated by duplication, which changes genome structures, tandem repeats may be a key process in the evolution of V. mimicus. Interestingly, many (59%) of the tandem repeats are in protein-encoding genes, which may exhibit higher mutation rates, allowing targeted sequence variation and thereby enabling a rapid response to challenging and hostile conditions in both the external environment and the human intestine.
Genomic rearrangement.
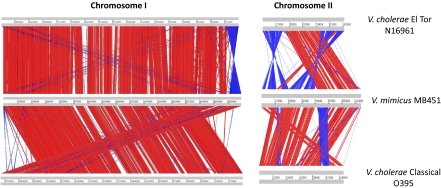
Our analysis of the V. mimicus MB451 genome, using MUMmer (26) and the Artemis Comparison Tool (27), suggested that evolution of the V. mimicus MB451 genome structure is marked by several intra- (Fig. S5) and interchromosomal (Fig. 2 and Fig. S3) rearrangements. It is apparent from the overall genomic comparison (Fig. S3) that V. mimicus MB451 is related more closely to V. cholerae and Vibrio sp. RC586 than to any of the other Vibrio spp. although a number of repeated inversions occur across the genome. These rearrangements appear to be involved in the generation of strain-specific CDSs and suggest that extensive genome plasticity is common among species of the genus Vibrio. Chromosome-wise comparison of V. mimicus with El Tor and classical biotypes of V. cholerae demonstrate that both chromosomes have greater synteny with the classical biotype of V. cholerae than with El Tor, and the overall gene content and position is much better conserved in C-I than in C-II (Fig. 2). Extensive genomic rearrangements (inversions, insertions, and deletions) obviously have occurred in C-II. These findings lead us to an important conclusion, namely, that C-II has a critical function in the evolution and genesis of speciation in the genus Vibrio. Moreover, the high degree of genome synteny, as well as the larger number of conserved ORFs shared with pre-seventh pandemic strains of V. cholerae, lead to the conclusion that, in the course of evolution, V. mimicus and V. cholerae probably diverged from a common ancestor having prototypic sixth pandemic clones of V. cholerae as their core. Once separated, the V. mimicus genome underwent extensive rearrangement, especially in C-II.
Fig. 2.
Linear pairwise comparison of the Vibrio mimicus genome by Artemis Comparison Toll. Regions with similarity are highlighted by connecting red or blue lines between the genomes; red lines indicate homologous blocks of sequence, and blue lines indicate inversions. Gaps indicate unique DNA. The gray bars represent forward and reverse strands.
Pathogenicity.
The genomes of MB451 and VM223 were found to possess virulence determinants previously known to occur in V. mimicus, such as CTX prophage and VPI, as well as a number of elements, such as VSP-II, MSHA, HlyA, PilE, and IntI4, which we identified in V. mimicus.
CTXΦ insertion site.
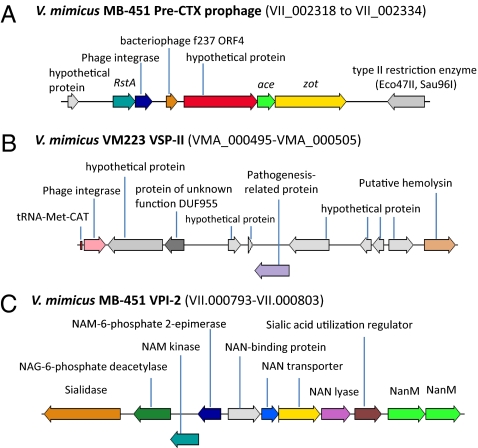
Inserted at the CTXΦ insertion locus on C-I of V. mimicus VM223 is a ca. 7.5-kb element encoding four ORFs (VMA_002135–38); three are annotated as hypothetical proteins and one as a phage protein. Inserted at this same locus in V. mimicus MB451 is a ca. 14-kb element encoding 16 ORFs (VII_002318–34), eight of which are annotated as hypothetical proteins. Interestingly, this element constitutes a pre-CTX prophage (Fig. 3A) evidenced by the presence of ORFs homologous to the zona occludens toxin, accessory cholera enterotoxin (Ace), phage-related replication protein (RstA), and phage-related integrase (RstB), which are found in strains of V. cholerae harboring pre-CTX phages. This phage-like element also encodes an ORF homologous to ORF9 of the filamentous bacteriophage f237, suggesting that the insertion locus for CTXΦ is conserved in V. mimicus and serves as an integration locus for various phages. The sequence of the putative prophage was compared with and found to be similar to that of environmental nontoxigenic V. cholerae; that is, 65–70% of the V. mimicus MB451 pre-CTX prophage had 80–83% sequence similarity with pre-CTX prophages of environmental V. cholerae non-O1/O139 strains AM-19226, 1587, MZO-3, VL426, 12129(1), and MZO-2. Moreover, 65% of the entire prophage has 82% identity with V. cholerae KSF-1ϕ, but similarity with other V. cholerae phages (such as VGJ Ace, fs1, VEJϕ, VSKK, VSK, and others) was not significant.
Fig. 3.
Pathogenicity islands found in the genome of V. mimicus.
VPI-2.
VPI-2 is a 57.3-kb gene cluster that encodes genes for neuraminidase and sialic acid metabolism and has characteristic features of a pathogenicity island (28). The genome of V. mimicus MB451 encodes a ca. 12.8-kb portion of VPI-2 (Fig. 3C) that contains genes responsible for sialic acid metabolism (homologous to VC1773-84 of V. cholerae N16961), surprisingly located on the C-II (VII_000793-VII_000803). Interestingly, in this segment of VPI-2, regions containing homologs of genes VC1758 (phage integrase)–VC1782, encoding the type I restriction modification system, and VC1785–VC1810, encoding the μ-phage, are deleted.
VSP II.
The genome of V. mimicus VM223 harbors a variant of VSP-II located on C-I (Fig. 3B). The 21.5-kb island encompasses 11 ORFs (VMA_000495–505) with an atypical G+C content of 40% and includes a phage integrase, protein of unknown function DUF955, pathogenesis-related protein, putative hemolysin, and several (63%) hypothetical proteins. Further scrutiny of this site revealed eight additional ORFs, including a DNA-repair protein, RadC, a transcriptional regulator, ribonuclease HI, and five hypothetical proteins, comprising a total of 19 ORFs encoded by V. mimicus VSP-II. In seventh pandemic isolates of V. cholerae O1 El Tor and O139, VSP-II is a 26.9-kb region (VC0490–VC0516), and 41% of V. mimicus VSP-II is identical to V. cholerae VSP-II. However, there are noticeable variations in the organization and content of ORFs in V. mimicus VSP-II compared with V. cholerae VSP-II.
Hemolysins.
V. mimicus genomes contained one copy of heat-labile (VII_000023 and VMA_001263), heat-stable (VII_000310 and VMA_001040), and enterotoxigenic (HlyA, VII_000877 and VMA_000630) hemolysin, a major virulence factor of V. mimicus (5). The heat-labile hemolysins share 97% nucleotide similarity with each other and 77% with V. cholerae El Tor hemolysin (HlyA), whereas the heat-stable hemolysins are 98% identical to each other and 76% identical to the thermostable hemolysin of V. cholerae, 70% identical to the Listonella anguillarum hemolysin (vah4), and 67% identical to the thermostable hemolysin of Vibrio vulnificus YJ016. Interestingly, no significant similarity was observed with Vibrio parahaemolyticus hemolysin (tdh). The hlyA gene of V. mimicus is 82% similar to V. cholerae hlyA (VCA0219). Both V. mimicus genomes contained 12 other hemolysins, five of which are located on C-II. This finding is in agreement with the presence of multiple hemolysins in the genomes of pathogenic and nonpathogenic Vibrio spp., including V. cholerae (23). Smith and Oliver (29) suggested these hemolysins play a role in the cold-shock response of V. vulnificus, and this suggestion certainly can be extended to V. mimicus.
Proteases.
V. mimicus protease (VMP or Vm-HA/protease) has been shown recently to modulate the activity of V. mimicus hemolysin by limited proteolysis (30). Because enhanced hemagglutinability surrogates strong bacterial cellular affinity for binding to host mucosa, VMP can be considered a significant component of V. mimicus virulence. VMP (VII_000553 and VMA_000854) of the V. mimicus genome showed 97% nucleotide similarity with each other and with the VMP of V. mimicus ES-39. VMP of VM223 and MB451 also showed significant similarity at both the nucleotide (80%) and amino acid level (88%) to HA/protease of V. cholerae. Both genomes also contained homologs (VII_000021; VMA_001265) of the prtV protease of V. cholerae on C-II, which recently has been shown to be a virulence factor in a Caenorhabditis elegans model (31). In addition, they possessed a number of other homologs of proteases, including a membrane-bound zinc metalloprotease (VII_001591; VMA_002784), protease IV (VII_001833; VMA_002551), and htpX protease (VII_002671; VMA_001839). Interestingly, the genome of V. mimicus MB451 encodes a second putative zinc metalloprotease (VII_001021), which is 97% identical (on the amino acid level) to the zinc metalloproteases of V. mimicus VM573 and VM603 and 77% identical to that of V. ulnificus CMCP6 and YJ016.
Pili and adherence.
Attachment is a critical step in pathogenesis, and both pili and flagella are important virulence factors in many pathogenic bacteria (32, 33). MSHA pilus (34), a type IV pilus (TFP), has been shown to play a role in adherence to plankton and in biofilm formation (35–37), but its role in intestinal colonization is not clear. V. mimicus genomes possess putative MSHA gene clusters (VII_003323-39; VMA_000581-91) which are of nearly equal length and are highly similar. Both are slightly smaller than that of V. cholerae, with the majority of the deleted sequence contained in the coding region of MshN. BLASTN analysis of V. mimicus MSHA demonstrated that these clusters are 77% identical with that of V. cholerae (VCO398-411), and their G+C content differs slightly from V. cholerae. Additionally, there are no apparent integrase or transposases defining this region as a pathogenicity island or suggesting another origin. Taken together, these results suggest that the MSHA gene cluster was acquired before V. cholerae and V. mimicus diverged from their most recent common ancestor. Because hemagglutination plays an important role in intestinal adherence of V. mimicus (38), the MSHA of V. mimicus may serve as a dual-function appendage, i.e., for intestinal adherence and attachment to plankton or other surfaces to form a biofilm. In addition, MSHA has been shown to serve as receptor for KSF-1Φ, VGJΦ, fs-1, fs-2, and 493 filamentous phages in V. cholerae (39–43). Therefore, the same family of phages may be expected to infect V. mimicus.
The V. mimicus genomes also harbored additional TFP genes. Both genomes contained a homolog of the type IV pilin gene, pilA (VII_001442; VMA_002922), which encodes subunits of fimbriae and often is expressed during human infection, and two copies of the tight adherence (tad) locus, with one copy on each chromosome (VMA_002363-70 and VII_002046-56 on C-1 and VMA_001394-402 and VII_000975-85 on C-II). A homolog of the TFP biogenesis protein PilE, a marker often used to assess presence of a TFP associated with virulence in V. cholerae, also was identified in the intergenic sequence between the type IV fimbrial biogenesis protein FimT (VII_002948) and the chaperone protein DnaJ (VII_002949) in V. mimicus VM223 and had 85% sequence similarity to V. cholerae pilE (VC0857).
Phylogenomics of V. mimicus.
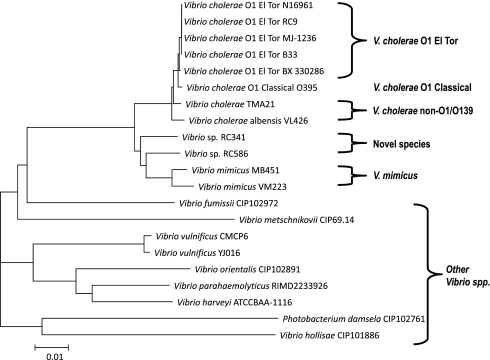
The phylogeny of V. mimicus is inferred from a neighbor-joining tree using homologous alignment of 75 conserved ORFs of different Vibrio species (Fig. 4). V. cholerae strains clustered together tightly, and the two V. mimicus strains branched separately from V. cholerae, clustering with the two novel Vibrio species (44), Vibrio spp. RC586 and RC341. Therefore, V. cholerae and V. mimicus may have evolved recently from a common ancestor and more likely are distinguished from each other by the presence or absence of mobile genetic elements, based on the high rate of insertion and deletion in the V. cholerae genome (23). A second genome-based neighbor-joining tree (Fig. S6), constructed using 925 highly conserved ORFs among V. cholerae and V. mimicus, demonstrates that V. mimicus is deeply rooted as independent branches at ancestral nodes, indicating evolutionary divergence from a progenitor of an ancestral V. cholerae. The evolutionary relationships inferred by this tree suggest that V. mimicus is more closely related to the Vibrio spp. RC586 and RC341, than to V. cholerae, consistent with results of the previous analysis Fig. 5). An evolutionary distance tree (Fig. S7), based on divergence of nucleotide sequences in different Vibrio genomes yielded results consistent with those shown in Fig. 4 and Fig. S6 and allows two important conclusions. First, the V. cholerae–V. mimicus clade forms a monophyletic group constituting a sister group of V. mimicus and Vibrio spp. RC586 and RC341, with V. cholerae as an outgroup. Second, the V. cholerae–V. mimicus clade diverged from a common ancestor before the two lineages diverged from each other. The first conclusion is strongly supported by ANI of the conserved ORFs, 84.5–85.5% with V. cholerae and 86–88% with RC341 and RC586 (Table S2); the second conclusion is supported by the evolutionary distance tree.
Fig. 4.
Neighbor-joining tree based on alignment of homologous sequences of 75 conserved ORFs. Bar represents 0.01 substitutions per site.
Conclusion
The results of the genome-sequence analysis reported here for V. mimicus provide insight into the biology of this organism, notably in refining and expanding our knowledge of the metabolism, virulence, evolution, and phylogeny of this organism. These V. mimicus genomes now can serve as reference for studies of communities in which Vibrio spp. are present. The presence of previously reported V. cholerae virulence regions (VPI and CTXΦ) in V. mimicus, as well as those identified in this study (VSP-II, MSHA, hlyA, pilE, and Intl4), and the greater similarity of these newly identified virulence regions with that of V. cholerae indicate recent interspecies lateral transfer between V. cholerae and V. mimicus and suggest that transfer of virulence factors among isolates is an ongoing process. The higher genomic relatedness of V. mimicus to the sixth pandemic biotype of V. cholerae and the acquisition of features (VSP-II, MSHA, HlyA, and Intl4) that are characteristic of the seventh pandemic biotype of V. cholerae provide insight into how genomic rearrangement can enhance virulence and environmental fitness. Finally, the genome of V. mimicus provides a starting point for understanding how a free-living, environmental bacterium can emerge as a human pathogen. It is anticipated that many of the genes identified as virulence genes will prove to be important for the fitness of the bacterium in its native aquatic environments, as evidenced by the presence of these genes in both the clinical and environmental strains and by the conservation of homologs in environmental V. cholerae genomes.
Materials and Methods
Genome Sequencing.
The genome of V. mimicus was sequenced, in part, by the Joint Genome Institute (JGI), and all general aspects of the sequencing performed at the JGI are available at http://www.jgi.doe.gov/. Draft sequences were obtained from a blend of Sanger and 454 sequences and involved paired-end Sanger sequencing on 5–8 kb plasmid libraries to 5× coverage and 20× coverage of 454 data. The Phred/Phrap/Consed software package (www.phrap.com) was used for both sequence assembly and quality assessment (45–47). After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Repeat resolution was performed using Dupfinisher (48). Gaps between contigs were closed by editing in Consed and by several targeted finishing reactions that included transposon bombs, primer walks on clones, primer walks on PCR products, and adapter PCR reactions. Gene-finding and annotation were achieved using the RAST server (49).
Comparative Genomics.
Genome-to-genome comparison was performed using three approaches as described by Chun et al. (23). To determine ANI and genetic distance between strains and to assign strains to species groups, a reciprocal best-match BLASTN analysis was performed for each genome. The average similarity between genomes was measured as the ANI of all conserved genes, as described by Konstantinidis and Tiedje (50).
Identification of GIs and Repeat Sequences.
The Island Viewer application (25), which uses three methods for GI prediction, IslandPick, IslandPath-DIMOB, and SIGI-HMM, was used for GI predictions. Because this application requires the genome to be completed, only the V. mimicus MB451 sequence was used for GI prediction, and the tandem repeat finder program (51) was used to identify the repeat sequences.
Phylogenetic Analyses Based on Genome Sequences.
A set of orthologs for each ORF of V. cholerae N16961 was obtained through comparison with different sets of strains and was individually aligned using the CLUSTALW2 program (30). The resultant multiple alignments were concatenated to generate genome-scale alignments that subsequently were used to reconstruct the neighbor-joining phylogenetic tree (52). The evolutionary model of Kimura (53) was used to generate the distance matrix, and the MEGA program (54) was used for phylogenetic analysis.
Supplementary Material
Acknowledgments
This study was supported by the Korea Science and Engineering Foundation National Research Laboratory Program Grant R0A-2005-000-10110-0 (to J.C.), National Institutes of Health Grant 1RO1A139129-01 (to R.R.C.), National Oceanic and Atmospheric Administration, Oceans and Human Health Initiative Grant S0660009 (to R.R.C.), by the Intelligence Community Post-Doctoral Fellowship Program (C.J.G), and by the Korean and Swedish governments (to International Vaccine Institute). Funding for genome sequencing was provided by the Office of the Chief Scientist and National Institute of Allergy and Infectious Diseases Microbial Sequencing Centers Grants N01-AI-30001 and N01-AI-40001.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013825107/-/DCSupplemental.
References
- 1.Davis BR, et al. Characterization of biochemically atypical Vibrio cholerae strains and designation of a new pathogenic species, Vibrio mimicus. J Clin Microbiol. 1981;14:631–639. doi: 10.1128/jcm.14.6.631-639.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd EF, Moyer KE, Shi L, Waldor MK. Infectious CTXPhi and the vibrio pathogenicity island prophage in Vibrio mimicus: Evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect Immun. 2000;68:1507–1513. doi: 10.1128/iai.68.3.1507-1513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byun R, Elbourne LD, Lan R, Reeves PR. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect Immun. 1999;67:1116–1124. doi: 10.1128/iai.67.3.1116-1124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhury MA, Yamanaka H, Miyoshi S, Aziz KM, Shinoda S. Ecology of Vibrio mimicus in aquatic environments. Appl Environ Microbiol. 1989;55:2073–2078. doi: 10.1128/aem.55.8.2073-2078.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhury MA, Aziz KM, Kay BA, Rahim Z. Toxin production by Vibrio mimicus strains isolated from human and environmental sources in Bangladesh. J Clin Microbiol. 1987;25:2200–2203. doi: 10.1128/jcm.25.11.2200-2203.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colwell RR, Huq A. Environmental reservoir of Vibrio cholerae. The causative agent of cholera. In: Wilson ME, Levins R, Spielman A, editors. Disease in Evolution: Global Changes and Emergence of Infectious Diseases. Vol. 740. New York: Ann. New York Acad. Sci; 1994. pp. 44–54. [DOI] [PubMed] [Google Scholar]
- 7.Campos E, et al. Vibrio mimicus diarrhea following ingestion of raw turtle eggs. Appl Environ Microbiol. 1996;62:1141–1144. doi: 10.1128/aem.62.4.1141-1144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury MA, Hill RT, Colwell RR. A gene for the enterotoxin zonula occludens toxin is present in Vibrio mimicus and Vibrio cholerae O139. FEMS Microbiol Lett. 1994;119:377–380. doi: 10.1111/j.1574-6968.1994.tb06916.x. [DOI] [PubMed] [Google Scholar]
- 9.Shi L, et al. Detection of genes encoding cholera toxin (CT), zonula occludens toxin (ZOT), accessory cholera enterotoxin (ACE) and heat-stable enterotoxin (ST) in Vibrio mimicus clinical strains. Microbiol Immunol. 1998;42:823–828. doi: 10.1111/j.1348-0421.1998.tb02357.x. [DOI] [PubMed] [Google Scholar]
- 10.Spira WM, Fedorka-Cray PJ. Purification of enterotoxins from Vibrio mimicus that appear to be identical to cholera toxin. Infect Immun. 1984;45:679–684. doi: 10.1128/iai.45.3.679-684.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishibuchi M, Seidler RJ. Medium-dependent production of extracellular enterotoxins by non-O-1 Vibrio cholerae, Vibrio mimicus, and Vibrio fluvialis. Appl Environ Microbiol. 1983;45:228–231. doi: 10.1128/aem.45.1.228-231.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gyobu Y, Kodama H, Uetake H. Production and partial purification of a fluid-accumulating factor of non-O1 Vibrio cholerae. Microbiol Immunol. 1988;32:565–577. doi: 10.1111/j.1348-0421.1988.tb01418.x. [DOI] [PubMed] [Google Scholar]
- 13.Nishibuchi M, Khaeomanee-iam V, Honda T, Kaper JB, Miwatani T. Comparative analysis of the hemolysin genes of Vibrio cholerae non-01, V. mimicus, and V. hollisae that are similar to the tdh gene of V. parahaemolyticus. FEMS Microbiol Lett. 1990;55:251–256. doi: 10.1016/0378-1097(90)90004-a. [DOI] [PubMed] [Google Scholar]
- 14.Chowdhury MA, Miyoshi S, Shinoda S. Purification and characterization of a protease produced by Vibrio mimicus. Infect Immun. 1990;58:4159–4162. doi: 10.1128/iai.58.12.4159-4162.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang JH, Lee JH, Park JH, Huh SH, Kong IS. Cloning and identification of a phospholipase gene from Vibrio mimicus. Biochim Biophys Acta. 1998;1394:85–89. doi: 10.1016/s0005-2760(98)00100-3. [DOI] [PubMed] [Google Scholar]
- 16.Shaw JF, et al. Nucleotide sequence of a novel arylesterase gene from Vibro mimicus and characterization of the enzyme expressed in Escherichia coli. Biochem J. 1994;298:675–680. doi: 10.1042/bj2980675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okujo N, Yamamoto S. Identification of the siderophores from Vibrio hollisae and Vibrio mimicus as aerobactin. FEMS Microbiol Lett. 1994;118:187–192. doi: 10.1111/j.1574-6968.1994.tb06824.x. [DOI] [PubMed] [Google Scholar]
- 18.Alam M, Miyoshi S, Maruo I, Ogawa C, Shinoda S. Existence of a novel hemagglutinin having no protease activity in Vibrio mimicus. Microbiol Immunol. 1994;38:467–470. doi: 10.1111/j.1348-0421.1994.tb01809.x. [DOI] [PubMed] [Google Scholar]
- 19.Mazel D, Dychinco B, Webb VA, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 20.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowe-Magnus DA, Guérout AM, Mazel D. Super-integrons. Res Microbiol. 1999;150:641–651. doi: 10.1016/s0923-2508(99)00127-8. [DOI] [PubMed] [Google Scholar]
- 22.Rowe-Magnus DA, et al. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc Natl Acad Sci USA. 2001;98:652–657. doi: 10.1073/pnas.98.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chun J, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA. 2009;106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasko DA, et al. The pangenome structure of Escherichia coli: Comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 2008;190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langille MGI, Brinkman FSL. IslandViewer: An integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25:664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carver TJ, et al. ACT: The Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 28.Jermyn WS, Boyd EF. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology. 2002;148:3681–3693. doi: 10.1099/00221287-148-11-3681. [DOI] [PubMed] [Google Scholar]
- 29.Smith B, Oliver JD. In situ and in vitro gene expression by Vibrio vulnificus during entry into, persistence within, and resuscitation from the viable but nonculturable state. Appl Environ Microbiol. 2006;72:1445–1451. doi: 10.1128/AEM.72.2.1445-1451.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 31.Vaitkevicius K, et al. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc Natl Acad Sci USA. 2006;103:9280–9285. doi: 10.1073/pnas.0601754103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bacterial pili: Molecular mechanisms of pathogenesis. Curr Opin Microbiol. 2000;3:65–72. doi: 10.1016/s1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 34.Marsh JW, Taylor RK. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J Bacteriol. 1999;181:1110–1117. doi: 10.1128/jb.181.4.1110-1117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiavelli DA, Marsh JW, Taylor RK. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl Environ Microbiol. 2001;67:3220–3225. doi: 10.1128/AEM.67.7.3220-3225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watnick PI, Fullner KJ, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alam M, Miyoshi S, Tomochika K, Shinoda S. Vibrio mimicus attaches to the intestinal mucosa by outer membrane hemagglutinins specific to polypeptide moieties of glycoproteins. Infect Immun. 1997;65:3662–3665. doi: 10.1128/iai.65.9.3662-3665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faruque SM, et al. Genomic sequence and receptor for the Vibrio cholerae phage KSF-1phi: Evolutionary divergence among filamentous vibriophages mediating lateral gene transfer. J Bacteriol. 2005;187:4095–4103. doi: 10.1128/JB.187.12.4095-4103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campos J, et al. VGJ phi, a novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTX phi. J Bacteriol. 2003;185:5685–5696. doi: 10.1128/JB.185.19.5685-5696.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honma Y, Ikema M, Toma C, Ehara M, Iwanaga M. Molecular analysis of a filamentous phage (fsl) of Vibrio cholerae O139. Biochim Biophys Acta. 1997;1362:109–115. doi: 10.1016/s0925-4439(97)00055-0. [DOI] [PubMed] [Google Scholar]
- 42.Ikema M, Honma Y. A novel filamentous phage, fs-2, of Vibrio cholerae O139. Microbiology. 1998;144:1901–1906. doi: 10.1099/00221287-144-7-1901. [DOI] [PubMed] [Google Scholar]
- 43.Jouravleva EA, et al. The Vibrio cholerae mannose-sensitive hemagglutinin is the receptor for a filamentous bacteriophage from V. cholerae O139. Infect Immun. 1998;66:2535–2539. doi: 10.1128/iai.66.6.2535-2539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haley BJ, et al. Comparative genomic analysis reveals evidence of two novel Vibrio species closely related to V. cholerae. BMC Microbiol. 2010;10:154. doi: 10.1186/1471-2180-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 46.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 47.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 48.Han CS, Chain P. Finishing repeat regions automatically with Dupfinisher. In: Arabnia HR, Valafar H, editors. International Conference on Bioinformatics and Computational Biology. Livermore, CA: CSREA Press; 2006. pp. 141–146. [Google Scholar]
- 49.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 53.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 54.Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.