Abstract
Heparanase is an endo-β-D-glucuronidase capable of cleaving heparan sulfate (HS) side chains at a limited number of sites, yielding HS fragments of still appreciable size. Importantly, heparanase activity correlates with the metastatic potential of tumor-derived cells, attributed to enhance cell dissemination as a consequence of HS cleavage and remodeling of the extracellular matrix (ECM) and basement membrane underlying epithelial and endothelial cells. Similarly, heparanase activity is implicated in neovascularization, inflammation and autoimmunity, involving migration of vascular endothelial cells and activated cells of the immune system. The cloning of a single human heparanase cDNA 10 years ago enabled researchers to critically approve the notion that HS cleavage by heparanase is required for structural remodeling of the ECM, thereby facilitating cell invasion. Progress in the field has expanded the scope of heparanase function and its significance in tumor progression and other pathologies. Notably, while heparanase inhibitors attenuated tumor progression and metastasis in several experimental systems, other studies revealed that heparanase also functions in an enzymatic activity-independent manner. Thus, inactive heparanase was noted to facilitate adhesion and migration of primary endothelial cells and to promote phosphorylation of signaling molecules such as Akt and Src, facilitating gene transcription (i.e., VEGF) and phosphorylation of selected Src substrates (i.e., EGF-receptor). The concept of enzymatic activity-independent function of heparanase gained substantial support by the recent identification of the heparanase C-terminus domain as the molecular determinant behind its signaling capacity. Identification and characterization of a human heparanase splice variant (T5) devoid of enzymatic activity and endowed with pro-tumorigenic characteristics, elucidation of a cross-talk between heparanase and other ECM-degrading enzymes, and identification of single nucleotide polymorphism associated with heparanase expression and increased risk of GVHD add other layers of complexity to heparanase function in health and disease.
Keywords: Heparanase, myeloma, head and neck carcinoma, signaling, splice variant, C-domain, MMP
INTRODUCTION
Proteoglycans are composed of core protein to which glycosaminoglycans (GAGs) side chains are covalently attached. GAGs are linear polysaccharides consisting of a repeating disaccharide generally of an acetylated amino sugar alternating with uronic acid. Units of N-acetylglucosamine and glucuronic/iduronic acid form heparan sulfate (HS). The polysaccharide chains are modified at various positions by sulfation, epimerization and N-acetylation, yielding clusters of sulfated disaccharides separated by low or non-sulfated regions [1, 2]. The sulfated saccharide domains provide numerous docking sites for a multitude of protein ligands, ensuring that a wide variety of bioactive molecules (i.e., cytokines, growth factors, enzymes, protease inhibitors, ECM proteins) bind to the cell surface and extracellular matrix (ECM) [3–6] and thereby function in the control of normal and pathological processes, among which are morphogenesis, tissue repair, inflammation, vascularization, and cancer metastasis [1–3]. Two main types of cell-surface HS proteoglycans (HSPG) core proteins have been identified: the transmembrane syndecan with four isoforms, carrying HS near their extracellular tips and occasionally also chondroitin sulfate chains near the cell surface [3], and the glycosylphosphatidyl inositol (GPI)-linked glypican with six isoforms, carrying several HS side chains near the plasma membrane and often an additional chain near the tip of its ectodomain [7]. Two major types of ECM-bound HSPG are found: agrin, abundant in most basement membranes, primarily in the synaptic region [8]; and perlecan, with a widespread tissue distribution and a very complex modular structure [9]. Accumulating evidence indicate that HSPGs act to inhibit cellular invasion by promoting tight cell-cell and cell-ECM interactions, and by maintaining the structural integrity and self assembly of the ECM [10, 11]. Notably, one of the characteristics of malignant transformation is down regulation of GAGs biosynthesis, especially of the HS chains [10, 11]. Low levels of cell surface HS also correlate with high metastatic capacity of many tumors. For example, reduced syndecan-1 levels on the cell surface of colon, lung, hepatocellular, breast and head & neck carcinomas was associated with increased tumor metastasis [10]. In other cases, syndecan-1 expression was nonetheless over-expressed, and appeared to promote metastasis [12]. This behavior is attributed mostly to HSPGs within the ECM, exemplified by the pro-tumorigenic function of shed syndecan-1in multiple myeloma [10, 13] (see below).
In addition to modulation of HSPGs levels, expression of enzymes involve in GAGs biosynthesis and modification is impaired during cell transformation. Hereditary multiple exostosis (HME) provided the first direct evidence linking aberrant HS structure to tumorigenesis. HME is an autosomal dominant disorder characterized by the presence of multiple bony outgrowths (exostoses), a consequence of mutation in EXT family members. These genes encode an enzyme (GlcA/GlcNAc transferase) required for chain elongation and synthesis of HS in the Golgi apparatus [14, 15]. Bone outgrowths as a result of mutation and inactivation of these enzymes imply their function as tumor-suppressors. HS can similarly be modified extracellularly by secreted enzymes such as heparan sulfate 6-O-endosulfatases (Sulfs) which selectively remove the 6-O-sulfate groups from HS. Human Sulf-1 (HSulf-1) appears to be miss-regulated in cancer; it is present in a variety of normal tissues but is down-regulated in cell lines originating from ovarian, breast, pancreatic, renal, and hepatocellular carcinomas [16]. Loss of HSulf-1 expression results in increased sulfation of HSPGs, sustained association of heparin-binding growth factors with their cognate receptors, and augmented downstream signaling. Expression of HSulf-1 in cell lines derived from head & neck carcinoma inhibits cell growth, motility and invasion in vitro [17]. Similarly, over-expression of HSulf-1 in CAG myeloma cells inhibits tumor xenograft development and the assembly of FGF-2 signaling complex on the cell surface [18], supporting its function as negative regulator of cancer.
Whereas the activity HSulf-1appeares to attenuate tumor progression, cleavage of HS by the endo-β-glucuronidase heparanase is strongly implicated in cell dissemination associated with tumor metastasis. Cloning of the heparanase gene 10 years ago [19–22] and the generation of specific tools (i.e., molecular probes, antibodies, siRNA) enabled researchers to critically approve the notion that HS cleavage by heparanase is required for structural remodeling of the ECM underlying tumor and endothelial cells, thereby facilitating cell invasion [23–25]. Progress in the field and the generation of genetic tools (i.e., heparanase transgenic and knockout mice) [26–29] has led in recent years to the discovery of new concepts which expand the scope of heparanase function and its significance in tumor progression and other pathologies.
In this review we discuss recent progress in heparanase research, focusing on enzymatic activity-dependent and independent functions mediated by defined protein domains and splice variants, and cross-talk between heparanase and proteases. Aspects such as heparanase gene regulation, proteolytic processing, cellular localization, and the development of heparanase inhibitors have been the subject of several recent review articles [23, 25, 30, 31] and will not be discussed in detail here.
Heparanase in tumor progression and metastasis
Enzymatic activity capable of cleaving glucuronidic linkages and releasing polysaccharide chains resistant to further degradation by the enzyme was first identified by Ogren and Lindahl [32]. The physiological function of this activity was initially implicated in degradation of macromolecular heparin to physiologically active fragments [32, 33]. The activity of the newly discovered endo-β-glucuronidase, referred to as heparanase, was shown soon after to be associated with the metastatic potential of tumor-derived cells such as B16 melanoma [34] and T-lymphoma [35]. These early observations gained substantial support when specific molecular probes became available shortly after cloning of the heparanase gene. Both over-expression and silencing of the heparanase gene clearly indicate that heparanase not only enhances cell dissemination, but also promotes the establishment of a vascular network that accelerates primary tumor growth and provides a gateway for invading metastatic cells [23, 25]. While these studies provided a proof-of-concept for the pro-metastatic and pro-angiogenic capacity of heparanase, the clinical significance of the enzyme in tumor progression emerged from a systematic evaluation of heparanase expression in primary human tumors. Immunohistochemistry, in situ hybridization, RT-PCR and real time-PCR analyses revealed that heparanase is up-regulated in essentially all human carcinomas examined [23, 25]. Notably, increased heparanase levels were most often associated with reduced patients’ survival post operation, increased tumor metastasis and higher microvessel density [23–25]. We choose to highlight the role of heparanase in human cancer by focusing on head & neck carcinoma and multiple myeloma as examples for solid and hematological malignancies.
Heparanase in head & neck carcinoma: signaling in motion
Squamous cell carcinoma of the head and neck (SCCHN) continues to be the sixth most common neoplasm in the world, where more than 500,000 new cases are projected annually [36]. Approximately 200,000 deaths occur yearly as the result of cancer of the oral cavity and pharynx, and the outcome has not improved significantly in the past 25 years [37]. Tumor metastases are common among patients with head & neck cancer with uncontrolled local or regional disease, and autopsy studies revealed 40–47% overall incidence of distant metastases [38, 39]. Applying immunohistochemistry, no staining of heparanase was detected in normal epithelium adjacent to the tumor lesions, likely due to methylation of the gene and its repression by p53 [40–43]. In contrast, heparanase up-regulation was found in the majority of head & neck [44], salivary gland [45], tongue [46], and oral [47] carcinomas. Notably, respective patients that exhibit no or weak heparanase staining are endowed with a favorable prognosis and prolonged survival post operation [44–46, 48]. For example, 70% of the patients with salivary gland carcinoma that stained negative for heparanase were still alive 300 months (25 years) following diagnosis, whereas none of patients stained strongly for heparanase survived at 300 months [45]. Somewhat surprising, heparanase up-regulation in head & neck and tongue carcinomas was associated with tumor larger in size [44, 46]. This association was also seen in hepatocellular, breast, and gastric carcinomas [49–51]. Likewise, heparanase over-expression enhanced [52–55], while local delivery of anti-heparanase siRNA inhibited the progression of tumor xenografts [56]. These results imply that heparanase function is not limited to tumor metastasis but is engaged in progression of the primary lesion.
Heparanase and tumor vascularization
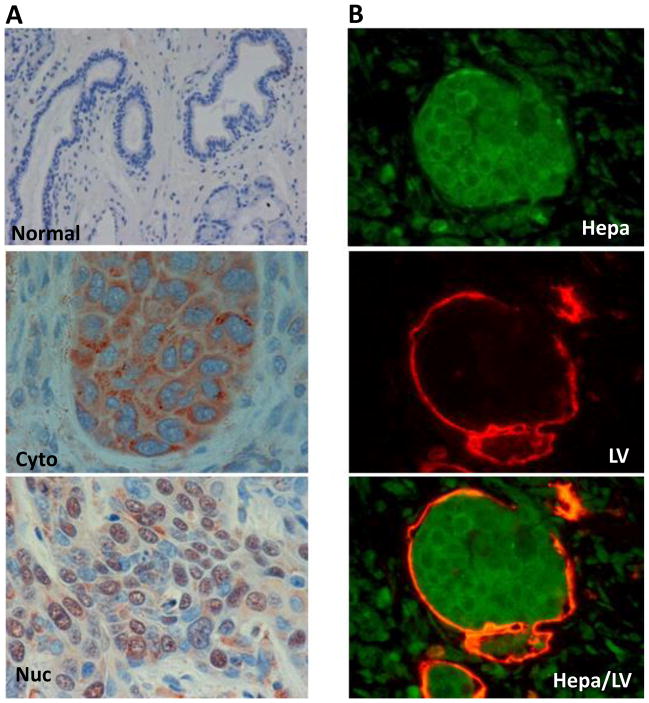
The cellular and molecular mechanisms underlying enhanced tumor growth by heparanase are only starting to be revealed. At the cellular level, both tumor cells and cells that comprise the tumor microenvironment (i.e., endothelial, fibroblasts, tumor-infiltrating immune cells) are likely to be affected by heparanase. Pro-angiogenic potency of heparanase was established clinically [23, 25, 31] and in several in vitro and in vivo model systems, including wound healing [29, 57], tumor xenografts [52, 55], Matrigel plug assay [57], and tube-like structure formation [58]. Moreover, micro vessel density was significantly reduced in tumor xenografts developed by Eb lymphoma cells transfected with anti-heparanase ribozyme [59]. The molecular mechanism by which heparanase facilitates angiogenic responses has traditionally been attributed primarily to the release of HS-bound growth factors such as VEGF-A and FGF-2 [60, 61], a direct consequence of heparanase enzymatic activity. In addition, enzymatically-inactive heparanase was noted to facilitate adhesion and migration of primary endothelial cells [58] and to promote phosphorylation of signaling molecules such as Akt and Src [53, 55, 58, 62, 63], the latter found responsible for VEGF-A induction following exogenous addition of heparanase or its over-expression [55]. Furthermore, heparanase was also noted to facilitate the formation of lymphatic vessels. In head & neck carcinoma, high levels of heparanase were associated with increased lymphatic vessel density (LVD), increased tumor cell invasion to lymphatic vessels (Fig. 1), and increased expression of VEGF-C [64], a potent mediator of lymphatic vessel formation [65]. Heparanase over-expression by melanoma, epidermoid, breast and prostate carcinoma cells induced a 3- to 5-fold elevation of VEGF-C expression in vitro, and facilitated lymph angiogenesis of tumor xenografts in vivo, whereas heparanase gene silencing was associated with decreased VEGF-C levels [64]. These results suggest that enhanced lymph angiogenesis by heparanase is not specific for head & neck carcinoma but rather is a common trait. Up-regulation of VEGF-C was greatly dependent on the cellular localization of heparanase. While localization of heparanase to the cytoplasm (representing secreted heparanase and predicting poor prognosis of cancer patients; Fig. 1, Cyto) was associated with increased VEGF-C staining, nuclear localization of heparanase (Fig. 1, Nuc), shown to correlate with a favorable prognosis of head & neck cancer patients [44], was associated with low levels of VEGF-C [64]. Similarly, localization of heparanase in the cell cytoplasm was associated with activation of the epidermal growth factor (EGF)-receptor (EGFR) in head & neck carcinoma [66].
Figure 1.
A. Immunohistochemical staining of heparanase in SCCHN tumor specimens Formalin-fixed, paraffin-embedded 5 micron sections of head & neck tumors were subjected to immunostaining of heparanase, applying anti-heparanase pAb #733. Shown are representative photomicrographs of positively stained specimens exhibiting cytoplasmic (Cyto, middle panel) and nuclear (Nuc, lower panel) heparanase localization. Normal looking tissue adjacent to the tumor lesion stained negative for heparanase (upper panel). Nuclear heparanase is associated with decreased levels of phospho-EGFR, lower lymph vessel density, and favorable prognosis of head & neck cancer patients (see text for details). B. Heparanase expression associates with tumor cell invasion into lymph vessels. Head & neck tumor specimen was stained with anti heparanase polyclonal (green, upper panel) and D2-40 monoclonal (a marker for human lymphatics; red, middle panel) antibodies, illustrating heparanase-positive tumor cells inside a lymphatic vessel lumen (merge, lower panel).
Heparanase and EGFR activation
Decorin, a CS/DSPG chondroitin/dermatan sulfate proteoglycan directly interacts with EGFR and this evokes a down regulation of the receptor and inhibition of its downstream signaling. The anti proliferative effect of decorin on cancer cells via EGFR is reviewed in this series by Iozzo and Schaefer [67] (review submitted to this MiniReview Series). On the other hand, EGFR phosphorylation is markedly increased in cells over-expressing heparanase or following its exogenous addition, while heparanase gene silencing is accompanied by reduced EGFR and Src phosphorylation levels [66]. Notably, EGFR activation was observed following the addition or over-expression of mutated, enzymatically inactive heparanase protein. Though inactive, double mutated (DM; Glu225, Glu343) [68] heparanase retains its high affinity towards HS and hence may facilitate signaling by ligation and activation of membrane HSPGs such as syndecan [69, 70]. This however appears not to be the case because heparanase deleted for its heparin binding domain (Δ10) [71] efficiently stimulated EGFR phosphorylation [66]. Notably, enhanced EGFR phosphorylation by heparanase was restricted to selected tyrosine residues (i.e., 845, 1173) thought to be direct targets of Src rather than a result of receptor autophosphorylation [72]. Indeed, enhanced EGFR phosphorylation of tyrosine residues 845 and 1173 in response to heparanase was abrogated in cells treated with Src inhibitors or anti-Src siRNA [66]. The functional significance of EGFR modulation by heparanase emerged by monitoring cell proliferation. Thus, heparanase gene silencing was accompanied by a decrease in cell proliferation, while heparanase over-expression resulted in enhanced cell proliferation and formation of larger colonies in soft agar, in a Src- and EGFR-dependent manner [66]. The clinical relevance of the heparanase-Src-EGFR pathway has been elucidated for head & neck carcinoma. Notably, heparanase expression in head & neck carcinomas correlated with phospho-EGFR immunostaining, and even more significant was the correlation between heparanase cellular localization (i.e., cytoplasmic vs. nuclear) and phospho-EGFR levels [66]. These studies provide a more realistic view of heparanase function in the course of tumor progression. Thus, while heparanase enzymatic activity has traditionally been implicated in tumor metastasis, the current view points to a multifaceted protein engaged in multiple aspects of tumor progression, combining enzymatic activity-dependent and independent activities of heparanase and affecting two systems critical for tumor progression, namely tumor vascularization and EGFR activation.
Signaling by heparanase C-domain
The concept of enzymatic activity-independent function of heparanase gained substantial support by the recent identification of the heparanase C-domain as the molecular determinant behind its signaling capacity. The existence of a C-terminus domain (C-domain) emerged from a prediction of the three dimensional structure of a single chain heparanase enzyme [73]. In this protein variant, the linker segment was replaced by three glycine-serine repeats (GS3), resulting in a constitutively active enzyme [74]. The structure obtained clearly illustrates a TIM-barrel fold, in agreement with previous predictions [68, 75]. Notably, the structure also delineates a C-terminus fold positioned next to the TIM-barrel fold [73]. The predicted heparanase structure led to the hypothesis that the seemingly distinct protein domains observed in the three dimensional model, namely the TIM-barrel and C-domain regions, mediate enzymatic and non-enzymatic functions of heparanase, respectively. Interestingly, cells transfected with the TIM-barrel construct (amino acids 36–417) failed to display heparanase enzymatic activity, suggesting that the C-domain is required for the establishment of an active heparanase enzyme, possibly by stabilizing the TIM-barrel fold [73]. Deletion and site directed mutagenesis further indicated that the C-domain plays a decisive role in heparanase enzymatic activity and secretion [73, 76, 77]. Notably, Akt phosphorylation was stimulated by cells over-expressing the C-domain (amino acids 413–543), while the TIM-barrel protein variant yielded no Akt activation compared with control, mock transfected cells [73]. These findings clearly indicate that the non-enzymatic signaling function of heparanase leading to activation of Akt is mediated by the C-domain. Notably, the C-domain construct lacks the 8 kDa segment (Gln36-Ser55) which, according to the predicted model, contributes one beta strand to the C-domain structure (reviewed by [78]). Indeed, Akt phosphorylation was markedly enhanced and prolonged in cells transfected with a mini gene comprising this segment linked to the C-domain sequence (8-C) [73, 78]. This finding further supports the predicted three dimensional model, indicating that the C-domain is indeed a valid functional domain responsible for Akt phosphorylation. The cellular consequences of C-domain over-expression were best revealed by monitoring tumor xenograft development. Remarkably, tumor xenografts produced by C-domain-transfected glioma cells grew faster and appeared indistinguishable from those produced by cells transfected with the full length heparanase in term of tumor size and angiogenesis, yielding tumors 6-fold bigger than control. In contrast, progression of tumors produced by TIM-barrel-transfected cells appeared comparable with control mock transfected cells [73, 78]. These results show, that in some tumor systems (i.e., glioma), heparanase facilitates primary tumor progression regardless of its enzymatic activity, while in others (i.e., myeloma) heparanase enzymatic activity dominates (see below). Enzymatic activity-independent function of heparanase is further supported by the recent identification of T5, a functional human splice variant of heparanase.
T5, a functional human heparanase splice variant
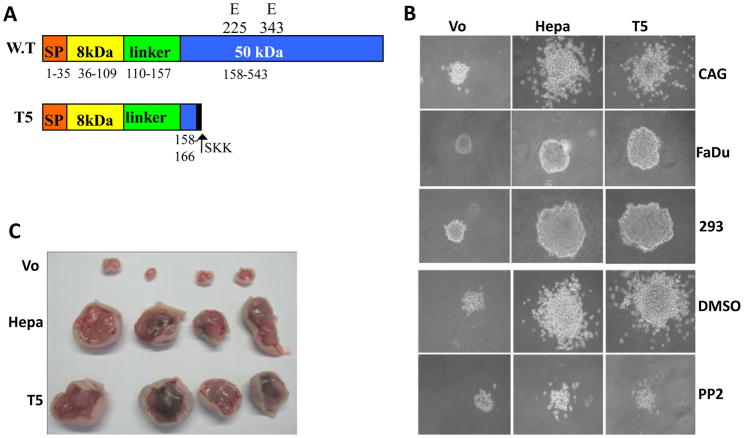
Almost all protein-coding genes contain introns that are removed in the nucleus by RNA splicing and are often alternatively spliced. Alternative splicing increases the coding capacity of the genome, generating multiple proteins from a single gene. The resulting protein isoforms frequently exhibit different biological properties that may play an essential role in tumorigenesis [79, 80]. A splice variant of human heparanase which lacks exon 5 has been described [81, 82]. This splice variant fails to get secreted and lacks enzymatic activity and its biological significance is yet unclear. Additional human heparanase splice variants have been predicted in silico [83] (Fig. 2A); the expression of one, termed T5, was found to be enriched in lung carcinoma and chronic myeloid leukemia compared with control tissue and cells. In this splice variant, 144 bp of intron 5 are joined with exon 4, resulting in a 169 amino acids protein that lacks enzymatic activity typical of heparanase [83]. Unlike previously identified splice variants of heparanase, T5 is secreted and facilitates Src phosphorylation [83]. Furthermore, Src phosphorylation was markedly reduced in cells treated with anti-T5 siRNA [83]. Over-expression of T5 by pharynx (FaDu), myeloma (CAG) and embryonic kidney (293) cells resulted in enhanced proliferation and larger colony formation in soft agar, which was attenuated by Src inhibitor (Fig. 2B) [83]. Likewise, T5 gene silencing was associated with reduced cell proliferation, indicating that endogenous levels of T5 and heparanase affect tumor cell proliferation. Moreover, development of tumor xenografts produced by heparanase- and T5-infected cells was markedly enhanced compared with xenografts generated by control cells (Fig. 2C) [83]. Tumors developed by T5 expressing cells exhibited a higher density of blood vessels decorated with smooth muscle actin (SMA)-positive cells (pericytes) [83], an indication of vessel maturation. The clinical relevance of T5 emerged from analysis of renal cell carcinoma biopsies, where T5 and heparanase expression appeared to be induced in 75% of the cases [83]. Thus, while inhibitors directed against the enzymatic activity of heparanase are being currently evaluated in clinical trials [84–87], T5 as well as the heparanase C-domain are not expected to be affected by these inhibitors. It appears therefore that a well defined enzymatic activity thought to be relatively easy to target, turned, at least in certain tumor systems, into a complex objective as more knowledge accumulates and the biology of the protein is being elucidated.
Figure 2. Heparanase splice variant, T5, endowed with pro-tumorigenic characteristics.
A. Schematic structure of wild type (WT) and heparanase splice variant, T5. SP-signal peptide; glutamic acids residues 225 and 343 critical for heparanase enzymatic activity, are detonated (see text for details). B. Colony formation in soft agar. Control (Vo) heparanase (Hepa)-, and T5-infected myeloma (CAG, upper panels), pharynx (FaDu, second panels) and embryonic kidney (293, third panels) cells (5×103 cells/dish) were mixed with soft agar and cultured for 3–5 weeks. CAG cells were similarly grown in the absence (DMSO; fourth panels) or presence of Src inhibitor (PP2, 0.4 nM; lower panels). Shown are representative photomicrographs of colonies at high (x100) magnification. C. Tumor xenograft development. Control (Vo), heparanase-, and T5-infected CAG myeloma cells were injected subcutaneously (1×106/0.1ml). At the end of the experiment on day 37, tumors were harvested and photographed.
Multiple myeloma: moving anti-heparanase therapy closer to reality
Multiple myeloma is the second most prevalent hematologic malignancy. This B-lymphoid malignancy is characterized by tumor cell infiltration of the bone marrow, resulting in severe bone pain and osteolytic bone disease. Although progress in the treatment of myeloma patients has been made over the last decade, the overall survival of patients is still poor.
In myeloma patients, heparanase enzymatic activity was elevated in the bone marrow plasma of 86% of patients examined [88], and gene array analysis showed elevated heparanase expression in 92% of myeloma patients [89]. Heparanase up-regulation in myeloma patients was associated with elevated microvessel density and syndecan-1 expression [88]. While heparanase is pro-angiogenic in myeloma, which is a common feature shared with solid tumors (II.1.1), heparanase regulation of syndecan-1 shedding has emerged as highly relevant to multiple myeloma progression. Syndecan-1 is particularly abundant in myeloma, and is the dominant and often the only HSPG present on the surface of myeloma cells [90]. Cell surface syndecan-1 promotes adhesion of myeloma cells and inhibits cell invasion in vitro [13]. In contrast, high levels of shed syndecan-1 are found in the serum of some myeloma patients and are associated with poor prognosis [91]. The multiple roles of syndecans in cancer progression and strategies for their targeting is presented in this series by Theocharis et al [92] (review submitted to this MiniReview Series). Shed syndecan-1 becomes trapped within the bone marrow ECM where it likely acts to enhance the growth, angiogenesis and metastasis of myeloma cells within the bone [13, 93, 94]. This is supported by the finding that enhanced expression of soluble syndecan-1 by myeloma cells promotes tumor growth and metastasis in a mouse model [13, 94]. Notably, heparanase up-regulates both the expression and shedding of syndecan-1 from the surface of myeloma cells [89, 95]. In agreement with this notion, heparanase gene silencing was associated with decreased levels of shed syndecan-1 [89]. Importantly, both syndecan-1 up-regulation and shedding require heparanase enzymatic activity, because over-expression of mutated inactive heparanase failed to stimulate syndecan-1 expression and shedding [95]. Syndecan-1 shedding was similarly augmented by the addition of recombinant active heparanase to CAG myeloma cells, and even more dramatic shedding was observed following the addition of bacterial heparinase III (heparitinase) [95]. These findings indicate that cleavage of HS by heparanase or heparinase III may render syndecan-1 more susceptible to proteases mediating the shedding of syndecan-1. However, it appears that heparanase may play an even more direct role in regulating shedding of syndecan-1, by facilitating the expression of proteases engaged in syndecan shedding.
Heparanase-MMP co-operation in myeloma progression
It was recently demonstrated that enhanced expression of heparanase leads to increased levels of MMP-9 (a syndecan-1 sheddase), while heparanase gene silencing resulted in reduced MMP-9 activity [96]. Up-regulation of MMP-9 expression has significant biological relevance because inhibition of MMP-9 reduces syndecan-1 shedding [96]. For the importance of syndecan shedding in diseases see the review of Manon-Jensen et al [97] (review submitted to this MiniReview Series). Moreover, not only MMP-9 but also urokinase-type plasminogen activator (uPA) and its receptor (uPAR), molecular determinants responsible for MMP-9 activation, are up-regulated by heparanase. These findings provided the first evidence for cooperation between heparanase and MMPs in regulating HSPGs on the cell surface and likely in the ECM, and are supported by the recent generation and characterization of heparanase knockout (KO) mice. HS chains isolated from these mice were longer, critically supporting the notion that heparanase is the only functional endoglycosidase capable of degrading HS [26]. Despite the complete lack of heparanase gene expression and enzymatic activity, heparanase-KO mice develop normally, are fertile, and exhibit no apparent anatomical or functional abnormalities [26]. Notably, heparanase deficiency was accompanied by a marked elevation of matrix metalloproteinase (MMP) family members such as MMP-2, MMP-9, and MMP-14, in an organ-dependent manner. Thus, MMP-14 levels were increased 8-fold in the liver of heparanase-KO mice compared with control littermates, while MMP-2 levels were increase 2.5-fold in the mammary gland [26], suggesting that MMPs provide tissue-specific compensation for heparanase deficiency. This is likely the reason for over-branching of the mammary gland in heparanase-KO mice [26], a phenotype also noted in heparanase transgenic mice [27]. Collectively, these results suggest that heparanase is intimately engaged in the regulation of gene transcription and acts as a master regulator of protease expression, mediating gene induction or repression depending on the biological setting.
The heparanase-syndecan axis is a target for therapy
Results from studies using several in vivo model systems support the notion that enzymatic activities responsible for syndecan-1 modification are valid targets for myeloma therapy. For example, enhanced expression of either HSulf-1 or HSulf-2 attenuated myeloma tumor growth [18]. Even a more dramatic inhibition of tumor growth was noted following administration of bacterial heparinase III (heparitinase) to SCID mice inoculated with either CAG myeloma cells or cells isolated from the bone marrow of myeloma patients [98]. Although heparinase III and human heparanase both degrade HS chains, their cleavage products are distinct. While heparinase III is a β-eliminase that extensively degrades HS, heparanase is an endo-β-D-glucuronidase whose substrate recognition sites were recently characterized [99]. Unlike the bacterial enzyme, heparanase cleaves HS more selectively and generates fragments that are 4–7 kDa in size, yielding strictly distinct outcomes in the context of tumor progression. While administration of heparinase III is associated with reduced tumor growth, heparanase activity is elevated in many hematological and solid tumors, correlating with poor prognosis and shorter post-operative survival rate (see II). Accordingly, inhibition of heparanase enzymatic activity is expected to suppress tumor progression. To examine this in myeloma, a chemically modified heparin, which is 100% N-acetylated and 25% glycol-split was tested. This flexible molecule is a potent inhibitor of heparanase enzymatic activity, lacks anti-coagulant activity typical of heparin, and does not displace ECM-bound FGF-2 or potentiate its mitogenic activity [30, 31, 100, 101]. The modified heparin profoundly inhibits the progression of tumor xenografts produced by myeloma cells [30, 98]. These studies support the notion that heparanase enzymatic activity not only facilitates tumor metastasis, but also promotes the progression of primary tumors.
CONCLUSIONS AND PERSPECTIVE
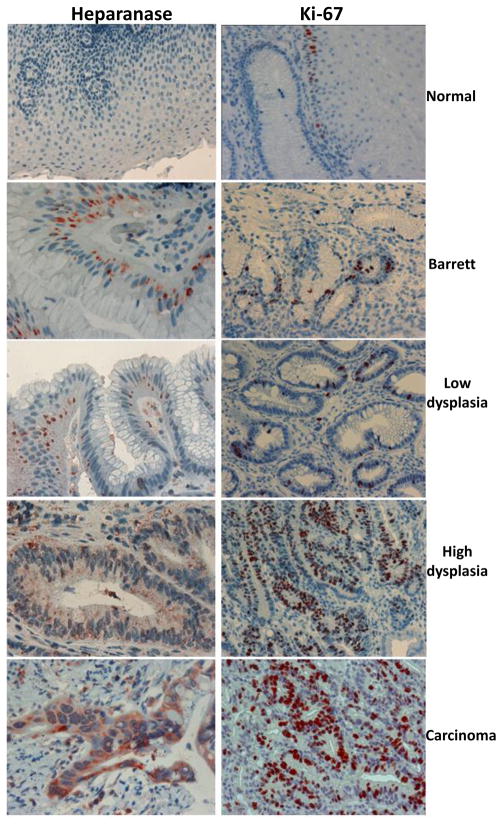
Although much has been learned in the last decade, the repertoire of heparanase functions in health and disease is only starting to emerge. Clearly, from activity implicated mainly in cell invasion associated with tumor metastasis, heparanase has turned into a multifaceted protein that appears to participate in essentially all major aspects of tumor progression. In this regard, evidence now support a concept by which growth of the primary tumor is fueled by circulating metastatic tumor cells [102, 103]. According to this notion, tumor cells are present in the circulation in large numbers even at the early stages of cancer and long before metastatic growth at distant sites can be detected [103]. These cells can reinfiltrate and promote growth and angiogenesis of the primary tumor [102]. The possible involvement of heparanase in tumor self-seeding is supported by the timing of its induction during tumorigenesis and its pro-metastatic function. Using the RIP-Tag2 tumor model, it was demonstrated that heparanase mRNA and protein are elevated upon the transition from normal to angiogenic islets, followed by a further increase when solid tumors were detected [104]. Furthermore, heparanase expression is elevated already at the early stages of human neoplasia. In the colon, heparanase gene and protein are expressed already at the stage of adenoma [105], and during esophageal carcinogenesis heparanase expression is induced in Barrett’s epithelium (Fig. 3), an early event that predisposes patients to formation of dysplasia which may progress to adenocarcinoma [106]. Tumor self-seeding also facilitates the recruitment of stromal components. While pro-angiogenic capacity of heparanase has been established, its likely impact on other components of the tumor microenvironment (i.e., fibroblasts, macrophages) awaits thorough investigation.
Figure 3. Immunohistochemical staining of esophageal specimens.
A. Formalin-fixed, paraffin-embedded 5 micron sections of normal (upper panel), Barrett’s (second panel), low grade (third panel), high-grade (fourth panel), and adenocarcinoma (lower panel) esophageal biopsies were subjected to immunostaining of heparanase, applying anti-heparanase pAb #733 (left panels) or anti Ki-67, a marker of cell proliferation (right panels).
Heparanase expression at the early stages of tumor initiation and progression, and by the majority of tumor cells (evident by a high extent of immunostaining), can be utilized to turn the immune system against the very same cells. Accumulating evidence suggest that peptides derived from human heparanase can elicit a potent anti-tumor immune response, leading to lysis of heparanase-positive human gastric (KATO III), colon (SW480), and breast (MCF-7) carcinoma cells, as well as hepatoma (HepG2) and sarcoma (U-2 OS) cells [107–109]. In contrast, no killing effect was noted towards autologous lymphocytes [107–109]. Notably, the development of tumor xenografts produced by B16 melanoma cells was markedly restrained in mice immunized with peptides derived from mouse heparanase (i.e., aa 398–405; 519–526) compared to a control peptide in both immunoprotection and immunotherapy approaches [109]. T-regulatory cells are frequently present in colorectal cancer patients; Interestingly, T-regulatory cells against heparanase could not be found [110]. Anti-heparanase immunotherapy is thus expected to be prolonged and more efficient due to the absence of T-suppressor cells. A related treatment approach is being tested in advanced metastasized breast cancer patients [111]. While this immunotherapeutic concept, together with available heparanase inhibitors, is hoped to advance cancer treatment, the identification of single nucleotide polymorphism (SNP) associated with heparanase expression and increased risk for GVHD following allogeneic stem cell transplantation [112–114] offers a genetic concept which can potentially be translated into patients’ diagnosis. Studies in these directions, identification of heparanase receptor(s) mediating its signaling function, and elucidation of heparanase route and function in the cell nucleus, will advance the field of heparanase research and reveal its significance in health and disease. Resolving the heparanase crystal structure will accelerate the development of effective inhibitory molecules and neutralizing antibodies paving the way for advanced clinical trials in patients with cancer and other diseases (i.e., colitis, psoriasis, diabetic nephropathy) involving heparanase.
Acknowledgments
We thank Prof. Benito Casu (‘Ronzoni’ Institute, Milan, Italy) for his continuous support and active collaboration. This work was supported by grants from the Israel Science Foundation (grant 549/06); National Institutes of Health (NIH) grants CA138535 (RDS) and CA106456 (IV); the Israel Cancer Research Fund (ICRF); and the Juvenile Diabetes Research Foundation (JDRF grant 1-2006-695). I. Vlodavsky is a Research Professor of the ICRF. We gratefully acknowledge the contribution, motivation and assistance of the research teams in the Hadassah-Hebrew University Medical Center (Jerusalem, Israel) and the Cancer and Vascular Biology Research Center of the Rappaport Faculty of Medicine (Technion, Haifa). We apologize for not citing several relevant articles, due to space limitation.
Abbreviations
- ECM
extracellular matrix
- HS
heparan sulfate
- HSPGs
heparan sulfate proteoglycans
- FGF
fibroblast growth factor
- EGF
epidermal growth factor
- EGFR
EGF-receptor
- GVHD
graft vs. host disease
- LVD
lymphatic vessel density
- MMP
matrix metalloproteinase
- VEGF
vascular endothelial growth factor
References
- 1.Iozzo RV, San Antonio JD. Heparan sulfate proteoglycans: heavy hitters in the angiogenesis arena. J Clin Invest. 2001;108:349–355. doi: 10.1172/JCI13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 3.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 4.Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl U, Li JP. Interactions between heparan sulfate and proteins-design and functional implications. Intl Rev Cell Mol Bio. 2009;276:105–159. doi: 10.1016/S1937-6448(09)76003-4. [DOI] [PubMed] [Google Scholar]
- 6.Whitelock JM, Iozzo RV. Heparan sulfate: a complex polymer charged with biological activity. Chem Rev. 2005;105:2745–2764. doi: 10.1021/cr010213m. [DOI] [PubMed] [Google Scholar]
- 7.Fransson LA, Belting M, Cheng F, Jonsson M, Mani K, Sandgren S. Novel aspects of glypican glycobiology. Cell Mol Life Sci. 2004;61:1016–1024. doi: 10.1007/s00018-004-3445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole GJ, Halfter W. Agrin: an extracellular matrix heparan sulfate proteoglycan involved in cell interactions and synaptogenesis. Persp Dev Neurobiology. 1996;3:359–371. [PubMed] [Google Scholar]
- 9.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 10.Sanderson RD. Heparan sulfate proteoglycans in invasion and metastasis. Semin Cell Dev Biol. 2001;12:89–98. doi: 10.1006/scdb.2000.0241. [DOI] [PubMed] [Google Scholar]
- 11.Timar J, Lapis K, Dudas J, Sebestyen A, Kopper L, Kovalszky I. Proteoglycans and tumor progression: Janus-faced molecules with contradictory functions in cancer. Semin Cancer Biol. 2002;12:173–186. doi: 10.1016/S1044-579X(02)00021-4. [DOI] [PubMed] [Google Scholar]
- 12.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 13.Sanderson RD, Yang Y. Syndecan-1: a dynamic regulator of the myeloma microenvironment. Clin Exp Metastasis. 2008;25:149–159. doi: 10.1007/s10585-007-9125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J Biol Chem. 1998;273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 15.McCormick C, Leduc Y, Martindale D, Mattison K, Esford LE, Dyer AP, Tufaro F. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat Genet. 1998;19:158–161. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- 16.Lai J, Chien J, Staub J, Avula R, Greene EL, Matthews TA, Smith DI, Kaufmann SH, Roberts LR, Shridhar V. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278:23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 17.Lai JP, Chien J, Strome SE, Staub J, Montoya DP, Greene EL, Smith DI, Roberts LR, Shridhar V. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene. 2004;23:1439–1447. doi: 10.1038/sj.onc.1207258. [DOI] [PubMed] [Google Scholar]
- 18.Dai Y, Yang Y, MacLeod V, Yue X, Rapraeger AC, Shriver Z, Venkataraman G, Sasisekharan R, Sanderson RD. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem. 2005;280:40066–40073. doi: 10.1074/jbc.M508136200. [DOI] [PubMed] [Google Scholar]
- 19.Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 20.Kussie PH, Hulmes JD, Ludwig DL, Patel S, Navarro EC, Seddon AP, Giorgio NA, Bohlen P. Cloning and functional expression of a human heparanase gene. Biochem Biophys Res Commun. 1999;261:183–187. doi: 10.1006/bbrc.1999.0962. [DOI] [PubMed] [Google Scholar]
- 21.Toyoshima M, Nakajima M. Human heparanase. Purification, characterization, cloning, and expression. J Biol Chem. 1999;274:24153–24160. doi: 10.1074/jbc.274.34.24153. [DOI] [PubMed] [Google Scholar]
- 22.Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 23.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Vlodavsky I, Elkin M, Abboud-Jarrous G, Levi-Adam F, Fuks L, Shafat I, Ilan N. Heparanase: one molecule with multiple functions in cancer progression. Connect Tissue Res. 2008;49:207–210. doi: 10.1080/03008200802143281. [DOI] [PubMed] [Google Scholar]
- 25.Vreys V, David G. Mammalian heparanase: what is the message? J Cell Mol Med. 2007;11:427–452. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zcharia E, Jia J, Zhang X, Baraz L, Lindahl U, Peretz T, Vlodavsky I, Li JP. Newly generated heparanase knock-out mice unravel co-regulation of heparanase and matrix metalloproteinases. PLoS ONE. 2009;4:e5181. doi: 10.1371/journal.pone.0005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zcharia E, Metzger S, Chajek-ShaulL T, Aingorn H, Elikn M, Friedmann Y, Weinstein T, Jin-Ping L, Lindahl U, Vlodavsky I. Transgenic expression of mammalian heparanase uncovers physiological functions of heparan sulfate in tissue morphogenesis, vascularization, and feeding behavior. FASEB J. 2004;18:252–263. doi: 10.1096/fj.03-0572com. [DOI] [PubMed] [Google Scholar]
- 28.Zcharia E, Philp D, Edovitsky E, Aingorn H, Metzger S, Kleinman HK, Vlodavsky I, Elkin M. Heparanase Regulates Murine Hair Growth. Am J Pathol. 2005;166:999–1008. doi: 10.1016/S0002-9440(10)62321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zcharia E, Zilka R, Yaar A, Yacoby-Zeevi O, Zetser A, Metzger S, Sarid R, Naggi A, Casu B, Ilan N, et al. Heparanase accelerates wound angiogenesis and wound healing in mouse and rat models. FASEB J. 2005;19:211–221. doi: 10.1096/fj.04-1970com. [DOI] [PubMed] [Google Scholar]
- 30.Casu B, Vlodavsky I, Sanderson RD. Non-anticoagulant heparins and inhibition of cancer. Pathophysiol Haemost Thromb. 2008;36:195–203. doi: 10.1159/000175157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: Structure, Biological Functions, and Inhibition by Heparin-Derived Mimetics of Heparan Sulfate. Curr Pharm Des. 2007;13:2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- 32.Ogren S, Lindahl U. Cleavage of macromolecular heparin by an enzyme from mouse mastocytoma. J Biol Chem. 1975;250:2690–2697. [PubMed] [Google Scholar]
- 33.Thunberg L, Backstrom G, Wasteson A, Robinson HC, Ogren S, Lindahl U. Enzymatic depolymerization of heparin-related polysaccharides. Substrate specificities of mouse mastocytoma and human platelet endo-beta-D-glucuronidases. J Biol Chem. 1982;257:10278–10282. [PubMed] [Google Scholar]
- 34.Nakajima M, Irimura T, DiFerrante D, DiFerrante N, Nicolson GL. Heparan sulfate degradation: relation to tumor invasion and metastatic properties of Mouse B 16 Melanoma sublines. Science (Wash DC) 1983;220:611–613. doi: 10.1126/science.6220468. [DOI] [PubMed] [Google Scholar]
- 35.Vlodavsky I, Fuks Z, Bar-Ner M, Ariav Y, Schirrmacher V. Lymphoma cells mediated degradation of sulfated proteoglycans in the subendothelial extracellular matrix: relation to tumor cell metastasis. Cancer Res. 1983;43:2704–2711. [PubMed] [Google Scholar]
- 36.Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328:184–194. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 37.Ford AC, Grandis JR. Targeting epidermal growth factor receptor in head and neck cancer. Head Neck. 2003;25:67–73. doi: 10.1002/hed.10224. [DOI] [PubMed] [Google Scholar]
- 38.Kotwall C, Sako K, Razack MS, Rao U, Bakamjian V, Shedd DP. Metastatic patterns in squamous cell cancer of the head and neck. Am J Surg. 1987;154:439–442. doi: 10.1016/0002-9610(89)90020-2. [DOI] [PubMed] [Google Scholar]
- 39.Zbaren P, Lehmann W. Frequency and sites of distant metastases in head and neck squamous cell carcinoma. An analysis of 101 cases at autopsy. Arch Otolaryngol Head Neck Surg. 1987;113:762–764. doi: 10.1001/archotol.1987.01860070076020. [DOI] [PubMed] [Google Scholar]
- 40.Baraz L, Haupt Y, Elkin M, Peretz T, Vlodavsky I. Tumor suppressor p53 regulates heparanase gene expression. Oncogene. 2006;25:3939–3947. doi: 10.1038/sj.onc.1209425. [DOI] [PubMed] [Google Scholar]
- 41.Ogishima T, Shiina H, Breault JE, Tabatabai L, Bassett WW, Enokida H, Li LC, Kawakami T, Urakami S, Ribeiro-Filho LA, et al. Increased heparanase expression is caused by promoter hypomethylation and up-regulation of transcriptional factor early growth response-1 in human prostate cancer. Clin Cancer Res. 2005;11:1028–1036. [PubMed] [Google Scholar]
- 42.Ogishima T, Shiina H, Breault JE, Terashima M, Honda S, Enokida H, Urakami S, Tokizane T, Kawakami T, Ribeiro-Filho LA, et al. Promoter CpG hypomethylation and transcription factor EGR1 hyperactivate heparanase expression in bladder cancer. Oncogene. 2005;24:6765–6772. doi: 10.1038/sj.onc.1208811. [DOI] [PubMed] [Google Scholar]
- 43.Shteper PJ, Zcharia E, Ashhab Y, Peretz T, Vlodavsky I, Ben-Yehuda D. Role of promoter methylation in regulation of the mammalian heparanase gene. Oncogene. 2003;22:7737–7749. doi: 10.1038/sj.onc.1207056. [DOI] [PubMed] [Google Scholar]
- 44.Doweck I, Kaplan-Cohen V, Naroditsky I, Sabo E, Ilan N, Vlodavsky I. Heparanase localization and expression by head and neck cancer: correlation with tumor progression and patient survival. Neoplasia. 2006;8:1055–1061. doi: 10.1593/neo.06577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ben-Izhak O, Kaplan-Cohen V, Ilan N, Gan S, Vlodavsky I, Nagler R. Heparanase expression in malignant salivary gland tumors inversely correlates with long-term survival. Neoplasia. 2006;8:879–884. doi: 10.1593/neo.06382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagler R, Ben-Izhak O, Cohen-Kaplan V, Shafat I, Vlodavsky I, Akrish S, Ilan N. Heparanase up-regulation in tongue cancer: tissue and saliva analysis. Cancer. 2007;110:2732–2739. doi: 10.1002/cncr.23095. [DOI] [PubMed] [Google Scholar]
- 47.Leiser Y, Abu-El-Naaj I, Sabo E, Akrish S, Ilan N, Peled M, Vlodavsky I. The prognostic value of heparanase expression and cellular localization in oral cancer. Head & Neck. 2010 doi: 10.1002/hed.21545. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bar-Sela G, Kaplan-Cohen V, Ilan N, Vlodavsky I, Ben-Izhak O. Heparanase expression in nasopharyngeal carcinoma inversely correlates with patient survival. Histopathology. 2006;49:188–193. doi: 10.1111/j.1365-2559.2006.02469.x. [DOI] [PubMed] [Google Scholar]
- 49.El-Assal ON, Yamanoi A, Ono T, Kohno H, Nagasue N. The Clinicopathological Significance of Heparanase and Basic Fibroblast Growth Factor Expressions in Hepatocellular Carcinoma. Clin Cancer Res. 2001;7:1299–1305. [PubMed] [Google Scholar]
- 50.Maxhimer JB, Quiros RM, Stewart R, Dowlatshahi K, Gattuso P, Fan M, Prinz RA, Xu X. Heparanase-1 expression is associated with the metastatic potential of breast cancer. Surgery. 2002;132:326–333. doi: 10.1067/msy.2002.125719. [DOI] [PubMed] [Google Scholar]
- 51.Tang W, Nakamura Y, Tsujimoto M, Sato M, Wang X, Kurozumi K, Nakahara M, Nakao K, Nakamura M, Mori I, et al. Heparanase: a key enzyme in invasion and metastasis of gastric carcinoma. Mod Pathol. 2002;15:593–598. doi: 10.1038/modpathol.3880571. [DOI] [PubMed] [Google Scholar]
- 52.Cohen I, Pappo O, Elkin M, San T, Bar-Shavit R, Hazan R, Peretz T, Vlodavsky I, Abramovitch R. Heparanase promotes growth, angiogenesis and survival of primary breast tumors. Int J Cancer. 2006;118:1609–1617. doi: 10.1002/ijc.21552. [DOI] [PubMed] [Google Scholar]
- 53.Doviner V, Maly B, Kaplan V, Gingis-Velitski S, Ilan N, Vlodavsky I, Sherman Y. Spatial and temporal heparanase expression in colon mucosa throughout the adenoma-carcinoma sequence. Mod Pathol. 2006;19:878–888. doi: 10.1038/modpathol.3800603. [DOI] [PubMed] [Google Scholar]
- 54.Yang Y, Macleod V, Bendre M, Huang Y, Theus AM, Miao HQ, Kussie P, Yaccoby S, Epstein J, Suva LJ, et al. Heparanase promotes the spontaneous metastasis of myeloma cells to bone. Blood. 2005;105:1303–1309. doi: 10.1182/blood-2004-06-2141. [DOI] [PubMed] [Google Scholar]
- 55.Zetser A, Bashenko Y, Miao H-Q, Vlodavsky I, Ilan N. Heparanase Affects Adhesive and Tumorigenic Potential of Human Glioma Cells. Cancer Res. 2003;63:7733–7741. [PubMed] [Google Scholar]
- 56.Lerner I, Baraz L, Pikarsky E, Meirovitz A, Edovitsky E, Peretz T, Vlodavsky I, Elkin M. Function of heparanase in prostate tumorigenesis: potential for therapy. Clin Cancer Res. 2008;14:668–676. doi: 10.1158/1078-0432.CCR-07-1866. [DOI] [PubMed] [Google Scholar]
- 57.Elkin M, Ilan N, Ishai-Michaeli R, Friedmann Y, Papo O, Pecker I, Vlodavsky I. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 2001;15:1661–1663. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 58.Gingis-Velitski S, Zetser A, Flugelman MY, Vlodavsky I, Ilan N. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. J Biol Chem. 2004;279:23536–23541. doi: 10.1074/jbc.M400554200. [DOI] [PubMed] [Google Scholar]
- 59.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase Gene Silencing, Tumor Invasiveness, Angiogenesis, and Metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- 60.Folkman J, Klagsbrun M, Sasse J, Wadzinski M, Ingber D, Vlodavsky I. A heparin-binding angiogenic protein--basic fibroblast growth factor--is stored within basement membrane. Am J Pathol. 1988;130:393–400. [PMC free article] [PubMed] [Google Scholar]
- 61.Vlodavsky I, Folkman J, Sullivan R, Fridman R, Ishai-Michaeli R, Sasse J, Klagsbrun M. Endothelial cell-derived basic fibroblast growth factor: synthesis and deposition into subendothelial extracellular matrix. Proc Natl Acad Sci U S A. 1987;84:2292–2296. doi: 10.1073/pnas.84.8.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ben-Zaken O, Gingis-Velitski S, Vlodavsky I, Ilan N. Heparanase induces Akt phosphorylation via a lipid raft receptor. Biochem Biophys Res Commun. 2007;361:829–834. doi: 10.1016/j.bbrc.2007.06.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong X, Jiang F, Kalkanis SN, Zhang ZG, Zhang X, Zheng X, Jiang H, Mikkelsen T, Chopp M. Increased chemotactic migration and growth in heparanase-overexpressing human U251n glioma cells. J Exp Clin Cancer Res. 2008;27:23. doi: 10.1186/1756-9966-27-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cohen-Kaplan V, Naroditsky I, Zetser A, Ilan N, Vlodavsky I, Doweck I. Heparanase induces VEGF C and facilitates tumor lymphangiogenesis. Int J Cancer. 2008;123:2566–2573. doi: 10.1002/ijc.23898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alitalo K, Carmeliet P. Molecular mechanisms of lymphangiogenesis in health and disease. Cancer Cell. 2002;1:219–227. doi: 10.1016/s1535-6108(02)00051-x. [DOI] [PubMed] [Google Scholar]
- 66.Cohen-Kaplan V, Doweck I, Naroditsky I, Vlodavsky I, Ilan N. Heparanase augments epidermal growth factor receptor phosphorylation: correlation with head and neck tumor progression. Cancer Res. 2008;68:10077–10085. doi: 10.1158/0008-5472.CAN-08-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iozzo RVSL. Proteoglycan roles in health and disease: Novel regulatory signaling mechanisms evoked by the small-leucine rich proteoglycans. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hulett MD, Hornby JR, Ohms SJ, Zuegg J, Freeman C, Gready JE, Parish CR. Identification of active-site residues of the pro-metastatic endoglycosidase heparanase. Biochemistry. 2000;39:15659–15667. doi: 10.1021/bi002080p. [DOI] [PubMed] [Google Scholar]
- 69.Beauvais DM, Rapraeger AC. Syndecans in tumor cell adhesion and signaling. Reprod Biol Endocrinol. 2004;2:3. doi: 10.1186/1477-7827-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tkachenko E, Rhodes JM, Simons M. Syndecans: new kids on the signaling block. Circ Res. 2005;96:488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- 71.Levy-Adam F, Abboud-Jarrous G, Guerrini M, Beccati D, Vlodavsky I, Ilan N. Identification and characterization of heparin/heparan sulfate binding domains of the endoglycosidase heparanase. J Biol Chem. 2005;280:20457–20466. doi: 10.1074/jbc.M414546200. [DOI] [PubMed] [Google Scholar]
- 72.Haskell MD, Slack JK, Parsons JT, Parsons SJ. c-Src tyrosine phosphorylation of epidermal growth factor receptor, P190 RhoGAP, and focal adhesion kinase regulates diverse cellular processes. Chemical Rev. 2001;101:2425–2440. doi: 10.1021/cr0002341. [DOI] [PubMed] [Google Scholar]
- 73.Fux L, Feibish N, Cohen-Kaplan V, Gingis-Velitski S, Feld S, Geffen C, Vlodavsky I, Ilan N. Structure-function approach identifies a COOH-terminal domain that mediates heparanase signaling. Cancer Res. 2009;69:1758–1767. doi: 10.1158/0008-5472.CAN-08-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nardella C, Lahm A, Pallaoro M, Brunetti M, Vannini A, Steinkuhler C. Mechanism of activation of human heparanase investigated by protein engineering. Biochemistry. 2004;43:1862–1873. doi: 10.1021/bi030203a. [DOI] [PubMed] [Google Scholar]
- 75.Abboud-Jarrous G, Rangini-Guetta Z, Aingorn H, Atzmon R, Elgavish S, Peretz T, Vlodavsky I. Site-directed mutagenesis, proteolytic cleavage, and activation of human proheparanase. J Biol Chem. 2005;280:13568–13575. doi: 10.1074/jbc.M413370200. [DOI] [PubMed] [Google Scholar]
- 76.Lai NS, Simizu S, Morisaki D, Muroi M, Osada H. Requirement of the conserved, hydrophobic C-terminus region for the activation of heparanase. Exp Cell Res. 2008;314:2834–2845. doi: 10.1016/j.yexcr.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 77.Simizu S, Suzuki T, Muroi M, Lai NS, Takagi S, Dohmae N, Osada H. Involvement of disulfide bond formation in the activation of heparanase. Cancer Res. 2007;67:7841–7849. doi: 10.1158/0008-5472.CAN-07-1053. [DOI] [PubMed] [Google Scholar]
- 78.Fux L, Ilan N, Sanderson RD, Vlodavsky I. Heparanase: busy at the cell surface. Trends Biochem Sci. 2009;34:511–519. doi: 10.1016/j.tibs.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim Biophys Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nasser NJ, Avivi A, Shushy M, Vlodavsky I, Nevo E. Cloning, expression, and characterization of an alternatively spliced variant of human heparanase. Biochem Biophys Res Commun. 2007;354:33–38. doi: 10.1016/j.bbrc.2006.12.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sato M, Amemiya K, Hayakawa S, Munakata H. Subcellular localization of human heparanase and its alternative splice variant in COS-7 cells. Cell Biochem Funct. 2008;26:676–683. doi: 10.1002/cbf.1492. [DOI] [PubMed] [Google Scholar]
- 83.Barash U, Cohen-Kaplan V, Arvatz G, Gingis-Velitski S, Levy-Adam F, Nativ O, Shemesh R, Ayalon-Sofer M, Ilan N, Vlodavsky I. A novel human heparanase splice variant, T5, endowed with protumorigenic characteristics. FASEB J. 2009;24:1239–1248. doi: 10.1096/fj.09-147074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Basche M, Gustafson DL, Holden SN, O’Bryant CL, Gore L, Witta S, Schultz MK, Morrow M, Levin A, Creese BR, et al. A phase I biological and pharmacologic study of the heparanase inhibitor PI-88 in patients with advanced solid tumors. Clin Cancer Res. 2006;12:5471–5480. doi: 10.1158/1078-0432.CCR-05-2423. [DOI] [PubMed] [Google Scholar]
- 85.Fairweather JK, Hammond E, Johnstone KD, Ferro V. Synthesis and heparanase inhibitory activity of sulfated mannooligosaccharides related to the antiangiogenic agent PI-88. Bioorg Med Chem. 2008;16:699–709. doi: 10.1016/j.bmc.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 86.Ferro V, Hammond E, Fairweather JK. The development of inhibitors of heparanase, a key enzyme involved in tumour metastasis, angiogenesis and inflammation. Mini Rev Med Chem. 2004;4:693–702. doi: 10.2174/1389557043403729. [DOI] [PubMed] [Google Scholar]
- 87.Lewis KD, Robinson WA, Millward MJ, Powell A, Price TJ, Thomson DB, Walpole ET, Haydon AM, Creese BR, Roberts KL, et al. A phase II study of the heparanase inhibitor PI-88 in patients with advanced melanoma. Invest New Drugs. 2008;26:89–94. doi: 10.1007/s10637-007-9080-5. [DOI] [PubMed] [Google Scholar]
- 88.Kelly T, Miao H-Q, Yang Y, Navarro E, Kussie P, Huang Y, MacLeod V, Casciano J, Joseph L, Zhan F, et al. High Heparanase Activity in Multiple Myeloma Is Associated with Elevated Microvessel Density. Cancer Res. 2003;63:8749–8756. [PubMed] [Google Scholar]
- 89.Mahtouk K, Hose D, Raynaud P, Hundemer M, Jourdan M, Jourdan E, Pantesco V, Baudard M, De Vos J, Larroque M, et al. Heparanase influences expression and shedding of syndecan-1, and its expression by the bone marrow environment is a bad prognostic factor in multiple myeloma. Blood. 2007;109:4914–4923. doi: 10.1182/blood-2006-08-043232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanderson RD, Yang Y, Suva LJ, Kelly T. Heparan sulfate proteoglycans and heparanase--partners in osteolytic tumor growth and metastasis. Matrix Biol. 2004;23:341–352. doi: 10.1016/j.matbio.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 91.Seidel C, Sundan A, Hjorth M, Turesson I, Dahl IM, Abildgaard N, Waage A, Borset M. Serum syndecan-1: a new independent prognostic marker in multiple myeloma. Blood. 2000;95:388–392. [PubMed] [Google Scholar]
- 92.Theocharis ADTG, Karamanos NK. Proteoglycan in Health and adisease: Novel proteoglycan roles in malignancy and their pharmacological targeting. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07800.x. [DOI] [PubMed] [Google Scholar]
- 93.Purushothaman A, Uyama T, Kobayashi F, Yamada S, Sugahara K, Rapraeger AC, Sanderson RD. Heparanase-enhanced shedding of syndecan-1 by myeloma cells promotes endothelial invasion and angiogenesis. Blood. 2010;115:2449–2457. doi: 10.1182/blood-2009-07-234757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang Y, Yaccoby S, Liu W, Langford JK, Pumphrey CY, Theus A, Epstein J, Sanderson RD. Soluble syndecan-1 promotes growth of myeloma tumors in vivo. Blood. 2002;100:610–617. doi: 10.1182/blood.v100.2.610. [DOI] [PubMed] [Google Scholar]
- 95.Yang Y, Macleod V, Miao HQ, Theus A, Zhan F, Shaughnessy JD, Jr, Sawyer J, Li JP, Zcharia E, Vlodavsky I, et al. Heparanase enhances syndecan-1 shedding: a novel mechanism for stimulation of tumor growth and metastasis. J Biol Chem. 2007;282:13326–13333. doi: 10.1074/jbc.M611259200. [DOI] [PubMed] [Google Scholar]
- 96.Purushothaman A, Chen L, Yang Y, Sanderson RD. Heparanase stimulation of protease expression implicates it as a master regulator of the aggressive tumor phenotype in myeloma. J Biol Chem. 2008;283:32628–32636. doi: 10.1074/jbc.M806266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Manon-Jensen TIY, Couchman JR. Proteoglycan roles in health and disease: The multiple roles of syndecan shedding. FEBS J. 2010 doi: 10.1111/j.1742-4658.2010.07798.x. [DOI] [PubMed] [Google Scholar]
- 98.Yang Y, MacLeod V, Dai Y, Khotskaya-Sample Y, Shriver Z, Venkataraman G, Sasisekharan R, Naggi A, Torri G, Casu B, et al. The syndecan-1 heparan sulfate proteoglycan is a viable target for myeloma therapy. Blood. 2007;110:2041–2048. doi: 10.1182/blood-2007-04-082495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peterson SB, Liu J. Unraveling the specificity of heparanase utilizing synthetic substrates. J Biol Chem. 2010;285:14504–14513. doi: 10.1074/jbc.M110.104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Casu B, Guerrini M, Guglieri S, Naggi A, Perez M, Torri G, Cassinelli G, Ribatti D, Carminati P, Giannini G, et al. Undersulfated and glycol-split heparins endowed with antiangiogenic activity. J Biol Chem. 2004;47:838–848. doi: 10.1021/jm030893g. [DOI] [PubMed] [Google Scholar]
- 101.Naggi A, Casu B, Perez M, Torri G, Cassinelli G, Penco S, Pisano C, Giannini G, Ishai-Michaeli R, Vlodavsky I. Modulation of the heparanase-inhibiting activity of heparin through selective desulfation, graded N-acetylation, and glycol splitting. J Biol Chem. 2005;280:12103–12113. doi: 10.1074/jbc.M414217200. [DOI] [PubMed] [Google Scholar]
- 102.Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH, Norton L, Massague J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leung CT, Brugge JS. Tumor self-seeding: bidirectional flow of tumor cells. Cell. 2009;139:1226–1228. doi: 10.1016/j.cell.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 104.Joyce JA, Freeman C, Meyer-Morse N, Parish CR, Hanahan D. A functional heparan sulfate mimetic implicates both heparanase and heparan sulfate in tumor angiogenesis and invasion in a mouse model of multistage cancer. Oncogene. 2005;24:4037–4051. doi: 10.1038/sj.onc.1208602. [DOI] [PubMed] [Google Scholar]
- 105.Friedmann Y, Vlodavsky I, Aingorn H, Aviv A, Peretz T, Pecker I, Pappo O. Expression of heparanase in normal, dysplastic, and neoplastic human colonic mucosa and stroma. Evidence for its role in colonic tumorigenesis. Am J Pathol. 2000;157:1167–1175. doi: 10.1016/S0002-9440(10)64632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brun R, Naroditsky I, Waterman M, Ben-Izhak O, Groisman G, Ilan N, Vlodavsky I. Heparanase expression by Barrett’s epithelium and during esophageal carcinoma progression. Mod Pathol. 2009;22:1548–1554. doi: 10.1038/modpathol.2009.115. [DOI] [PubMed] [Google Scholar]
- 107.Chen T, Tang XD, Wan Y, Chen L, Yu ST, Xiong Z, Fang DC, Liang GP, Yang SM. HLA-A2-restricted cytotoxic T lymphocyte epitopes from human heparanase as novel targets for broad-spectrum tumor immunotherapy. Neoplasia. 2008;10:977–986. doi: 10.1593/neo.08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tang XD, Liang GP, Li C, Wan Y, Chen T, Chen L, Yu ST, Xiong Z, Fang DC, Wang GZ, et al. Cytotoxic T lymphocyte epitopes from human heparanase can elicit a potent anti-tumor immune response in mice. Cancer Immunol Immunother. doi: 10.1007/s00262-010-0829-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tang XD, Wan Y, Chen L, Chen T, Yu ST, Xiong Z, Fang DC, Liang GP, Yang SM. H-2Kb-restricted CTL epitopes from mouse heparanase elicit an antitumor immune response in vivo. Cancer Res. 2008;68:1529–1537. doi: 10.1158/0008-5472.CAN-07-5965. [DOI] [PubMed] [Google Scholar]
- 110.Bonertz A, Weitz J, Pietsch DH, Rahbari NN, Schlude C, Ge Y, Juenger S, Vlodavsky I, Khazaie K, Jaeger D, et al. Antigen-specific Tregs control T cell responses against a limited repertoire of tumor antigens in patients with colorectal carcinoma. J Clin Invest. 2009;119:3311–3321. doi: 10.1172/JCI39608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schuetz F, Ehlert K, Ge Y, Schneeweiss A, Rom J, Inzkirweli N, Sohn C, Schirrmacher V, Beckhove P. Treatment of advanced metastasized breast cancer with bone marrow-derived tumour-reactive memory T cells: a pilot clinical study. Cancer Immunol Immunother. 2009;58:887–900. doi: 10.1007/s00262-008-0605-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ostrovsky O, Korostishevsky M, Levite I, Leiba M, Galski H, Vlodavsky I, Nagler A. Association of heparanase gene (HPSE) single nucleotide polymorphisms with hematological malignancies. Leukemia. 2007;21:2296–2303. doi: 10.1038/sj.leu.2404821. [DOI] [PubMed] [Google Scholar]
- 113.Ostrovsky O, Korostishevsky M, Shafat I, Mayorov M, Ilan N, Vlodavsky I, Nagler A. Inverse correlation between HPSE gene single nucleotide polymorphisms and heparanase expression: possibility of multiple levels of heparanase regulation. J Leukoc Biol. 2009;86:445–455. doi: 10.1189/jlb.1208735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ostrovsky O, Shimoni A, Rand A, Vlodavsky I, Nagler A. Genetic variations in the heparanase gene (HPSE) associate with increased risk of GVHD following allogeneic stem cell transplantation: effect of discrepancy between recipients and donors. Blood. 2010;115:2319–2328. doi: 10.1182/blood-2009-08-236455. [DOI] [PMC free article] [PubMed] [Google Scholar]