Abstract
Background
DNA promoter methylation is a signature for silencing of tumor suppressor genes. Most widely used methods to detect DNA methylation involve three separate independent processes that include DNA extraction, bisulfite conversion and methylation detection through PCR, such as methylation specific PCR (MSP). This method includes many disconnected steps with loss of material potentially reducing the analytical sensitivity required for analysis of challenging clinical samples.
Methods
Methylation-on-Beads (MOB) is a new technique that integrates DNA extraction, bisulfite conversion and PCR in a single tube by using silica superparamagnetic beads (SSBs) as a common DNA carrier that facilitates cell debris removal and buffer exchange throughout the entire process. In addition, PCR buffer was used to directly elute bisulfite treated DNA from SSBs for subsequent target amplifications. The sensitivity of MOB was evaluated by methylation analysis of p16INK4a promoter in serum DNA of lung cancer patients and compared with conventional methods.
Result
Methylation analysis beginning with DNA extraction, followed by bisulfite conversion and MSP was successfully carried out in a single-tube within 9 hours. Median pre-PCR DNA yield was 6.6 fold higher in MOB when compared to conventional techniques. Further, MOB allowed for increased diagnostic sensitivity in analysis of p16INK4a promoter in patient serum by successfully detecting methylation in 74% of cancer patients versus the 45% detected using conventional techniques.
Conclusion
MOB successfully combined three processes into a single-tube thereby allowing for ease in handling and increased throughput in detection. Increased pre-PCR yield in MOB allowed for efficient, diagnostically sensitive methylation detection.
Keywords: promoter hypermethylation, DNA methylation, nanotechnology, magnetic beads, methylation detection, circulating DNA
The most well characterized epigenetic changes are the heritable transcriptional silencing of tumor suppressor genes (TSGs) by aberrant CpG DNA hypermethylation of their promoters(1, 2). The effect of such promoter methylation is similar to loss-of-function genetic mutations and has been observed at well-characterized TSGs that cause inherited forms of cancer when mutated in the germline events(3–5). Since methylation-based gene inactivation can occur very early during cancer progression, even before mutations are observed, detecting DNA methylation may be useful for early cancer detection (6–8). In recent years, several approaches have been designed to detect and differentiate methylated sequences in normal versus cancer tissues. First generation methods were primarily based on the use of methylation-sensitive restriction enzymes followed by southern blotting(9). Second generation methods include approaches that are focused on either discovering differentially methylated regions in normal versus cancer tissues or analyzing the methylation profile of candidate TSGs(8, 10, 11). These techniques can be broadly classified into a) CpG detection methods including MSP, qMSP and nested MSP, b) detailed analysis of specific CpG methylation patterns, for example in bisulfite sequencing and, c) genome wide approaches using array based detection such as Illumina(12) or gene expression analyses to identify genes that are expressed on reversal of epigenetic modifications by pharmacological agents(13). Most of the above methods include DNA extraction followed by sodium bisulfite conversion(14) of the denatured template DNA. DNA extraction typically involves chemical lysis of cells followed by organic solvent extraction and ethanol precipitation (PC) requiring both centrifugation and air drying(15). Extracted DNA is subject to sodium bisulfite conversion (Bst), which requires denaturation of genomic DNA, deamination of unmethylated cytosines with high concentration of sodium bisulfite followed by desulfonation using a strong base. Temperature, pH, and salt concentration all require careful calibration and efficient bisulfite conversion is recommended for 12–16h(11). High yield and quality of DNA as well as proper efficiency in bisulfite treatment are pre-requisites for these techniques to function well.
Given these multiple steps, conventional methods are relatively laborious compared to solid substrate extraction methods, wherein DNA is known to bind to silica surface in chaotropic salt solutions such as those containing iodide or perchlorate salt(16). While solid substrate methods have been implemented to simplify the process, methylation analysis is still a disjointed process wherein DNA extraction, bisulfite conversion and PCR amplification are carried out in separate tubes. Methylation-on-Beads (MOB) addresses this problem as a single-tube methylation detection method (Supplemental Data Fig. 1). MOB begins with cell lysate or patient samples mixed with silica superparamagnetic beads (SSBs)(17) in a chaotropic guanidine HCl in citric acid buffer solution, which promotes binding of DNA to SSBs. Other macromolecules and cell debris remain unbound in the solution and are then removed by extracting the liquid phase. Additional washing steps with alcohol are required to ensure the DNA purity for further analysis. The bound DNA is then eluted in low ionic strength buffer and used for the next step in this process (Supplemental Data Table 1 and Supplemental Methods). MOB can be completed, from DNA isolation to methylation analysis using MSP, qMSP or MS-qFRET(18), in 9h.
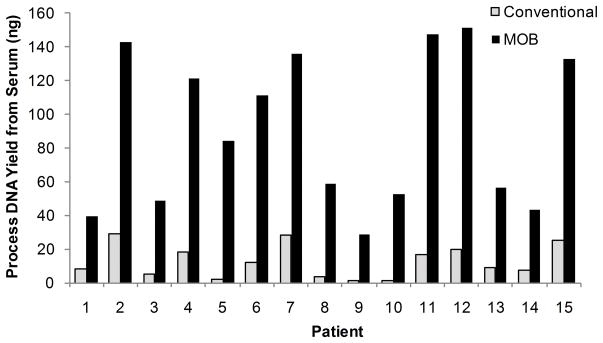
Pre-PCR DNA yields are enhanced in MOB due to a combination of processes; large surface area of SSBs allows large amounts of DNA to be captured, minimization of wash/binding steps decreases DNA loss at each step, and single-tube processing reduces DNA loss during tube transfers. Insufficient DNA yield represents a challenge for the development of blood-based biomarker detection systems. To illustrate the advantage of a single-tube process, we compared pre-PCR yield and methylation detection with MOB to PC/Bst/MSP. Representative comparison for 15 serum samples from lung cancer patients (7 Stage I, 3 Stage II, 5 Stage III) analyzed is shown in Fig. 1. MOB recovery was higher than PC/Bst for each patient serum sample (median increase = 6.61). Extraction yields were higher in MOB when compared to commercial kits meant for DNA extraction (Supplemental Data Fig. 2a). The analysis was extended to 10 samples including fresh tissue and paraffin embedded tissue from normal patients, fresh tumors from cancer patients and sputum samples. Median DNA yield increase of 7.8, 5.3, 6.4 and 7.5 respectively was observed using MOB when compared to PC/Bst (Supplemental Data Fig. 2b).
Figure 1.
Pre-PCR DNA yield from serum is compared for MOB and PC/Bst. Yields have been normalized for 25 μL input of serum. Median increase of 6.6 times was seen for MOB when compared to PC/Bst.
Apart from being a single-tube process, a unique feature of this technique includes combining deamination and desulfonation, a simplification from the conventional approach that requires binding and wash steps in between. By minimizing the number of binding and wash steps, DNA yield is further increased, while assay time is also reduced (data not shown). In addition, the technique contains silica superparamagnetic beads (SSBs) within the tube for both the bisulfite conversion process and PCR. In order to demonstrate that the presence of SSBs do not hinder bisulfite conversion, real-time MSP was used to evaluate p16INK4a promoter methylation, and 5 triplicate reactions were examined with input DNA from varying bisulfite treatment durations (0h, 1h, 3h, 4h, 8h), and compared to the control using 16 hrs of conventional bisulfite treatment (Supplemental Methods). Results indicate that 4 h of bisulfite treatment is sufficient for conversion, and that the presence of beads does not alter the conversion process (Supplemental Data Fig. 3). Further, the ability to generate precise real time quantitative methylation results and PCR products illustrate that beads are also not detrimental to PCR.
Sources of DNA in serum are still unknown, but likely to include both circulating tumor cells and free DNA released from tumor masses. Assessment of methylation in serum or plasma can therefore be a useful tool for early detection of cancer. Several studies have illustrated hypermethylation-associated inactivation of p16INK4a as an early and frequent event in NSCLCs (SCC, 60–80%; adenocarcinoma, 30–45%) and other cancers(19–21). While most of these studies have utilized MSP as an analytical tool to assess gene methylation, extending such an analysis to clinically usable serum/ blood-based tests has been limited by the lack of sensitivity of previous methods. In order to address directly whether improving DNA yields can affect methylation detection, we compared methylation of p16INK4a promoter in 49 patient serum samples (18 normal and 31 cancer) in a blinded study using both MOB and PC/Bst/MSP. The 31 tumor samples were pre-selected from patients diagnosed with lung cancer who were also methylated for p16INK4a promoter in corresponding tumors. Primers and methods are described in detail (Supplemental Data Table 2, Supplemental Methods). While p16INK4a methylation was detected in 14/31 patients with lung cancer using conventional DNA extraction, conventional bisulfite treatment and MSP, using MOB, we were able to detect p16INK4a methylation in 23/31of these patients (Supplemental Data Table 3).
When samples used for methylation analysis contain large amounts of DNA (cell lines, tumors etc), a single-tube analysis of entire input amount may be unnecessary. Instead, MOB allows for storage of either extracted DNA or bisulfite treated DNA that can be used for future downstream analysis. In addition, multiple reactions in parallel are feasible by directly splitting the magnetic beads into several different tubes. Using MOB on colorectal cancer cell line RKO, with extracted DNA yields ranging from 20 to 60 μg, we demonstrated that splitting DNA bound to SSB into 10 different tubes still provided successful MSP analysis for p16INK4a, p15INKb, ASC/TMS1 promoters (data not shown).
In this a magnetically actuated single-tube methylation analysis system using silica superparamagnetic beads (SSBs), since reagents are siphoned into/out of the single tube in a similar manner and binding/elution processes are consistent, the process should be automatable and compatible with commercial available robots using magnetic capture. The introduction of SSBs simplifies sample handling and bypasses the use of liquid transfer, air drying, and centrifugation, and increases yields when compared to conventional methods. SSBs present in the tube do not hinder bisulfite conversion, MSP or other methods including Ms-qFRET(18) which can further enhance analytical sensitivity through nanotechnology-based detection (Supplemental Data Fig. 4). By minimizing binding and wash steps by combining deamination and desulfonation, further efficiency can be achieved. Since the process allows for completion in as little as 9 h, the method presents a viable way for clinically implementing methylation analysis.
Supplementary Material
Bailey-Supplementary Figure 1
Supplemental Data Figure 2. a) DNA extraction methods are compared for 2 serum samples and results are normalized to yields from 20 μL. MOB is compared with commercial DNA extraction kits from Qiagen, Epigentek and Invitrogen using 200 μL serum obtained from same patient volunteers. While Phenol Choloroform/ Ethanol precipitation (PC) results have not been shown in the above chart, our results consistently indicate slightly better yields using PC than Qiagen Kit. b) DNA Yields are compared pre-PCR for MOB and PC/Bst for fresh-tissue, paraffin-embedded tissue, tumor and sputum samples. MOB yields are higher than the conventional methodsfor all samples. Median increases for MOB when compared to PC/Bst are 7.8, 5.3, 6.4 and 7.5 for fresh-tissue, paraffin, tumor and sputum respectively. Yields are normalized for 20 μL input of sample (Supplemental Methods).
Supplemental Data Figure 3. Real time MSP analysis of bisulfite treatment DNA using MOB and the 16 h conventional bisulfite treated DNA from peripheral blood normal lymphocytes (NL) (control). Average Ct value is obtained from triplicate reactions using p16INK4a M primers. Ct values demonstrate that only 4 h is required for the MOB technique for efficient and equivalent bisulfite conversion.
Supplemental Data Figure 4. Representative illustration of MS-qFRET curve for p16INK4a for serum DNA from patient 3 and a water control (no DNA added). Successful detection through MOB indicatesthe compatibility of MOB with MS-qFRET.
Supplementary Table 1. MOB reagents and buffers are provided with appropriate concentrations.
Supplementary Table 2. Sequence of primers used for MSP and MS-qFRET analysis.
Supplementary Table 3. Methylation analysis of p16INK4a promoter compared for MOB and PC/Bst/MSP in serum DNA from normal and cancer patients. MOB was able to detect 23 out of 31 cancerous samples while PC/Bst/MSP detected only 14 out of 31.
Acknowledgments
Financial support was provided by NCI the National Cancer Institute grants SPORE CA058184 and R21-CA120742-01, and National Science Foundation grants 0546012, 0730503 and 0725528. VJB acknowledges the Hodson Trust and the Siebel Scholars Foundation.
Abbreviations
- PCR
Polymerase chain reaction
- MSP
Methylation specific PCR
- MOB
Methylation-on-Beads
- SSB
Silica superparamagnetic beads
- TSG
Tumor-suppressor genes
- PC
Phenol chloroform extraction/Ethanol precipitation
- Bst
Conventional bisulfite treatment
- IVD
in vitro methylated DNA
- NL
normal lymphocyte
- MS-qFRET
Methylation specific quantum dot fluorescence resonance energy transfer
Footnotes
Human Genes Used:
General Use: p16INK4a, HUGO: CDKN2A
General Use: p15INK4b, HUGO: CDKN2B
General Use: ASC/TMS1, HUGO: PYCARD
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–7. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 6.Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 8.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–66. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 9.Bird AP, Southern EM. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978;118:27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- 10.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylatio-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–93. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–31. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 14.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphreys GO, Willshaw GA, Anderson ES. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975;383:457–63. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- 16.Melzak KA, Sherwood CS, Turner RFB, Haynes CA. Driving forces for DNA adsorption to silica in perchlorate solutions. Journal of Colloid and Interface Science. 1996;181:635–44. [Google Scholar]
- 17.Predoi D, Clerac R, Raileanu M, Crisan M, Zaharescu M. Structural and magnetic properties of FexOy nanoparticles dispersed in silica matrix. Journal of Optoelectronics and Advanced Materials. 2007;9:1495–8. [Google Scholar]
- 18.Bailey VJ, Easwaran H, Zhang Y, Griifiths E, Beliknsy S, Herman JG, et al. MS-qFRET: A quantum dot-based method for analysis of DNA methylation. Genome Research. 2009;19:1455–61. doi: 10.1101/gr.088831.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JPJ, Davidson NE, et al. Inactivation of the cdkn2/p16/mts1 gene is frequently associated with aberrant dna methylation in all common human cancers. Cancer Research. 1995;55:4525–30. [PubMed] [Google Scholar]
- 20.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumor-suppressor p16/cdkn2/mts1 in human cancers. Nature Medicine. 1995;1:686–92. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 21.Wong IHN, Lo YMD, Zhang J, Liew CT, Ng MHL, Wong N, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Research. 1999;59:71–3. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bailey-Supplementary Figure 1
Supplemental Data Figure 2. a) DNA extraction methods are compared for 2 serum samples and results are normalized to yields from 20 μL. MOB is compared with commercial DNA extraction kits from Qiagen, Epigentek and Invitrogen using 200 μL serum obtained from same patient volunteers. While Phenol Choloroform/ Ethanol precipitation (PC) results have not been shown in the above chart, our results consistently indicate slightly better yields using PC than Qiagen Kit. b) DNA Yields are compared pre-PCR for MOB and PC/Bst for fresh-tissue, paraffin-embedded tissue, tumor and sputum samples. MOB yields are higher than the conventional methodsfor all samples. Median increases for MOB when compared to PC/Bst are 7.8, 5.3, 6.4 and 7.5 for fresh-tissue, paraffin, tumor and sputum respectively. Yields are normalized for 20 μL input of sample (Supplemental Methods).
Supplemental Data Figure 3. Real time MSP analysis of bisulfite treatment DNA using MOB and the 16 h conventional bisulfite treated DNA from peripheral blood normal lymphocytes (NL) (control). Average Ct value is obtained from triplicate reactions using p16INK4a M primers. Ct values demonstrate that only 4 h is required for the MOB technique for efficient and equivalent bisulfite conversion.
Supplemental Data Figure 4. Representative illustration of MS-qFRET curve for p16INK4a for serum DNA from patient 3 and a water control (no DNA added). Successful detection through MOB indicatesthe compatibility of MOB with MS-qFRET.
Supplementary Table 1. MOB reagents and buffers are provided with appropriate concentrations.
Supplementary Table 2. Sequence of primers used for MSP and MS-qFRET analysis.
Supplementary Table 3. Methylation analysis of p16INK4a promoter compared for MOB and PC/Bst/MSP in serum DNA from normal and cancer patients. MOB was able to detect 23 out of 31 cancerous samples while PC/Bst/MSP detected only 14 out of 31.