Abstract
Hibernomas are benign tumors with morphological features resembling brown fat. They consistently display cytogenetic rearrangements, typically translocations, involving chromosome band 11q13. Here we demonstrate that these aberrations are associated with concomitant deletions of AIP and MEN1, tumor suppressor genes that are located 3 Mb apart and that underlie the hereditary syndromes pituitary adenoma predisposition and multiple endocrine neoplasia type I. MEN1 and AIP displayed a low expression in hibernomas whereas the expression of genes up-regulated in brown fat—PPARA, PPARG, PPARGC1A, and UCP1—was high. Thus, loss of MEN1 and AIP is likely to be pathogenetically essential for hibernoma development. Simultaneous loss of two tumor suppressor genes has not previously been shown to result from a neoplasia-associated translocation. Furthermore, in contrast to the prevailing assumption that benign tumors harbor relatively few genetic aberrations, the present analyses demonstrate that a considerable number of chromosome breaks are involved in the pathogenesis of hibernoma.
Keywords: soft tissue tumor, lipoma, adipocytic tumor, adipose tissue, SNP array
Hibernoma is a benign neoplasm with morphological features highly similar to brown adipose tissue (BAT) (1). In contrast to white adipose tissue (WAT), which stores energy, BAT enables energy from oxidized lipids to dissipate as heat. This ability is dependent on the expression of uncoupling protein 1 (UCP1), a mitochondrial proton transporter that uncouples electron transport from ATP production. Morphological similarities between BAT and hibernoma include a typical yellow to brown appearance partially as a result of their rich vascularization (1). Microscopically, the BAT/hibernoma cells show a multivacuolated cytoplasm, numerous mitochondria, and a centrally located nucleus. Intermingled with the brown fat cells of hibernoma are varying proportions of mature, univacuolated white adipocytes with a peripherally located nucleus. The percentage of white adipocytes can be high and tumors with only small clusters of brown fat are referred to as lipoma-like. To avoid misdiagnosing hibernomas as ordinary lipomas or, more importantly, liposarcomas, clinical and morphological data can be complemented by cytogenetic analysis. The presence of translocations affecting 11q13 with few or no other aberrations is a karyotypic signature of hibernoma. FISH analysis has indicated that these rearrangements are more complex than expected from the karyotypes (2). Both hemi- and homozygous deletions have been described in the affected region, although so far without conclusive results regarding the target gene(s) (3). In the present study, we wished to determine the genetic pathways associated with hibernoma development.
Results
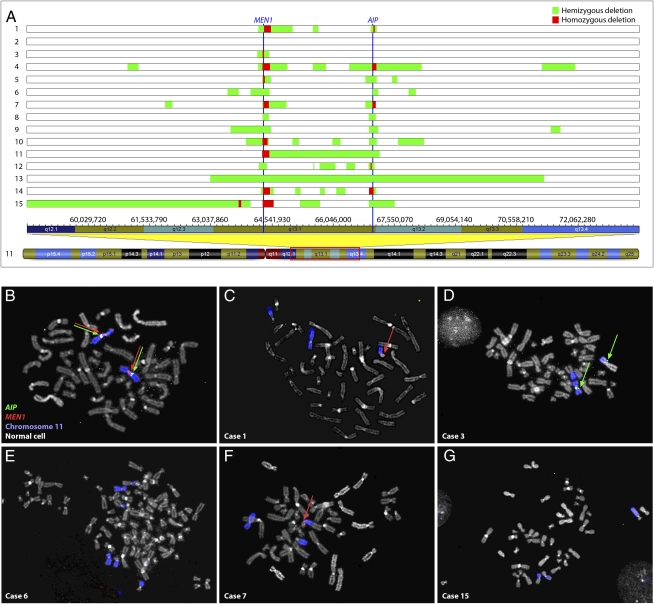
Fifteen hibernomas were available for genetic analyses (clinical information is presented in Table S1 and representative morphological features are shown in Fig. S1). Chromosome banding had been performed in eight of the cases and rearrangement of 11q13, or the neighboring 11q21 band, was found in all of them (Table S1). Genomic copy numbers were analyzed using SNP arrays and deletions in 11q13 were detected in all but one of the 15 cases (Table S2). Apart from these deletions, few or no other aberrations were identified. One additional recurrent deletion was detected in 14q11 in two cases, both of which presented translocations involving this chromosome band. Also included in the SNP analyses were normal blood DNA samples from four of the patients with hibernoma; none of them displayed the aberrations detected in the corresponding tumor samples (Table S2). Deletions in 11q13 primarily clustered around the regions covering MEN1 and AIP (Fig. 1, Fig. S2, and Table S2). Losses of these genes were confirmed by multiplex ligation-dependent probe amplification (MLPA) analysis (Fig. S3). Using this technique, the tumor lacking alterations by SNP array (case 2) also displayed loss of MEN1 and AIP. Furthermore, FISH analysis on metaphase spreads from short-term cultured tumor cells could be performed in five cases (cases 1, 3, 6, 7, and 15). This technique confirmed the SNP array findings for MEN1 in three cases and for AIP in two cases (Table 1 and Fig. 1). In the remaining cases, the results from SNP array and FISH analyses were discrepant. When SNP array data were interpreted as showing hemizygous loss of the genes, FISH analysis displayed homozygous deletion of AIP in three cases and of MEN1 in one case. This is likely explained by the fact that when tumor cells are mixed with stromal cells FISH analysis is more accurate than the corresponding SNP array. In case 7, homozygous deletion of MEN1 was detected by SNP array but could not be confirmed by FISH, as the homozygous deletion was too small to be detected by the fosmid clone used for the FISH analyses.
Fig. 1.
Genomic losses in hibernoma detected by SNP array and FISH analyses. (A) Global DNA copy numbers were evaluated by SNP array analysis and all cases with an aberrant SNP array profile displayed deletions in 11q13. In case 2, the genomic profile was normal. The vertical blue lines represent the MEN1 and AIP genes. Eight tumors showed homozygous deletion of MEN1 and three showed homozygous loss of AIP. The remaining cases demonstrated hemizygous deletion of MEN1 and all but one showed hemizygous loss of AIP. The genomic positions of all alterations are available in Table S2 and the aberrations affecting MEN1 and AIP are presented in detail in Fig. S2. Fluorescence in situ hybridization analysis with fosmid probes covering MEN1 and AIP was used to detect deletions in tumor cells, identified by rearrangements of chromosome 11. (B) As controls, signals from the probes were readily observed in normal cells from the same cases. (C) In case 1, homozygous loss of AIP and hemizygous deletion of MEN1 were detected, (D) case 3 showed homozygous loss of MEN1, (E) case 6 displayed homozygous deletions of both genes, (F) case 7 showed hemizygous deletion of MEN1 and homozygous loss of AIP, and (G) in case 15 both genes were homozygously deleted. The discrepancies between the SNP array and FISH analyses can be explained by differences in resolution of the techniques as well as normal cell contamination.
Table 1.
DNA copy number aberrations of MEN1 and AIP
|
MEN1 |
AIP |
|||||||
| Case | SNP | MLPA | FISH | Conclusion* | SNP | MLPA | FISH | Conclusion* |
| 1 | Hemi del | Normal | Hemi del | Homo del† | Hemi del | Del | Homo del | Homo del |
| 2 | Normal‡ | Del | NA | Del | Normal† | Del | NA | Del |
| 3 | Homo del | Del | Homo del | Homo del | Normal | Normal | Normal | Normal |
| 4 | Homo del | Del | NA | Homo del | Homo del | Del | NA | Homo del |
| 5 | Homo del | Del | NA | Homo del | Hemi del | Del | NA | Del |
| 6 | Hemi del | Del | Homo del | Homo del | Hemi del | Del | Homo del | Homo del |
| 7 | Homo del | Del | Hemi del | Homo del | Homo del | Del | Homo del | Homo del |
| 8 | Hemi del | Del | NA | Del | Hemi del | Del | NA | Del |
| 9 | Hemi del | Del | NA | Del | Hemi del | Del | NA | Del |
| 10 | Homo del | Del | NA | Homo del | Hemi del | Del | NA | Del |
| 11 | Homo del | Del | NA | Homo del | Hemi del | Del | NA | Del |
| 12 | Hemi del | Del | NA | Del | Hemi del | Del | NA | Del |
| 13 | Hemi del | Del | NA | Del | Hemi del | Del | NA | Del |
| 14 | Homo del | Del | NA | Homo del | Homo del | Del | NA | Homo del |
| 15 | Homo del | NA | Homo del | Homo del | Hemi del | NA | Homo del | Homo del |
del, deletion affecting at least one allele; Hemi, hemizygous; Homo, homozygous; NA, not analyzed.
*The resolution of the techniques as well as the consequence of normal cell contamination differs between SNP array, MLPA and FISH analyses, explaining the discrepancies in classifying deletions as homo- or hemizygous.
†In case 1, a homozygous deletion of MEN1 was detected by break-point cloning (Fig. 2).
‡The genomic SNP array analysis showed a normal profile in case 2, likely as a result of normal cell contamination.
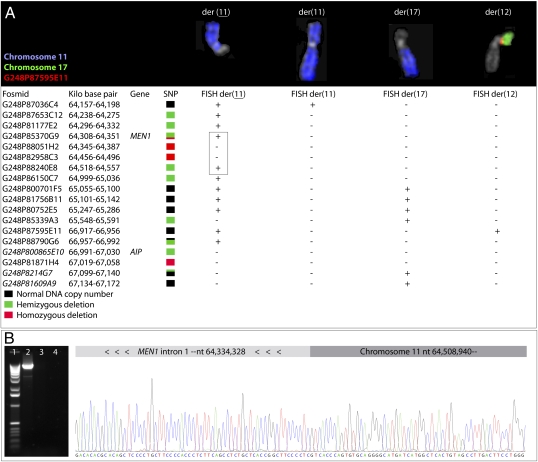
To map further the deletions associated with translocations affecting chromosome band 11q13 in hibernomas, FISH was performed in case 1 using 18 different fosmid clones (Fig. 2A). This analysis revealed complex translocation and deletion events that could be explained only partly by the t(11;17;12)(q13;q12;p13) detected at banding analysis. Both the der(12) and the der(17) chromosomes displayed genetic material from band 11q13, the der(11) chromosome had at least two interstitial deletions, and the karyotypically normal chromosome 11 was rearranged with loss of a segment greater than 3 Mb. Combined, the rearrangements and deletions must have involved at least 11 different chromosome breaks. Two of the breakpoints were cloned by using long-range PCR (Fig. 2B). One break was located in intron 1 of MEN1 and the other was positioned 180 kb upstream of this gene. Thus, exon 1 of MEN1 was homozygously lost also in this case. In summary, the whole or parts of MEN1 and AIP were homozygously deleted in 10 and six of 15 cases, respectively. One case showed normal copy number of AIP and in the remaining cases at least one of the alleles of both genes was deleted (Table 1). In all cases in which homozygous losses of MEN1 and/or AIP could not be confirmed, the coding regions of the genes were sequenced without detecting any mutations.
Fig. 2.
Mapping of chromosomal rearrangements and cloning of genomic breakpoints. (A) FISH analysis of case 1 using 18 fosmid clones detected genetic material from 11q13 on the der(12) and der(17) chromosomes, as well as loss of 11q13 material from both homologues of chromosome 11 [denoted der(11) and der(11), respectively]. The FISH analysis confirmed the deletion pattern detected by SNP array analysis, with the exception of three fosmid clones (in italics). Combined, the translocations and deletions affecting the four derivative chromosomes must have required at least 11 different chromosome breaks. Two of the breakpoints on the der(11) chromosome (indicated by the box) were cloned by using long-range PCR. The forward and reverse primers were positioned in the retained regions on opposite sides of the deletion at 64 Mb. (B) The amplified approximate 10 kb product (lane 2) was sequenced using these primers and the ends of the fragment corresponded to the expected regions. Sequencing analyses using two primers located in MEN1 showed a fusion of nt 64,334,328 in MEN1 intron 1 to nt 64,508,940 (National Center for Biotechnology Information build 36, hg18), resulting in homozygous loss of MEN1 exon 1 in this case. No amplification products were detected in PCR reactions using normal DNA as template (lane 3) or without DNA (lane 4). Lane 1 represents a 1-kb ladder.
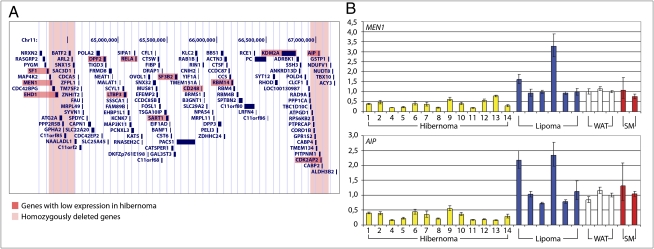
Transcriptional effects of the genomic losses were assessed in all but one of the hibernomas by using global gene expression analysis. As references, 22 lipomas, three WAT, and two skeletal muscle (SM) samples were included. The expression of all genes located in the 3 Mb region in 11q13 to which the deletions clustered was extracted from the gene expression data set (Fig. 3A). This included 132 genes from NRXN2 in 11q13.1 to ALDH3B2 in 11q13.2 (genomic position 64.13–67.20 Mb). Thirteen genes showed a significantly lower expression level in hibernoma compared with lipoma and WAT (P < 3.7 × 10−4; Dataset S1 and Fig. 3A). Four of these—MEN1, EHD1, AIP, and CDK2AP2—were located in regions affected by homozygous deletions. To evaluate if these four genes were differentially expressed in white compared with brown fat, gene expression signatures from mouse white and brown preadipocytes were downloaded from the Gene Expression Omnibus (GEO) public database (accession no. GSE7032) (4). Although the expression of EHD1 was significantly lower in brown preadipocytes compared with white preadipocytes (P < 3.7 × 10−4; Dataset S1), this was not the case for MEN1, AIP, and CDK2AP2. Although little is known about CDK2AP2, this gene is an unlikely target for deletion because activation rather than loss of function of cyclin-dependent kinases is associated with tumor development (5). The low transcript levels of MEN1 and AIP in hibernoma compared with lipoma and WAT were confirmed by real-time quantitative PCR (RT-qPCR; Fig. 3B). Both genes had a lower expression in all hibernoma samples compared with the controls (P < 0.001). As there was no difference in gene expression levels between cases with hemi- and homozygous deletion of the genes, the detected expression likely derives from contaminating normal cells. Furthermore, as brown adipocytes may be derived from a progenitor cell expressing myoblast markers, the transcript levels of MEN1 and AIP were also evaluated in SM samples (6). In this tissue, the genes showed an expression similar to the levels in lipoma and WAT (Fig. 3B). Genes highly expressed in brown fat—PPARA, PPARG, PPARGC1A, and UCP1—all showed a high expression in hibernoma compared with lipoma and WAT (P < 3.7 × 10−4; Dataset S1).
Fig. 3.
Expression analysis of genes located in 11q13. (A) The 3-Mb region in 11q13 to which the deletions clustered—comprising the deletion hotspots around MEN1 and AIP and the region in between—harbors 132 genes, 13 of which displayed a significantly lower expression level in hibernomas compared with lipomas and WAT (Mann–Whitney U test, Bonferroni corrected P < 3.7 × 10−4, Dataset S1). Of these, MEN1, EHD1, AIP, and CDK2AP2 are located in regions affected by homozygous deletions. (B) RT-qPCR confirmed the significantly lower levels of MEN1 and AIP in hibernomas compared with lipomas and WAT (Mann–Whitney U test, P < 0.001). SM showed similar expression levels for these genes as lipomas and WAT. Error bars indicate ranges.
Discussion
Hibernomas typically present balanced translocations between chromosome band 11q13 and a variety of partner chromosomes (1). Often, it is the sole cytogenetic change. Here, we show by whole-genome DNA copy number and global gene expression profiling as well as directed molecular and FISH analyses that the translocations are associated with deletion and transcriptional down-regulation of the MEN1 and AIP genes. The genes are situated in 11q13 3 Mb apart, with several seemingly unaffected genes located in between. Among the 15 tumors investigated, homozygous losses of MEN1 and AIP were detected in 10 and six cases, respectively. Of these, two cases showed homozygous loss of only part of MEN1; the remaining tumors showed loss of the entire loci. In all but one of the remaining cases, at least one of the alleles of both genes was deleted. As shown here in the cases in which FISH analyses were possible, normal cell contamination and cryptic losses may have prevented the detection of homozygous deletions in these cases. This can explain why no mutations were identified when the coding regions of MEN1 and AIP were sequenced. There was no difference in MEN1 and AIP expression levels between cases with confirmed homozygous losses and those with seemingly hemizygous deletions, suggesting that both homologues of the genes were targeted by deletion or otherwise silenced in the latter group as well.
As the deletions were not continuous and usually homozygous, the rearrangements must have involved several events and included the homologue not involved in translocation. This is supported by the detailed FISH analysis of case 1. Although this case displayed a seemingly balanced three-way translocation involving chromosomes 11, 12, and 17 as the sole cytogenetic change, FISH analysis showed complex rearrangements of chromosome 11 as well as the derivative chromosomes 12 and 17. Genetic material from 11q13 was detected on all four chromosomes and 11 chromosome breaks could be identified. Two of the breakpoints were cloned revealing loss of MEN1 exon 1, which had not been detected by any of the other methods used. Based mostly on cytogenetic information, benign tumors are believed to harbor relatively simple genetic aberrations. The present analyses of hibernoma show that benign tumors also may harbor very complex rearrangements. Furthermore, the large number of breakpoints as well as the variation in both centromeric and telomeric borders argue against the involvement of specific, break-prone sequences in the origin of the deletions affecting MEN1 and AIP.
Balanced cytogenetic abnormalities classically result in increased expression of a gene in one of the breakpoints or in the creation of a chimeric gene (7). Sometimes these gene fusions display alternating partners. However, in hibernoma, the breakpoints in 11q13 are scattered over a 10-Mb region, and the translocations affecting chromosome 11 consistently involve different partner chromosomes. Combined, these findings do not support the formation of a gene fusion. The genomic aberrations instead result in recurrent deletions of two nonadjacent tumor suppressors, and such targeting of two separate loci by seemingly balanced translocations has not been described before to our knowledge. The alterations are likely to be pathogenetically essential as the rearrangements of 11q13 are cytogenetic hallmarks of hibernoma and in all investigated cases result in losses and transcriptional down-regulation of MEN1 and AIP.
The two target genes encode well known tumor suppressors involved in hereditary tumor-predisposing syndromes. Mutations in AIP are responsible for the familial disease pituitary adenoma predisposition. The encoded protein (aryl hydrocarbon receptor interacting protein) affects the subcellular localization of the transcription factor aryl hydrocarbon receptor and is also known to interact with other proteins discussed later. Menin, the protein product of MEN1, is located primarily in the nucleus and interacts with proteins involved in transcriptional regulation and the control of genome stability (8). Constitutional mutations in MEN1 are the cause of the hereditary syndrome multiple endocrine neoplasia type I (MEN1). In addition to endocrine tumors, patients with MEN1 syndrome have displayed lipomas (or hibernomas) with subsequent loss of the WT MEN1 allele (9, 10). The exact incidence of benign adipose tumors in patients with MEN1, or for that matter in the general population, is not known, as these neoplasms are painless and slow-growing and do not always require treatment. There are, however, reports suggesting that lipomatous tumors are overrepresented in this syndrome (9, 11).
Through protein interactions, menin and AIP can be directly connected to pathways of vital importance for brown fat development. AIP is believed to function as a repressor of peroxisome proliferator-activated receptor (PPAR)-α and menin was recently shown to interact with PPAR-γ (12, 13). The latter protein is highly expressed in both brown and white adipocytes. In brown adipocytes, PPAR-γ interacts with the PPAR-γ coactivator 1-α (PPARGC1A) to induce UCP1 (14). Expression of PPAR-α is preferentially found in brown adipocytes and may be rate-limiting for brown adipogenesis (15). This protein also interacts with PPARGC1A to coactivate the transcription of UCP1 (16). In line with this, PPARA, PPARG, PPARGC1A, and particularly UCP1 all showed a high expression in hibernoma compared with lipoma and WAT. Thus, it is plausible that elimination of menin and AIP contributes to the transformation of hibernoma precursor cells by directing the gene expression profile toward a brown adipocytic phenotype, resulting in the induction of UCP1. An explanation for the dual targeting of MEN1 and AIP may be that PPAR-γ and PPAR-α act synergistically to achieve large differences in UCP1 expression, as previously suggested (17). As displayed in Fig. S1, high protein levels of UCP1 are characteristic for multivacuolated hibernoma cells. Unfortunately, there are no established hibernoma cell lines available, precluding studies in this cell type regarding the aforementioned interactions and molecular effects. In support of the importance of the PPAR proteins are the findings that the adipose tissue of rats treated with PPAR-α/γ agonists display morphologic changes highly similar to the features associated with hibernoma (18). The number of adipocytes is increased in both WAT and BAT following such treatment, and this is often associated with lobular formation of the tissues. A lobulated growth pattern is also found in hibernoma (1). Histological alterations after PPAR-α/γ agonist treatment include primarily microvacuolation of the white adipocytes and macrovacuolation of the brown adipocytes, features characteristic of hibernoma cells (19).
Materials and Methods
Tumor Samples and Chromosome Banding.
Biopsy specimens from 15 hibernomas were included in the study, and clinical information is available in Table S1. Fresh tumor samples had been processed for G-banding analysis in eight of the cases. Karyotypes of cases 1 through 6 have been published before (2, 20–22).
DNA and RNA Extraction.
DNA and RNA were extracted from fresh frozen tumor biopsies using the DNeasy Tissue Kit including the optional RNaseH treatment and the RNeasy lipid tissue kit, according to the manufacturer's instructions (Qiagen). Quality and concentration of the extracted material were measured by using a 2100 Bioanalyzer (Agilent Technologies) and NanoDrop ND-1000 (Thermo Fisher Scientific).
Whole-Genome DNA Copy Number Analysis.
Global DNA copy number analyses were performed using SNP array analysis. Tumor DNA was hybridized onto Illumina Human 1M-Duo v3.0 BeadChip (cases 1–14) and Illumina Human Omni-Quad BeadChip (case 15; Illumina), following standard protocols supplied by the manufacturer. DNA from normal blood and the corresponding tumor samples were analyzed in cases 1, 2, 4, and 6 by using the Human CNV370-Quad v3.0 BeadChip. Data analysis was done by using BeadStudio software (Illumina). SNP array data are available in GEO under accession number GSE19040.
MLPA.
Deletions of the MEN1 and AIP genes were investigated by MLPA with use of the SALSA MLPA kit P244 AIP-MEN1 according to the manufacturer's instructions (MRC-Holland). Normal blood DNA from cases 1, 2, 4, and 6 were used as controls for normal copy number. The data were intranormalized by dividing the peak area/height of each fragment by the total area/height of only the reference probes, excluding reference probes located in 11q13. Subsequently, this intranormalized probe ratio in each sample was divided by the average intranormalized probe ratio of all reference samples.
FISH.
FISH on metaphase chromosomes was used to investigate deletions of MEN1 and AIP in cases 1, 3, 6, 7, and 15. For this purpose, fosmid clones G248P85370G9 (covering MEN1) and G248P800865E10 (covering AIP) were used (BACPAC Resources Center). In case 1, translocation and deletion breakpoints in 11q13 were investigated by using fosmid clones presented in Fig. 2 (BACPAC Resources Center). Abnormal cells were identified by whole chromosome paint probes specific for chromosomes 11 and 17 (Vysis). FISH was performed as described previously (23).
Cloning of Genomic Breakpoints.
Long-range PCR using the primer pair 5′-AAATGCAGCCCAATTTCATC (forward) and 5′-CAGCCTCCTTCCCTCTTCTT (reverse) was performed according to the manufacturer's instructions (Qiagen). The amplified fragment was sequenced using the aforementioned primers as well as the primers 5′-ACCCCCTTCTCGAGGATAGA and 5′-CCAGGGTCCGCTAAGGTT.
Genomic Sequencing of MEN1 and AIP.
The coding regions of MEN1 and AIP were amplified by using primers presented in Table S3. PCR amplification of AIP was performed as described (24), and PCR protocols for both genes are available upon request. Sequencing was done by Molecular Cloning Laboratories, and data analysis was performed by using Mutation Surveyor software (SoftGenetics).
Global Gene Expression Profiling.
Global gene expression analyses using Affymetrix Human Gene 1.0 ST Arrays were performed in all cases except case 3, according to the manufacturer's instructions (Affymetrix). As controls, RNA from 22 lipomas, three WAT samples [part AM7956 (Applied Biosystems), lot 7120146 (Clontech Laboratories), lot A608327 (BioChain Institute)], and three SM samples (lots 8062503A and 7080016; Clontech Laboratories) were included. Gene expression signatures from mouse white and brown preadipocytes were downloaded from GEO under accession number GSE7032 (4). Expression data were normalized, background-corrected, and summarized by using the RMA algorithm implemented in the Affymetrix Expression Console version 1.0 software.
RT-qPCR Analysis.
The relative expressions of MEN1 (Hs00365720_m1) and AIP (Hs00610222_m1) were investigated in all cases except cases 3 and 15 using RT-qPCR and the TaqMan Gene Expression Assays (Applied Biosystems). Also included in the analysis were six of the lipoma, WAT, and SM samples used as controls in the global gene expression analysis. As an endogenous control, the expression level of TBP (part 4333769T) was quantified in all samples and calculations were done using the comparative CT method (i.e., ΔΔCT method). All reactions were performed in triplicate and assayed on a 7500 real-time PCR system (Applied Biosystems). The rationale for using TBP as endogenous control was its uniform expression in all samples at global gene expression analysis (Table S4).
Protein Detection.
Four-micrometer sections were cut from formalin-fixed, paraffin-embedded blocks and dried at 60 °C for 1 h. After dewaxing and rehydration, the sections were treated with 10 mM citrate buffer in a microwave oven for 10 min for antigen retrieval. The immunohistochemical staining with rabbit anti-UCP1 (anti–UCP-1 U6382, 1:500; Sigma-Aldrich) was performed in an automated immunostainer (Autostainer plus; Dako) by using the biotin–streptavidin–peroxidase method with diaminobenzidine as the chromogen (REAL Detection System, peroxidase/DAB+, rabbit/mouse; Dako). Mayer hematoxylin was used for counterstaining.
Statistical Analyses.
Statistical analyses were done using the Mann–Whitney U test, with the P values adjusted for multiple testing by Bonferroni correction.
Supplementary Material
Acknowledgments
We thank Y. Jin and H. Svensson for technical assistance and acknowledge the help with the microarray analyses from the Swegene Centre for Integrative Biology at Lund University. Professors L. Aaltonen and M. Nordenskjöld are gratefully acknowledged for supplying PCR protocols for the AIP and MEN1 genes. This work was supported by the Magnus Bergvall Foundation, Royal Physiographic Society (Lund, Sweden), Swedish Cancer Society, and Swedish Research Council.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE19040).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013512107/-/DCSupplemental.
References
- 1.Miettinen MM, Fanburg-Smith JC, Mandahl N. In: World Health Organization: Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. Fletcher CDM, Unni KK, Mertens F, editors. Lyon: IARC Press; 2002. pp. 33–34. [Google Scholar]
- 2.Gisselsson D, Höglund M, Mertens F, Dal Cin P, Mandahl N. Hibernomas are characterized by homozygous deletions in the multiple endocrine neoplasia type I region. Metaphase fluorescence in situ hybridization reveals complex rearrangements not detected by conventional cytogenetics. Am J Pathol. 1999;155:61–66. doi: 10.1016/S0002-9440(10)65099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maire G, et al. 11q13 alterations in two cases of hibernoma: large heterozygous deletions and rearrangement breakpoints near GARP in 11q13.5. Genes Chromosomes Cancer. 2003;37:389–395. doi: 10.1002/gcc.10223. [DOI] [PubMed] [Google Scholar]
- 4.Timmons JA, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci USA. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 6.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 8.Karhu A, Aaltonen LA. Susceptibility to pituitary neoplasia related to MEN-1, CDKN1B and AIP mutations: An update. Hum Mol Genet. 2007;16(Spec No 1):R73–R79. doi: 10.1093/hmg/ddm036. [DOI] [PubMed] [Google Scholar]
- 9.Vortmeyer AO, Böni R, Pak E, Pack S, Zhuang Z. Multiple endocrine neoplasia 1 gene alterations in MEN1-associated and sporadic lipomas. J Natl Cancer Inst. 1998;90:398–399. doi: 10.1093/jnci/90.5.398. [DOI] [PubMed] [Google Scholar]
- 10.Dong Q, et al. Loss of heterozygosity at 11q13: Analysis of pituitary tumors, lung carcinoids, lipomas, and other uncommon tumors in subjects with familial multiple endocrine neoplasia type 1. J Clin Endocrinol Metab. 1997;82:1416–1420. doi: 10.1210/jcem.82.5.3944. [DOI] [PubMed] [Google Scholar]
- 11.Vidal A, Iglesias MJ, Fernández B, Fonseca E, Cordido F. Cutaneous lesions associated to multiple endocrine neoplasia syndrome type 1. J Eur Acad Dermatol Venereol. 2008;22:835–838. doi: 10.1111/j.1468-3083.2008.02578.x. [DOI] [PubMed] [Google Scholar]
- 12.Sumanasekera WK, Tien ES, Turpey R, Vanden Heuvel JP, Perdew GH. Evidence that peroxisome proliferator-activated receptor alpha is complexed with the 90-kDa heat shock protein and the hepatitis virus B X-associated protein 2. J Biol Chem. 2003;278:4467–4473. doi: 10.1074/jbc.M211261200. [DOI] [PubMed] [Google Scholar]
- 13.Dreijerink KM, et al. The multiple endocrine neoplasia type 1 (MEN1) tumor suppressor regulates peroxisome proliferator-activated receptor gamma-dependent adipocyte differentiation. Mol Cell Biol. 2009;29:5060–5069. doi: 10.1128/MCB.01001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 15.Xue B, et al. Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. J Lipid Res. 2007;48:41–51. doi: 10.1194/jlr.M600287-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Barberá MJ, et al. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J Biol Chem. 2001;276:1486–1493. doi: 10.1074/jbc.M006246200. [DOI] [PubMed] [Google Scholar]
- 17.Xue B, Coulter A, Rim JS, Koza RA, Kozak LP. Transcriptional synergy and the regulation of Ucp1 during brown adipocyte induction in white fat depots. Mol Cell Biol. 2005;25:8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long GG, Reynolds VL, Dochterman LW, Ryan TE. Neoplastic and non-neoplastic changes in F-344 rats treated with Naveglitazar, a gamma-dominant PPAR alpha/gamma agonist. Toxicol Pathol. 2009;37:741–753. doi: 10.1177/0192623309343775. [DOI] [PubMed] [Google Scholar]
- 19.Manieri M, Murano I, Fianchini A, Brunelli A, Cinti S. Morphological and immunohistochemical features of brown adipocytes and preadipocytes in a case of human hibernoma. Nutr Metab Cardiovasc Dis. 2009;20:567–574. doi: 10.1016/j.numecd.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 20.Mertens F, et al. Hibernomas are characterized by rearrangements of chromosome bands 11q13-21. Int J Cancer. 1994;58:503–505. doi: 10.1002/ijc.2910580408. [DOI] [PubMed] [Google Scholar]
- 21.Bartuma H, et al. Assessment of the clinical and molecular impact of different cytogenetic subgroups in a series of 272 lipomas with abnormal karyotype. Genes Chromosomes Cancer. 2007;46:594–606. doi: 10.1002/gcc.20445. [DOI] [PubMed] [Google Scholar]
- 22.Bartuma H, et al. Expression levels of HMGA2 in adipocytic tumors correlate with morphologic and cytogenetic subgroups. Mol Cancer. 2009;8:36. doi: 10.1186/1476-4598-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahlén A, et al. Clustering of deletions on chromosome 13 in benign and low-malignant lipomatous tumors. Int J Cancer. 2003;103:616–623. doi: 10.1002/ijc.10864. [DOI] [PubMed] [Google Scholar]
- 24.Vierimaa O, et al. Pituitary adenoma predisposition caused by germline mutations in the AIP gene. Science. 2006;312:1228–1230. doi: 10.1126/science.1126100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.