Abstract
Terpene volatiles play important roles in plant-organism interactions as attractants of pollinators or as defense compounds against herbivores. Among the most common plant volatiles are homoterpenes, which are often emitted from night-scented flowers and from aerial tissues upon herbivore attack. Homoterpene volatiles released from herbivore-damaged tissue are thought to contribute to indirect plant defense by attracting natural enemies of pests. Moreover, homoterpenes have been demonstrated to induce defensive responses in plant–plant interaction. Although early steps in the biosynthesis of homoterpenes have been elucidated, the identity of the enzyme responsible for the direct formation of these volatiles has remained unknown. Here, we demonstrate that CYP82G1 (At3g25180), a cytochrome P450 monooxygenase of the Arabidopsis CYP82 family, is responsible for the breakdown of the C20-precursor (E,E)-geranyllinalool to the insect-induced C16-homoterpene (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene (TMTT). Recombinant CYP82G1 shows narrow substrate specificity for (E,E)-geranyllinalool and its C15-analog (E)-nerolidol, which is converted to the respective C11-homoterpene (E)-4,8-dimethyl-1,3,7-nonatriene (DMNT). Homology-based modeling and substrate docking support an oxidative bond cleavage of the alcohol substrate via syn-elimination of the polar head, together with an allylic C-5 hydrogen atom. CYP82G1 is constitutively expressed in Arabidopsis stems and inflorescences and shows highly coordinated herbivore-induced expression with geranyllinalool synthase in leaves depending on the F-box protein COI-1. CYP82G1 represents a unique characterized enzyme in the plant CYP82 family with a function as a DMNT/TMTT homoterpene synthase.
Keywords: floral scent, herbivory, terpene biosynthesis
Plants interact with the environment by producing a variety of chemical compounds. In particular, volatile compounds emitted from flowers and vegetative plant tissues serve as attractants for pollinators or exert defensive activities against herbivores, thereby contributing to plant survival and reproductive success. Among the most common plant volatiles are the irregular C16-homoterpene (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene (TMTT) and its C11-analog (E)-4,8-dimethyl-1,3,7-nonatriene (DMNT), both of which are widespread floral odor constituents contributing to the “white-floral image” of night scented flowers (1). Moreover, TMTT and DMNT are released in response to herbivore attack from the foliage of gymnosperms (2) and numerous angiosperms, both monocots and dicots (3–7). Several studies have indicated a role of homoterpene volatiles in the attraction of herbivore predators in indirect plant defense. For example, de Boer et al. (8) demonstrated that TMTT influenced the foraging behavior of predatory mites when emitted in the presence of other induced volatiles from lima bean leaves infested by spider mites (Tetranychus urticae). In addition, treatment of lima bean with the terpenoid pathway inhibitor fosmidomycin, which severely reduced the emission of homoterpenes, led to a reduced attraction of the predatory mite Phytoseiulus persimilis (9).
In Arabidopsis, emission of TMTT and other volatiles is induced upon leaf damage by the crucifer-specialists Pieris rapae and Plutella xylostella (10, 11). The volatile blend was shown to attract the parasitic wasp Cotesia rubecula, which parasitizes P. rapae larvae and, therefore, led to increased plant fitness (10, 12). Although Arabidopsis leaves release none or negligible amounts of DMNT under natural conditions, olfactometer experiments with transgenic Arabidopsis plants, which were constitutively emitting DMNT and its precursor (E)-nerolidol, demonstrated the ability of the two volatile compounds to attract P. persimilis (13).
Homoterpenes may also exert other defensive activities, such as the direct repulsion of aphids (14). Moreover, homoterpene emission from Arabidopsis is induced upon infection by Pseudomonas syringae DC3000 (15) and after fungal elicitor treatment (11). Finally, studies in lima bean revealed the ability of TMTT to induce the expression of defense genes in plant–plant interactions (5).
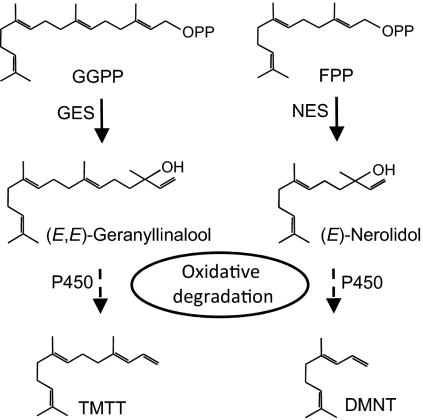
Despite the wide occurrence of homoterpenes in floral odors and volatile blends induced by biotic stress, knowledge of the biosynthesis of these compounds has been fragmentary. The first committed step in the formation of TMTT is the conversion of the central C20-diterpene precursor geranylgeranyl diphosphate to the tertiary alcohol (E,E)-geranyllinalool (Fig. 1). A geranyllinalool synthase (GES) has been identified from Arabidopsis (11) and terpene synthases catalyzing the analogous conversion of the C15-prenyldiphosphate farnesyl diphosphate to (E)-nerolidol in DMNT formation have been characterized (3, 7). Experiments with stable-isotope precursors suggested a subsequent oxidative degradation of (E,E)-geranyllinalool and (E)-nerolidol to their respective homoterpenes (Fig. 1) (16). Analogous biosynthetic pathways involving the oxidative C-C bond cleavage of a tertiary alcohol have been described for the dealkylation of 22-hydroxy-cholesterol into androstenolone (17) and the formation of the furanocoumarin psoralen from its precursor (+)-marmesin in Apiaceae (Ammi majus) (18). Because these pathways are catalyzed by one or two enzymes of the cytochrome P450 monooxygenase (P450) family, it was assumed that P450-type enzymes may catalyze the final degradation steps in homoterpene biosynthesis (Fig. 1).
Fig. 1.
Proposed biosynthetic pathways for the formation of volatile homoterpenes in plants. GGPP, geranylgeranyl diphosphate; FPP, farnesyl diphosphate; TMTT, 4,8,12-trimethyltrideca-1,3,7,11-tetraene; DMNT, 4,8-dimethyl-1,3,7-nonatriene; GES, geranyllinalool synthase; NES, nerolidol synthase; P450, cytochrome P450 monooxygenase.
Here, we report that CYP82G1 (At3g25180), encoding a P450 enzyme of the Arabidopsis thaliana CYP82 family, is responsible for the conversion of (E,E)-geranyllinalool to TMTT. CYP82G1 is expressed constitutively in flowers and coexpressed locally with GES at the sites of insect feeding damage. We further show that the recombinant CYP82G1 enzyme is able to convert (E)-nerolidol, the C15-analog of geranyllinalool, to the respective C11-terpene DMNT and that substrate specificity is dependent on the position of the hydroxyl group and configuration of the prenyl chain. CYP82G1 is unique as a characterized enzyme in the plant CYP82 family that functions in homoterpene volatile formation.
Results
CYP82G1 Is Coexpressed with GES upon Elicitor Treatment.
As a strategy to identify putative P450s involved in TMTT biosynthesis, we performed in silico screening of P450 gene candidates, which are coexpressed with GES encoding the precursor enzyme geranyllinalool synthase (Table S1). Besides P450s, putative coexpressed flavin-dependent monooxygenases, dioxygenases, and peroxidases, were considered as possible gene candidates because their role in TMTT formation could not be entirely excluded at the beginning of the study. A total of 16 genes with the highest coexpression coefficients were selected for further analysis (Table S1).
We then performed semiquantitative RT-PCR to examine transcripts of the selected genes in detached rosette leaves treated with alamethicin, an ion-channel–forming peptide elicitor from the fungus Trichoderma viride (19) (Fig. S1A). Because alamethicin induces the expression of GES and the formation of TMTT (11), enhanced mRNA levels of candidate genes coexpressed with GES were expected in comparison with basal transcription levels in control leaves. Gene transcript analysis was also performed in plant lines constitutively expressing GES under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Fig. S1A). These plants were previously shown to produce geranyllinalool and TMTT at ratios of 1:2 to 1:5 (11), indicating expression of the genes responsible for the conversion of geranyllinalool to TMTT. As an additional screening strategy, we tested whether the transcription of candidate genes was dependent on the F-box protein COI1, a central regulator in jasmonate-dependent responses. Plants constitutively expressing GES (11) and crossed into the coi1 background produced geranyllinalool but no TMTT (Fig. S1B), demonstrating COI1 dependency of the final steps in TMTT biosynthesis. Therefore, no expression of candidate genes was expected in these lines. A comparison of transcripts of all 16 genes in the different plants under the described conditions resulted in the selection of two gene candidates as best matches for the expected transcript profile (Fig. S1A): the putative P450 gene At3g25180 (CYP82G1) and the putative flavin-dependent monooxygenase At1g19250 (20).
CYP82G1 Gene Knockout Plants Do Not Produce TMTT and Their Phenotype Is Complemented by the Constitutive Expression of CYP82G1.
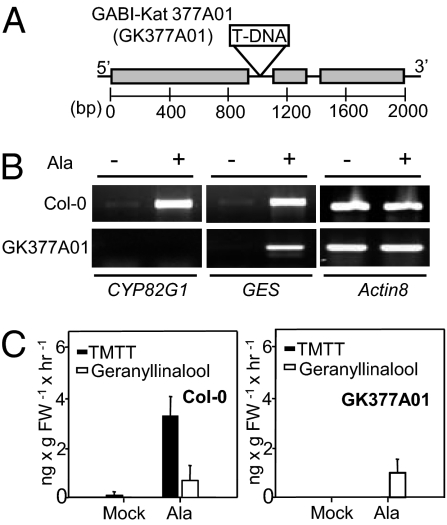
To determine the function of CYP82G1 in the formation of TMTT in vivo, a corresponding gene knockout line, GABI-Kat (GK) 377A01 (21), was analyzed. GK377A01 plants carry a T-DNA insertion in the first intron of the CYP82G1 gene (Fig. 2A). When leaves of homozygous GK377A01 plants were treated with alamethicin, no CYP82G1 transcript was detected by RT-PCR (Fig. 2B) compared with wild-type plants. In contrast, induction of the GES transcript was found in wild-type and CYP82G1 knockout plants, confirming a successful elicitation by alamethicin (Fig. 2B). In agreement with these results, the CYP82G1 mutant produced geranyllinalool but no TMTT in response to alamethicin treatment (Fig. 2C and Fig. S1C). Interestingly, emission levels of geranyllinalool in alamethicin-treated CYP82G1 knockout plants were similar to those in wild-type plants and no major accumulation of geranyllinalool was observed, suggesting feedback regulatory mechanisms in the formation of the TMTT precursor. We also tested for the formation of TMTT in alamethicin-treated knockout lines of At1g19250 (salk_026163) and two other genes (salk_114795 for At3g55970 and salk_073705 for At5g05600), which showed no expression in Pro35S:GES × coi1 plants (Fig. S1A and Table S1). None of the mutants showed disrupted TMTT formation (Fig. S1D).
Fig. 2.
Emission of TMTT and expression of CYP82G1 in wild type and CYP82G1 knockout plants in response to mock- and alamethicin-treatment. (A) Schematic of the CYP82G1 gene and position of the T-DNA insertion in the GK377A01 line. Gray boxes represent exons, and introns are shown by the black line. The position of the T-DNA insertion in intron 1 is indicated. (B) Semiquantitative RT-PCR analysis of CYP82G1 and GES (geranyllinalool synthase) gene transcripts in rosette leaves of Col-0 and GK377A01 plants in response to 30 h of mock- and alamethicin (Ala)-treatment. Actin8 transcripts were analyzed as a control. (C) Quantitative analysis of geranyllinalool and TMTT emission from wild-type and CYP82G1 mutants treated as described in B. Numbers are means ± SEM (n = 3).
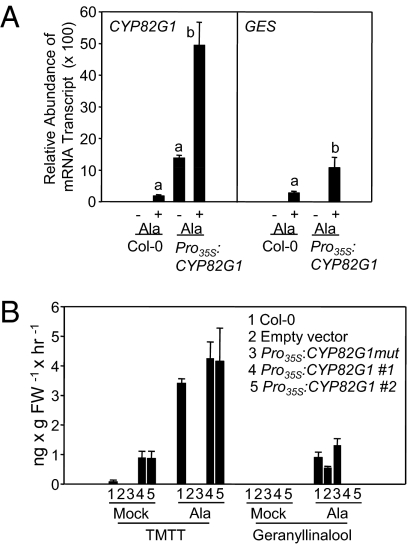
To complement the GK377A01 phenotype, a 1,548-bp CYP82G1 cDNA (GenBank NM_113423) was expressed constitutively under the control of the CaMV 35S promoter in the GK377A01 mutant background. Expression of CYP82G1 in the transgenic lines (Fig. 3A and Fig. S1E) restored the formation of TMTT (Fig. 3B). In the absence of an elicitor, emission of TMTT from Pro35S:CYP82G1 plants was higher than TMTT background emissions from wild-type plants (Fig. 3B), indicating an increased conversion of basal levels of geranyllinalool. Interestingly, treatment of leaves of CYP82G1-expressing plants with alamethicin caused higher levels of CYP82G1 transcript (and to some extent GES transcript) compared with mock-controls, suggesting an increased mRNA stability under these conditions (Fig. 3A). In contrast, the rise in alamethicin-induced TMTT emission was only ≈1.5-fold over that of induced wild-type plants (Fig. 3B), which further indicates additional regulatory mechanisms in TMTT formation based on substrate limitation or feedback control. Nevertheless, the higher emission rates of TMTT in elicitor-treated CYP82G1-expressing lines correlated with undetectable levels of geranyllinalool in contrast to wild-type plants (Fig. 3B), suggesting an efficient conversion of geranyllinalool to TMTT by the constitutively expressed CYP82G1 enzyme.
Fig. 3.
Complementation analysis of the CYP82G1 T-DNA insertion line GK377A01. (A) Quantitative RT-PCR analysis of relative CYP82G1 and GES transcript levels in Col-0 and Pro35S:CYP82G1 plants with and without alamethicin (Ala) treatment. Transcript levels of Col-0 mock controls were arbitrarily set to 1. The average GES mRNA level in mock controls of transgenic plants was 11.1 (not visible at the selected y-axis scale). Different letters above bars indicate significant differences among the means calculated with one-way analysis of variance (ANOVA) and Tukey-Kramer HSD test for CYP82G1 (P < 0.01) and GES (P < 0.05). Values were normalized to transcript levels of protein phosphatase 2A and are means ± SEM of three individual wild type plants or transgenic plants of three independent lines (#1–3). (B) Quantification of TMTT and geranyllinalool emission from wild type and Pro35S:CYP82G1 plants of two independent lines. Lines carrying the empty expression vector or an expression construct with a truncated CYP82G1 gene were used as controls (Fig. S1E). Numbers are means ± SEM from at least three individual plants. Differences in TMTT emission between Pro35S:CYP82G1 plants and wild-type plants were not statistically significant.
In addition to the full-length ORF of CYP82G1, a 1,121-bp cDNA encoding a CYP82G1 protein truncated by 150 amino acids at the carboxy-terminus was expressed in the GK377A01 mutant background (Fig. S1E). Complementation with this truncated CYP82G1 protein, which lacked the heme-binding domain (FGSGRRSCPG) and the highly conserved PERF region (Fig. S2A), did not result in the production of TMTT (Fig. 3B).
CYP82G1 Functions as a TMTT and DMNT Synthase.
To verify the biochemical function of CYP82G1 in vitro, the ORF of the CYP82G1 cDNA encoding 515 amino acids was cloned into the yeast expression vector YEp352 under the control of the constitutive alcohol dehydrogenase 1 (ADH1) promoter (22). Microsomes were isolated from transformed yeast WAT11 cells and tested for P450 enzyme activity using (3RS)-(E,E)-geranyllinalool as the substrate. As expected, TMTT was detected as the enzyme product in the presence of the P450 cofactor NADPH (Fig. S3A). No TMTT was found in assays with microsomes extracted from yeast carrying the empty YEp352 vector.
To probe the substrate specificity of CYP82G1, enzyme assays were also conducted with the smaller C15- and C10-analogs of geranyllinalool, (E)-nerolidol and linalool. (E)-Nerolidol [>95% (3S)-enantiomer] was converted into DMNT (Fig. S3B and Table S2), although no C6-degradation product (4-methyl-1,3-pentadiene) was detected from racemic linalool or (R)-(−)-linalool (Table S2).
Conversion of (E,E)-geranyllinalool and (E)-nerolidol to their respective homoterpene products could also be achieved by in vivo assays and headspace volatile analysis of the transformed yeast cultures (Table S2). A C4-cleavage product (but-1-en-3-one) resulting from the breakdown of (E,E)-geranyllinalool or (E)-nerolidol was not observed, neither in vitro nor in vivo, and none of the previously proposed ketone intermediates, C18-farnesylacetone and C13- geranylacetone (16), were detected in comparison with empty vector control assays.
In addition to (E)-nerolidol and linalool, we tested eight other putative substrates using in vitro and in vivo assays (Table S2). With the exception of the in vivo formation of small amounts of DMNT and TMTT in the presence of (E,E)-farnesylacetone and (E)-geranylacetone, none of the selected compounds was converted into the homoterpene products (Table S2). The trace amounts of TMTT and DMNT found by incubation with (E,E,E)-geranylgeraniol and (E,E)-farnesol, respectively, can most likely be attributed to contamination by (E,E)-geranyllinalool and (E)-nerolidol.
We further determined basic kinetic properties of CYP82G1 for the substrates (3RS)-(E,E)-geranyllinalool and (3S)-(E)-nerolidol (Table 1). Apparent Km values were similar for both substrates and comparable to the apparent Km of A. majus psoralen synthase, which catalyzes an analogous cleavage reaction of (+)-marmesin (18). In contrast, catalytic efficiencies for (E)-nerolidol and (E,E)-geranyllinalool were 30- to 70-fold lower than that reported for psoralen synthase. A more detailed comparison of kinetic parameters between the single (3S)- or (3R)-enantiomers of (E)-nerolidol and (E,E)-geranyllinalool was not possible because of substrate unavailability.
Table 1.
Kinetic parameters of the CYP82G1 recombinant enzyme
| Substrate | Km (μM) | Vmax (pkat/mg) | kcat (s−1) | kcat/Km (s−1/μM) |
| (3RS)-(E,E)-geranyllinalool | 2.68 ± 0.71 | 16.37 ± 0.78 | 0.11 ± 0.01 | 0.05 ± 0.01 |
| (3S)-(E)-nerolidol | 1.84 ± 0.11 | 29.09 ± 1.39 | 0.20 ± 0.01 | 0.11 ± 0.01 |
Values shown are means ± SEM of three replicates. Km, Michaelis-Menten constant; Vmax, maximal velocity; kcat, turnover number.
CYP82G1 is Expressed in Inflorescences and in Leaves upon Insect Feeding.
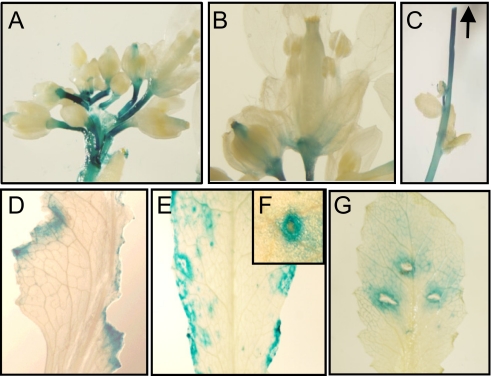
To allow for a more detailed analysis of the tissue-specific expression of CYP82G1, we performed histochemical CYP82G1 promoter-beta-glucuronidase (GUS) assays. In Arabidopsis plants stably transformed with a GUS vector construct carrying a 2.6-kb CYP82G1 promoter fragment, GUS staining occurred in flower peduncles, in the receptacle of developing and mature flowers, and in the stigma of mature opening flower buds (Fig. 4 A and B). GUS staining was also observed in stems with GUS activity gradually increasing toward the inflorescence (Fig. 4C). Although no GUS staining was detected in undamaged leaves of mature plants, reporter-gene activity was induced within 24 to 48 h of feeding damage by P. xylostella larvae (Fig. 4D). GUS staining occurred locally in a 0.3-inch wide zone around the site of damage (Fig. 4D). A similar response was found in leaves infested with thrips (Fig. 4 E and F), and after 24 h of mechanical wounding (Fig. 4G).
Fig. 4.
Histochemical analysis of CYP82G1 promoter activity. A 2.6-kb fragment of the CYP82G1 promoter was cloned upstream of the GUS reporter gene. ProCYP82G1:GUS gene expression pattern in inflorescences (A and B), and stems (C). GUS activity was induced locally at sites of feeding damage by P. xylostella (D), and thrips (E, Inset F), as well as by mechanical wounding (G). The arrow indicates the part of the shoot, which is closest to the inflorescence. Images are representative for at least five independent transgenic lines.
Induction of CYP82G1 in response to feeding by P. xylostella larvae was confirmed at the gene transcript level using quantitative real-time PCR (Fig. S4A). Transcription of CYP82G1 was also found upon infection with P. syringae DC3000, which induces TMTT emission and expression of GES (11, 15) (Fig. S4A). Transcript levels were in both cases lower than those in alamethicin-treated leaves in comparison with the respective mock-controls (Fig. S4A). In contrast to the induction of CYP82G1 transcripts by biotic stress and elicitor-treatment, only a transient accumulation of CYP82G1 mRNA similar to that of GES was found after mechanical damage, which is in agreement with the absence of TMTT emission after 24 h of mechanical wounding (11) (Fig. S4B). The observed GUS activity after mechanical wounding is a result of the stability of the transiently induced GUS protein.
Discussion
We have identified CYP82G1 from Arabidopsis as a P450 enzyme that catalyzes the final step in the biosynthesis of the common plant homoterpene volatiles TMTT and DMNT. The CYP82G1 amino acid sequence contains the highly conserved heme-binding domain, as well as the P450-specific PERF motif and the proline-rich domain required for efficient protein folding (Fig. S2A). Phylogenetic analysis revealed highest similarity of CYP82G1 to CYP82L2 from Populus trichocarpa (53% amino acid sequence identity) and to CYP82L3 from papaya (52% identity) within the CYP82 family of the CYP71 clan (Fig. S2B). No biochemical function has been ascribed to the latter proteins and to most of the other members of the CYP82 family except for tobacco CYP82E4v1 and CYP82E5v2, which catalyze an oxidative N-demethylation reaction in the conversion of nicotine to nornicotine (23, 24) and Arabidopsis CYP82C2 and CYP82C4, which can hydroxylate the substrate 8-methoxypsoralen (25). Given the common emission of homoterpenes among a large number of angiosperms, including tobacco (26), Medicago (6), and poplar (27, 28), it is possible that other members of the CYP82 family (Fig. S2B) are involved in homoterpene biosynthesis. Interestingly, the CYP82 family is absent in monocots (29), indicating an independent convergent evolution of P450s with functions in homoterpene biosynthesis in monocots and dicots.
The recombinant CYP82G1 enzyme showed narrow substrate specificity for (E,E)-geranyllinalool and (E)-nerolidol. No enzymatic product was found for the (Z)-stereoisomer of nerolidol, for primary prenylalcohols, or unsaturated substrate analogs (Table S2), indicating a critical role for the position of the hydroxyl group and the configuration and degree of saturation of the aliphatic chain for substrate binding and conversion. The latter result is in agreement with feeding experiments in lima bean using several partially saturated analogs of (E)-nerolidol (30).
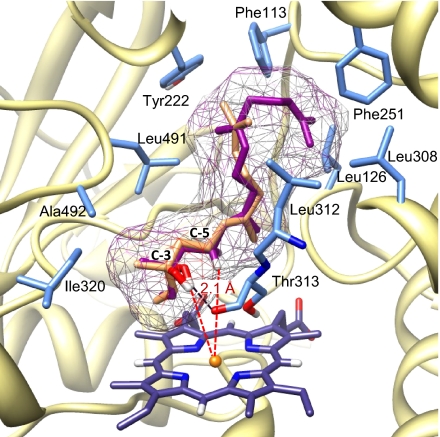
To further predict the binding mode of the two substrates, the CYP82G1 fold was modeled on structural cores and regions of templates derived from four mammalian P450 structures (2F9Q, 3CZH, 3E6I, 2Q9F) (SI Materials and Methods), which were selected based on highest amino acid sequence similarity (∼42%) to the CYP82G1 protein. Although these P450s catalyze primarily hydroxylation reactions of different substrates (SI Materials and Methods), several of them accommodate substrates of hydrophobic nature in hydrophobic active-site cavities. The generated CYP82G1 model was supported by the identification of 10 putative substrate-binding residues (Fig. 5), all of which are positioned in five substrate recognition sites (SRS1, -2, -3, -4, -6) previously predicted for Arabidopsis P450s (31) (Fig. S2A). Molecular docking of (E,E)-geranyllinalool and (E)-nerolidol showed that both substrates occupied the same position in the enzyme binding site with the hydroxyl group at C-3, forming a strong hydrogen bond to the carbonyl oxygen of Thr313 (Fig. 5). Positioning of the hydrophobic chain of both substrates was supported by several hydrophobic residues lining the active site cavity (Fig. 5). The model further indicated that the position of the allylic hydrogen atoms at C-5 of (E,E)-geranyllinalool and (E)-nerolidol and the hydroxyl group at C-3 relative to the reactive iron-oxo heme moiety (Fig. 5) supports an oxidative-bond cleavage reaction proceeding by a syn-elimination (β-elimination) mechanism, as previously reported by Boland et al. (16). In contrast, a docked position for linalool with the least-favorite binding energy among the compounds studied made the hydrogen atoms at C-5 inaccessible to molecular oxygen (Fig. S5A).
Fig. 5.
Position of (3S)-(E,E)-geranyllinalool (purple) and (3S)-(E)-nerolidol (orange) in the active site of the CYP82G1 homology model. The main interacting residues including hydrophobic active site residues are illustrated. According to the best-ranked docking mode for both substrates, a strong hydrogen bond is formed between the hydroxyl groups at C-3 and the carbonyl oxygen of Thr313 with a distance of 2.1 Å (black dashed line). A hydrogen at C-5 and the hydroxyl group at C-3 have equal distances of 5.3 Å (red dashed lines) to the Fe atom of the heme group.
Further analysis to explore structural elements possibly contributing to the oxidative degradation activity of the enzyme will be required. The tertiary alcohol substrate may be converted by a one-step reaction into an equimolar mixture of the homoterpene product and the C4-carbonyl fragment but-1-en-3-one (Fig. S5B, Path a), which is analogous to the conversion of the furanocoumarin (+)-marmesin into psoralen and acetone catalyzed by CYP71AJ1 from A. majus (18). Similar to results of previous experiments applying deuterium-labeled (E)-nerolidol to floral tissues or lima bean leaves (16, 17, 26), we did not detect a C4-carbonyl cleavage product, which might be because of its high chemical reactivity. In an alternative reaction, (E)-nerolidol or (E,E)-geranyllinalool may first be degraded into a C11-geranyl- or C18-farnesylacetone intermediate by removal of a C2-moiety, as suggested previously (17), before a second C-C bond cleavage yielding the final homoterpene product and acetate (30) (Fig. S5B, Path b). This pathway is in part analogous to steroid dealkylation reactions (17). In our experiments, geranylacetone and farnesylacetone were not detected as products of CYP82G1 and both compounds were rather inefficient substrates for conversion into homoterpenes. Docking of both substrates suggested an antiorientation of the carbonyl-group relative to the C-4 hydrogen (Fig. S5A), which does not allow syn-elimination and may explain the lack of enzyme activity. On the other hand, it is possible that the active site of the enzyme might undergo conformational changes after initial binding of the alcohol substrate, which may allow subsequent cleavage of a ketone intermediate.
The demonstrated enzymatic conversion of both (E,E)-geranyllinalool and (E)-nerolidol to TMTT and DMNT, respectively, indicates that a single P450 enzyme can produce both homoterpenes. Although emission of DMNT from Arabidopsis leaves is absent or marginal because of the lack of an (E)-nerolidol synthase activity, many plants produce both DMNT and TMTT as components of insect-induced or floral volatile blends. The induced expression of CYP82G1 is tightly coordinated with that of GES in a local-wound response depending on COI1 as the central regulator of jasmonate-mediated reactions. Interestingly, the expression of GES and CYP82G1 overlap only partially in the peduncles and receptacles of flowers, which may be a reason why only trace emissions of homoterpenes have been observed in Arabidopsis flowers (32).
Materials and Methods
Plant Material and Growth Conditions.
A. thaliana ecotype Columbia wild-type plants and gene-knockout mutants were grown in potting mix under short-day conditions (10-h light/14-h dark) at 22 °C and ∼150 μmol m−2·s−1 PAR. The GABI-377A01 At3g25180 gene-knockout mutant was purchased from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk) and the other gene-knockout lines were from the ABRC Stock Center (http://www.arabidopsis.org). Pro35S:GES plants were generated as described previously (11). Additional information is given in SI Materials and Methods.
Plant Treatments.
Plants used for treatments were 6- to 7-wk-old. Treatments of leaves with the elicitor alamethicin and with larvae of P. xylostella were performed with some modification, as described previously (11). For infection with P. syringae, leaves were sprayed with a P. syringae pv. tomato DC3000 cell suspension (OD600 = 0.01) according to Yan et al. (33). For mechanical wounding, leaves were wounded with a sterile needle (for GUS assays) or standard pliers (for RT-PCR) at independent sites. For RNA isolation, only the wounded area of six sites was harvested and immediately frozen at 0 to 24 h after treatment. Further details are provided in SI Materials and Methods.
Volatile Collection and Analysis.
Volatiles from alamethicin-treated leaves were collected for 28 h (7-h light period I, 14-h dark, 7-h light period II) on 5-mg charcoal traps using a closed-loop stripping method, as described previously (11, 32). Volatiles eluted from the traps were analyzed by GC-MS. For qualitative and quantitative analysis of volatiles from P450 in vitro or in vivo enzyme assays, automated solid-phase microextraction (SPME)-GC-MS was performed using a Shimadzu AOC-5000 autoinjector. Further details are described in SI Materials and Methods.
Transcript-Profile Analysis of CYP82G1 and Selected Candidate Genes.
For semiquantitative and quantitative RT-PCR analysis of gene transcripts, total RNA was isolated from leaves with the TRI REAGENT (Molecular Research Center, Inc.). First-strand cDNA was synthesized from 3 μg of DNase-treated total RNA using an oligo(dT) primer and SuperScript II reverse transcriptase (Invitrogen). Detailed information on semiquantitative and real-time PCR analysis is given in SI Materials and Methods.
Plant Transformation and Screening of Transformants.
Stable plant transformation was conducted with Agrobacterium tumefaciens strain GV3101 using the floral vacuum-infiltration method (34). Transformed seedlings were screened according to Harrison et al. (35) on 1/2 MS medium containing 0.8% sucrose and 50 μg/mL kanamycin.
Genetic Complementation Analysis.
The coding region of CYP82G1 (1,548 bp) was amplified by RT-PCR from total RNA extracted from alamethicin-treated leaves and transferred to the binary destination vector pK7WG2 (36). As a negative control of the complementation experiment, a heme-domain truncated CYP82G1 gene fragment (1,121 bp) was amplified and cloned into the same vector. Transformation of Arabidopsis and selection of kanamycin-resistant transgenic plants was performed as described above. Additional details are provided in SI Materials and Methods.
Yeast Expression and Enzyme Assay.
The full-length CYP82G1 cDNA was cloned into the modified YEp352 gateway vector under control of the constitutive ADH1 promoter (22). Expression of CYP82G1 was performed in the yeast WAT11 strain following the procedure described previously (22). CYP82G1 enzyme activity was measured in vivo or in vitro by incubating a 5-mL culture or 1-mL microsomal reaction in 20- or 10-mL PTFE/Silicon Septa screw cap glass vials, respectively, in the presence of different putative substrates (Table S2). Volatiles in the headspace of the vial were collected and analyzed by automated SPME-GC-MS as described under Volatile Analysis in SI Materials and Methods. Volatiles were sampled by SPME rather than organic solvent extracted or trapped by small-scale closed-loop stripping, as none of the latter methods proved to be successful for recovering measurable amounts of enzymatic product. Km and Vmax values were calculated by using the HYPER 1.01 software (J. S. Easterby, University of Liverpool, United Kingdom). Additional details are described in SI Materials and Methods.
Histochemical Assay.
A CYP82G1 promoter-GUS reporter gene fusion construct was generated by amplifying a 2.6-kb CYP82G1 promoter fragment (ProCYP82G1) upstream of the start codon from genomic DNA with primers C and D (Table S3). The ProCYP82G1 PCR product was inserted into the TOPO-pENTR vector (Invitrogen) and then recombined into the binary pKGWFS7.0 vector (36). Transgenic plants were generated as described above. Histochemical GUS staining was performed as previously described (37).
Comparative Modeling of CYP82G1 and Docking of Substrates.
Four high-resolution mammalian P450, structures with highest sequence similarity to CYP82G1 (PDB: 2F9Q, 3CZH, 3E6I, 2Q9F) were selected for multiple template comparative modeling using Modeler 9v7 (38). Details of homology modeling and substrate docking are described in SI Materials and Methods and Table S4.
Supplementary Material
Acknowledgments
We thank Eran Pichersky for helpful comments and Janet Webster for editing the manuscript, Wilhelm Boland (The Max Planck Institute for Chemical Ecology, Jena, Germany) for providing standards for DMNT and TMTT, Tony Shelton and Hilda Collins (Cornell University, Ithaca, NY) for providing Plutella xylostella larvae, Joe Chappell (University of Kentucky, Lexington, KY) for making available the modified YEp352 vectors, and Daniele Werck-Reichhart (Centre National de la Recherche Scientifique, Strasbourg, France) for providing yeast WAT11 cells. This work was supported by a US Department of Agriculture Cooperative State Research, Education, and Extension Service National Research Initiative Grant 2007-35318-18384 and by funds from Virginia Tech (to D.T.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009975107/-/DCSupplemental.
References
- 1.Kaiser R. In: Perfumes: Art, Science and Technology. Müller PM, Lamparsky D, editors. London: Elsevier Applied Science; 1991. pp. 213–250. [Google Scholar]
- 2.Su J-W, Zeng J-P, Qin X-W, Ge F. Effect of needle damage on the release rate of Masson pine (Pinus massoniana) volatiles. J Plant Res. 2009;122:193–200. doi: 10.1007/s10265-008-0203-7. [DOI] [PubMed] [Google Scholar]
- 3.Degenhardt J, Gershenzon J. Demonstration and characterization of (E)-nerolidol synthase from maize: A herbivore-inducible terpene synthase participating in (3E)-4,8-dimethyl-1,3,7-nonatriene biosynthesis. Planta. 2000;210:815–822. doi: 10.1007/s004250050684. [DOI] [PubMed] [Google Scholar]
- 4.Kant MR, Ament K, Sabelis MW, Haring MA, Schuurink RC. Differential timing of spider mite-induced direct and indirect defenses in tomato plants. Plant Physiol. 2004;135:483–495. doi: 10.1104/pp.103.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arimura G, et al. Herbivory-induced volatiles elicit defence genes in lima bean leaves. Nature. 2000;406:512–515. doi: 10.1038/35020072. [DOI] [PubMed] [Google Scholar]
- 6.Arimura G, et al. Herbivore-induced terpenoid emission in Medicago truncatula: Concerted action of jasmonate, ethylene and calcium signaling. Planta. 2008;227:453–464. doi: 10.1007/s00425-007-0631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouwmeester HJ, Verstappen FWA, Posthumus MA, Dicke M. Spider mite-induced (3S)-(E)-nerolidol synthase activity in cucumber and lima bean. The first dedicated step in acyclic C11-homoterpene biosynthesis. Plant Physiol. 1999;121:173–180. doi: 10.1104/pp.121.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Boer JG, Posthumus MA, Dicke M. Identification of volatiles that are used in discrimination between plants infested with prey or nonprey herbivores by a predatory mite. J Chem Ecol. 2004;30:2215–2230. doi: 10.1023/b:joec.0000048784.79031.5e. [DOI] [PubMed] [Google Scholar]
- 9.Mumm R, Posthumus MA, Dicke M. Significance of terpenoids in induced indirect plant defence against herbivorous arthropods. Plant Cell Environ. 2008;31:575–585. doi: 10.1111/j.1365-3040.2008.01783.x. [DOI] [PubMed] [Google Scholar]
- 10.Van Poecke RMP, Posthumus MA, Dicke M. Herbivore-induced volatile production by Arabidopsis thaliana leads to attraction of the parasitoid Cotesia rubecula: Chemical, behavioral, and gene-expression analysis. J Chem Ecol. 2001;27:1911–1928. doi: 10.1023/a:1012213116515. [DOI] [PubMed] [Google Scholar]
- 11.Herde M, et al. Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect-induced volatile C16-homoterpene TMTT. Plant Cell. 2008;20:1152–1168. doi: 10.1105/tpc.106.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Loon JJA, de Boer JG, Dicke M. Parasitoid-plant mutualism: Parasitoid attack of herbivore increases plant reproduction. Entomol Exp Appl. 2000;97:219–227. [Google Scholar]
- 13.Kappers IF, et al. Genetic engineering of terpenoid metabolism attracts bodyguards to Arabidopsis. Science. 2005;309:2070–2072. doi: 10.1126/science.1116232. [DOI] [PubMed] [Google Scholar]
- 14.Bruce TJA, et al. cis-Jasmone induces Arabidopsis genes that affect the chemical ecology of multitrophic interactions with aphids and their parasitoids. Proc Natl Acad Sci USA. 2008;105:4553–4558. doi: 10.1073/pnas.0710305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attaran E, Rostás M, Zeier J. Pseudomonas syringae elicits emission of the terpenoid (E,E)-4,8,12-trimethyl-1,3,7,11-tridecatetraene in Arabidopsis leaves via jasmonate signaling and expression of the terpene synthase TPS4. Mol Plant Microbe Interact. 2008;21:1482–1497. doi: 10.1094/MPMI-21-11-1482. [DOI] [PubMed] [Google Scholar]
- 16.Boland W, Gäbler A, Gilbert M, Feng Z. Biosynthesis of C11 and C16 homoterpenes in higher plants; Stereochemistry of the C-C-bond cleavage reaction. Tetrahedron. 1998;54:14725–14736. [Google Scholar]
- 17.Donath J, Boland W. Biosynthesis of acyclic homoterpenes in higher plants parallels steroid hormone metabolism. J Plant Physiol. 1994;143:473–478. [Google Scholar]
- 18.Larbat R, et al. Molecular cloning and functional characterization of psoralen synthase, the first committed monooxygenase of furanocoumarin biosynthesis. J Biol Chem. 2007;282:542–554. doi: 10.1074/jbc.M604762200. [DOI] [PubMed] [Google Scholar]
- 19.Engelberth J, et al. Ion channel-forming alamethicin is a potent elicitor of volatile biosynthesis and tendril coiling. Cross talk between jasmonate and salicylate signaling in lima bean. Plant Physiol. 2001;125:369–377. doi: 10.1104/pp.125.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartsch M, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosso MG, et al. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi S, et al. Metabolic engineering of sesquiterpene metabolism in yeast. Biotechnol Bioeng. 2007;97:170–181. doi: 10.1002/bit.21216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavilano LB, Siminszky B. Isolation and characterization of the cytochrome P450 gene CYP82E5v2 that mediates nicotine to nornicotine conversion in the green leaves of tobacco. Plant Cell Physiol. 2007;48:1567–1574. doi: 10.1093/pcp/pcm128. [DOI] [PubMed] [Google Scholar]
- 24.Siminszky B, Gavilano L, Bowen SW, Dewey RE. Conversion of nicotine to nornicotine in Nicotiana tabacum is mediated by CYP82E4, a cytochrome P450 monooxygenase. Proc Natl Acad Sci USA. 2005;102:14919–14924. doi: 10.1073/pnas.0506581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruse T, et al. In planta biocatalysis screen of P450s identifies 8-methoxypsoralen as a substrate for the CYP82C subfamily, yielding original chemical structures. Chem Biol. 2008;15:149–156. doi: 10.1016/j.chembiol.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Boland W, Feng Z, Donath J, Gäbler A. Are acyclic C11 and C16 homoterpenes plant volatiles indicating herbivory? Naturwissenschaften. 1992;79:368–371. [Google Scholar]
- 27.Blande JD, Tiiva P, Oksanen E, Holopainen JK. Emission of herbivore-induced volatile terpenoids from two hybrid aspen (Populus tremula × tremuloides) clones under ambient and elevated ozone concentrations in the field. Glob Change Biol. 2007;13:2538–2550. [Google Scholar]
- 28.Arimura G, Huber DPW, Bohlmann J. Forest tent caterpillars (Malacosoma disstria) induce local and systemic diurnal emissions of terpenoid volatiles in hybrid poplar (Populus trichocarpa × deltoides): cDNA cloning, functional characterization, and patterns of gene expression of (-)-germacrene D synthase, PtdTPS1. Plant J. 2004;37:603–616. doi: 10.1111/j.1365-313x.2003.01987.x. [DOI] [PubMed] [Google Scholar]
- 29.Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S. Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 2004;135:756–772. doi: 10.1104/pp.104.039826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gäbler A, Boland W, Preiss U, Simon H. Stereochemical studies on homoterpene biosynthesis in higher plants; Mechanistic, phylogenetic, and ecological aspects. Helv Chim Acta. 1991;74:1773–1789. [Google Scholar]
- 31.Rupasinghe S, Schuler M. Homology modeling of plant cytochrome P450s. Phytochem Rev. 2006;5:473–505. [Google Scholar]
- 32.Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E. Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 2005;42:757–771. doi: 10.1111/j.1365-313X.2005.02417.x. [DOI] [PubMed] [Google Scholar]
- 33.Yan S, et al. Role of recombination in the evolution of the model plant pathogen Pseudomonas syringae pv. tomato DC3000, a very atypical tomato strain. Appl Environ Microbiol. 2008;74:3171–3181. doi: 10.1128/AEM.00180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 35.Harrison SJ, et al. A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods. 2006;2:19. doi: 10.1186/1746-4811-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 37.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martí-Renom MA, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.