Abstract
Fluorescent proteins have revolutionized cell biology by allowing researchers to non-invasively peer into the inner workings of cells and organisms. While the most common applications of fluorescent proteins are to image expression, localization, and dynamics of protein chimeras, there is a growing interest in using fluorescent proteins to create biosensors for minimally invasive imaging of concentrations of ions and small molecules, the activity of enzymes, and changes in the conformation of proteins in living cells. This tutorial review provides an overview of the progress made in the development of fluorescent protein-based biosensors to date.
1. Introduction
Two decades ago intrinsically fluorescent proteins (FPs), defined as homologues of the archetypal Aequorea victoria jellyfish green FP (avGFP), were an obscure and poorly understood biochemical oddity; today they are used daily in thousands of labs around the planet as powerful tools for studying dynamic processes in live cells. In recognition of the importance of FPs in modern biological research, the 2008 Nobel Prize in Chemistry was awarded to Osamu Shimomura, Martin Chalfie, and Roger Y. Tsien; three researchers who made key contributions to the discovery and popularization of FPs. Specifically, Shimomura had been the first to identify the presence of avGFP in extracts of Aequorea jellyfish.1 Chalfie was the first to demonstrate that the polypeptide chain of avGFP (obtained by expression of the gene cloned by Prasher in 1992)2 was self-sufficient to perform the reactions necessary for formation of the intrinsic chromophore and could thus be functionally expressed in animals other than jellyfish.3 Tsien is, arguably, the researcher who appreciated the full potential of avGFP first and contributed the most to its popularization through his successful engineering of improved and hue-shifted variants of FPs.4 Tsien also made a series of early and important contributions to the development of various types of genetically encoded biosensors,5,6 paving the way for much of the work that will be discussed in this review.
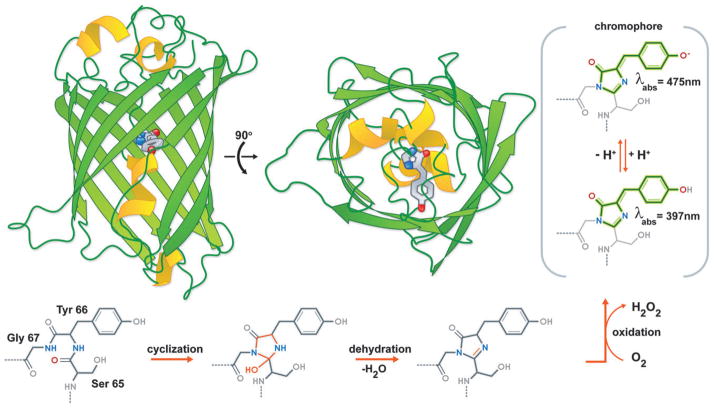
To fully appreciate just what makes FPs such useful tools in life science research, we must first consider their atomic structure. X-ray crystal structures of avGFP7 and its homologues from reef Anthozoan species revealed that these proteins share a ‘β-can’ fold comprised of an 11-stranded β-sheet polypeptide wrapped into a pseudosymmetric cylinder (Fig. 1). At the centre of the avGFP β-can is the visible wavelength 4-(p-hydroxybenzylidene)-5-imidazolidinone fluorophore, autocatalytically generated from the protein’s own amino acids. The fluorophore of avGFP is derived from a tripeptide sequence composed of a serine, a tyrosine, and a glycine (Fig. 1).8 In a series of three chemical reactions, which include cyclization of the main chain, elimination of a molecule of water, and oxidation by molecular oxygen, this tripeptide is converted into the chromophore that is the defining feature of the FP family of proteins.
Fig. 1.
The protein and chromophore structure of avGFP. Two orthogonal views of the avGFP structure (PDB ID. 1EMA) with the intrinsic chromophore shown in stick representation. Also shown is the chromophore formation mechanism.
Together with its engineered variants and homologues, avGFP has exerted a profound effect on biological science. Indeed, it is difficult to point to an area of modern biology that has not been facilitated, if not revolutionized, by the advent of FPs. In certain areas, such as the study of protein dynamics and interactions in live cells, FPs were ‘game-changing’ technology that made many experiments, which would otherwise be technically impractical, quite feasible. This review will briefly introduce the key developments and applications of FPs for the study of protein dynamics in live cells. In such applications FPs are ‘passive’ participants in the experiment, ideally acting only as a benign label with steadfast fluorescent properties.
The primary focus of the review will be on another ‘game-changing’ application of FPs: namely their use in the creation of genetically encoded biosensors that can be used to image complex biochemical processes occurring in live cells. In these applications, the engineered FP-containing construct is an ‘active’ participant in the experiment as it is engaging with the endogenous molecules under investigation and reporting on this engagement with a modulation of its fluorescent properties.
2. The toolbox of FP variants
In nature, FPs first arose more than 500 million years ago in a common ancestor shared by species as diverse as lancelets, Copepoda, and jellyfish.9 While the complete scope of the biological functions of FPs remains an open question, they seem to clearly have important roles in light signalling and protection from harmful radiation. It is apparent that the normal biological roles of FPs are quite different from the applications of FPs in the research laboratory. Accordingly, protein engineering has been used to improve the practical utility of the wild-type avGFP and its naturally occurring homologues. Although much progress was made in this regard through the mid-to-late 1990s, substantial efforts continue to this day. One of the first and most significant improvements was the engineering of an enhanced avGFP (EGFP) variant that matured more efficiently at 37 °C.4 In parallel efforts, avGFP was engineered to fluoresce with hues of blue, cyan, and yellow. This small palette of colour variants, known as BFP, CFP, and YFP, respectively, greatly expanded the range of possible applications of the growing family of FP variants.4 In particular, the availability of a selection of colours enabled simultaneous imaging of multiple fusion proteins in live cells to identify protein pairs that did, or did not, colocalize to specific intracellular compartments. The recent development of ‘sub-diffraction limit’ imaging techniques based on engineered FPs should effectively enable colocalization to be used for the analysis of protein–protein interactions.10,11
One of the most significant and unexpected advances in FP technology was the discovery and cloning of a variety of avGFP homologues, with fluorescence hues of cyan, yellow, orange, and red, from reef Anthozoa.12 In avGFP homologues with red fluorescent chromophores (i.e., red FPs (RFPs)), the basic mechanism of chromophore formation is retained with one additional elaboration. Following formation of the avGFP-type chromophore, the Cα–N bond of the residue that aligns with Ser65 of avGFP is oxidized with consumption of another equivalent of molecular oxygen (refer to Fig. 1).13 This oxidation produces an acylimine extension of the conjugated system that lowers the excitation and emission energy of the chromophore such that it becomes red, rather than green, fluorescent. Unfortunately, these wild-type reef Anthozoa FPs could be applied to only a limited subset of live cell imaging applications due to their oligomeric structure, slow and/or incomplete folding, slow and/or incomplete chromophore formation, and inadequate brightness. Monomeric variants of coral FPs suited for a greater variety of live cell imaging applications have been engineered by a combination of rational protein engineering and directed evolution.14 Moreover, a colourful palette of monomeric FPs with fluorescent hues that span the visible spectrum have been produced.15
An intriguing new class of FPs that emerged over the last several years is characterized by the ability to undergo illumination-dependent photochemical reactions or isomerizations that change the spectral properties of the chromophore. FPs that undergo such reactions can be thought of as optical highlighters that can be turned on (and sometimes o.) by illumination at specific ‘switch’ wavelengths that differ from the excitation wavelength.15 These tools are often referred to as photoactivatable, photoconvertable, or photoswitchable FPs, depending on the particular mechanism by which the illumination-dependent change occurs. Optical highlighter FPs have enabled numerous new applications including imaging of sub-populations of cells during organism development, imaging of fast protein dynamics, and imaging with sub-diffraction limit resolution.10,11
3. ‘Passive’ applications of FPs
Relative to competing fluorophore technologies such as synthetic dyes and quantum dots, the fact that FPs can be genetically introduced into cells, tissues, or whole organisms is a unique and overwhelming advantage for the vast majority of live cell imaging applications. FPs are frequently used as a whole cell marker or a reporter of gene promoter activation. Recently, however, researchers have become more interested in imaging the localization and dynamics of a FP chimera, in which the gene encoding the FP is fused in frame with the gene encoding a second protein of primary interest. Fortunately FPs are very robust with respect to genetic fusions to their N- and C-termini and thus can be genetically fused to any one of a practically unlimited variety of protein targets in order to determine its subcellular localization. In cases where neither the N- nor the C-terminus is suitable for fusing a FP (either due to interference with function, or because the terminal fusions do not provide sufficient signal), it has been possible to insert the FP at other locations within the protein of interest, especially surface exposed loops.16 By combining imaging of either standard or ‘highlighter’ FP fusions with sophisticated localized photobleaching or protein highlighting, detailed investigations of protein dynamics can be undertaken.17 As a consequence, photobleaching and photo-activation devices and software have become a standard component of many confocal microscope systems.
Needless to say, the availability of a broad palette of FP hues makes it possible to image the localization and dynamics of multiple FP chimeras in a single cell. In one impressive demonstration of multiple FP imaging, six different hues of FP were imaged using a single excitation laser line and spectral unmixing.18 Using widefield fluorescence microscopy with bandpass filter sets, imaging of three or four different hues of FP has become a relatively straightforward endeavour. One important biological question addressed using multiple FP imaging is whether two particular proteins of interest localize to similar locations within the cell. Taking this approach to its logical extreme, the localization of more than 4000 different yeast proteins fused to an avGFP variant has been analysed and categorized with respect to their colocalization with each of 11 distinct RFP-fused localization markers.19 Similar analyses covering the whole genomes of plants and humans are expected to follow suit.
3.1 Translocation-based biosensors
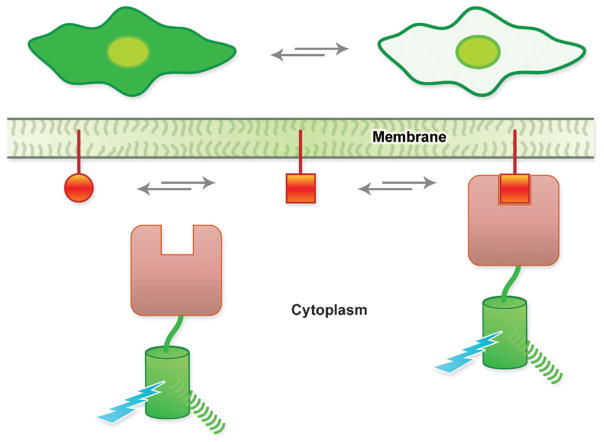
Translocation-based biosensors consist of the genetic fusion of at least one FP to a protein domain that exhibits specific binding to a lipid, such as a phosphatidylinositol or diacylglycerol, that is transiently generated at the membrane in response to a signal (Fig. 2).20 Such biosensors are particularly useful for dissecting the dynamics of complex signalling pathways. For example, the translocation of an avGFP-fused pleckstrinhomology domain of mammalian phospholipase C δ (PHPLCδ) can serve as a probe of dynamic changes in the relative concentrations of PtdIns(4,5)P2 and Ins(1,4,5)P3. PHPLCδ binds to both the membrane-anchored PtdIns(4,5)P2 and the freely soluble Ins(1,4,5)P3. When concentrations of Ins(1,4,5)P3 are low, avGFP–PHPLCδ fusions remain associated with the membrane. As levels of Ins(1,4,5)P3 rise, the PtdIns(4,5)P2 is displaced from the binding site on the protein and avGFP–PHPLCδ is released from themembrane. Accordingly, the dynamics of Ins(1,4,5)P3 at the membrane, generated through the catalytic action of phospholipase C, can be imaged in single cells in real time. By fusing a FRET pair of FPs, rather than a single FP, to the translocating domain it is possible to augment the intensiometric signal of a translocation-based biosensor with a ratiometric change in fluorescent intensity.21 When bound to the membrane the overall concentration of the FPs is increased relative to the cytoplasm and FPs from adjacent protein constructs are brought close enough to exhibit FRET. Translocation biosensors are used to systematically identify signalling cascades via high throughput screens.20
Fig. 2.
Schematic representation of the mechanism of a translocation-based biosensor. At the top of the figure is shown the expected result when imaging a cell expressing a translocation-based biosensor.
4. ‘Active’ applications of FPs
4.1 Imaging of protein–protein proximity and interactions
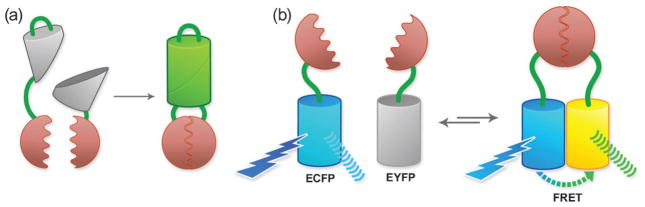
While the experimental observation of colocalization is obviously a necessary condition for two proteins to be involved in a specific interaction, it is by no means a sufficient condition. The resolution of standard fluorescence imaging is not better than 100s of nanometres, and so proteins that colocalize may not in fact be interacting physically. However, FPs do enable methods for probing the distance between proteins at a much finer resolution, approaching the dimensions of proteins themselves. For example, FPs can be split into two halves and fused to potential interaction partners.22 These free FP-halves do not appear to readily associate at low effective molarity (Fig. 3a). However, when present at a high effective molarity, such as when they are brought into close proximity by interaction between their fusion partners, a functional FP is reconstituted. At high concentrations, the inherent affinity of the FP-halves for each other (as opposed to the association of the fusion partners) may result in false positive signals. Moreover, the effectively irreversible reconstitution of GFP makes it generally unsuitable for analyses of complex disassembly.
Fig. 3.
Methods of detecting protein–protein interactions using FPs. (a) FP complementation. (b) FRET for detecting protein–protein interactions.
An alternative method for interrogating whether or not two proteins are within several nanometres of each other, and therefore likely involved in a protein–protein interaction, involves the use of Förster resonance energy transfer (FRET; sometimes also defined as fluorescence resonance energy transfer; Fig. 3b).23 FRET is based on a non-radiative quantum mechanical process, which neither requires collisions between the molecules nor produces heat. FRET is the distance- and orientation-dependent radiationless transfer of excitation energy from a shorter wavelength (more blue-shifted) donor fluorophore to a longer wavelength (more red-shifted) acceptor chromophore. Although the physical basis of FRET requires a good deal of theory to explain, it manifests itself in a very simple and practical manner. If two fluorophores are close enough together in space (i.e., less than 10 nm), the higher energy fluorophore passes its excited state energy to the lower energy fluorophore via a dipole–dipole interaction. Interestingly, FRET does not require the acceptor to be fluorescent. Typically, FRET is measured as sensitized emission from the acceptor, that is, by measuring light produced by emission of the acceptor when only the donor is excited. The ratio of acceptor to donor fluorescence is typically used as a surrogate for actual FRET efficiency in live cell imaging. This can simply be achieved by filter-based assays. Alternatively, FRET efficiency can be inferred from the rate of photobleaching of the donor or acceptor, which will be modulated when the distance and orientation between donor and acceptor change. More sophisticated approaches include analysis of the lifetime of donor-fluorescence or anisotropy decay measurements. Generally speaking, the use of FRET for detection of protein– protein interactions in live cells is highly qualitative. Recently, however, some researchers have begun attempting to use FRET for quantitative determination of protein–protein dissociation constants (Kd) in live cells.24
The BFP–EGFP combination was the first pair of engineered FPs that had the appropriate spectral overlap to enable their use as a FRET pair. (Criteria for selecting the optimal FRET pair: (i) FRET requires spectral overlap between the donor emission and excitation of the acceptor; at the same time, excitation of the acceptor by donor excitation light should be minimal to avoid “bleed through”; (ii) spectra of fluorophores must be in a range that coincides with low endogenous fluorescence (autofluorescence)).4 The BFP–EGFP pair was soon superseded by the still popular CFP–YFP combination, which has formed the reporter element for the vast majority of FRET-based biosensors to date. Although a variety of additional FRET pairs have been reported in the literature, few of these newer pairs have any substantial advantages relative to the CFP–YFP pair. One trend in recent years has been to investigate the pairing of yellow or green fluorescent donor FPs with orange and red fluorescent acceptors in FRET pairs. Several research groups have recently undertaken thorough comparisons of the various orange FPs and RFPs as FRET acceptors (and in some cases, donors).25,26 They have found that certain FRET pairs (including both yellow–red and orange–red pairs) are a promising alternative to CFP–YFP if used for FRET imaging methods that only rely on donor quench (i.e., donor fluorescence lifetime imaging (FLIM)). However, it is apparent that FRET pairs with monomeric RFPs as acceptors are less than ideal when used with imaging methods that depend on having strong sensitized emission from the acceptor. Fortunately, efforts to create ever brighter monomeric RFPs continue,27 and it is almost certain that variants with improved properties (i.e., high quantum yield) that will make them more suitable as FRET acceptors will be developed within the next few years. It must be noted that, despite their limitations, monomeric RFPs can still be useful for detecting protein–protein interactions in live cells. For example, even the first generation monomeric RFP (mRFP1)14 has proven to be a useful acceptor for an EGFP donor for detection of a variety of different protein–protein interactions in live cells.
4.2 Biosensors: versatile tools for imaging biochemistry in live cells
In the broadest sense, molecular fluorescent biosensors are a highly diverse class of biosensors that, by definition, exploit the intrinsic molecular recognition specificity of a biomolecule to modulate the fluorescent intensity or hue of a fluorophore or a pair of fluorophores. The primary advantages of such biosensor designs are their exquisite sensitivity, versatility, and technical simplicity. In the case of biosensors based on FPs, there is the added advantage of being entirely genetically encoded and thus amenable to sensing applications in specific subcellular compartments of individual cells in culture, tissues, or even whole animals. Accordingly, a major effort within the field of FP research has been invested in the engineering of molecular fluorescent biosensors that are active reporters of the cell environment rather than simple passive reporters of gene expression or fusion protein localization.28 One recent example that illustrates the versatility of FPs for active imaging applications is a system for the precise marking of cell cycle that has been applied in a variety of contexts ranging from cultured cells to transgenic animals.29 For the sake of this review, we focus on the subclass of biosensors that are composed of single polypeptide chains incorporating at least one FP. These FP-based biosensors undergo either a dynamic change in intrinsic fluorescence intensity or fluorescence hue in response to a biologically-relevant stimulus. A list of published biosensor constructs is provided in Supplementary Table 1 available online.‡
4.2.1 FRET-based biosensors
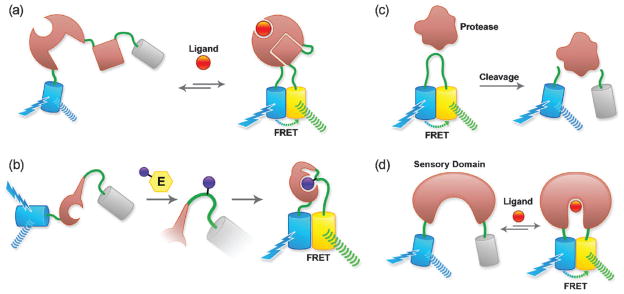
The single most fruitful approach to the development of FP-based biosensors has been to exploit intramolecular FRET, and create single polypeptides composed of a ‘sensing’ domain sandwiched between a donor FP and an acceptor FP.28 The sensing domain has two essential functions. First, it must provide the specificity of the biosensor for the biochemical signal of interest. Second, it must respond to this biochemical signal with a change in conformation that effectively modulates the FRET efficiency (dependent on inter-fluorophore distance and relative orientation) between the two fused FPs. In practice, the change in FRET efficiency between the low-FRET and high-FRET states must be large enough to provide a robust change in the intensity ratio Ia/Id (or vice versa), where Id and Ia are the fluorescence intensities in the donor and acceptor emission channels upon excitation of the donor. A wide variety of designs of sensing domains for FRET-based biosensors have been investigated. Four prototypical designs are schematically represented in Fig. 4a–d. In these designs the sensing domain is either: a fusion of two proteins that participate in a small-molecule-dependent interaction (Fig. 4a); a fusion of a peptide substrate and a binding domain that interact only when the substrate is post-translationally modified by an enzyme (Fig. 4b); a substrate for an enzyme, here a protease (Fig. 4c); or a protein that undergoes a conformational change when bound to a particular target molecule (Fig. 4d).
Fig. 4.
FRET-based biosensors. (a) Biosensors based on a ligand-dependent protein–protein interaction. Cameleons (based on a fusion of calmodulin and M13) and GTPase biosensors (based on a fusion of the GTPase and its effector) fall into this category. (b) Post-translational modification biosensor (i.e., for a kinase). (c) Protease substrate-type biosensor. (d) Biosensor based on conformational change of a single protein (i.e., based on periplasmic binding proteins).
The first demonstration that FPs variants could be used for the construction of biosensors based on single polypeptide chains was published in 1996 by Tsien and coworkers.30 In this original work, the biosensor was a relatively simple construct consisting of a BFP and a GFP linked by a trypsin cleavable linker. FRET-based biosensors (or substrates) for proteolytic enzyme activity continue to be an important player in the FP toolbox. One area of research where they are particularly useful is in the study of caspase activation during apoptosis.26 A recent trend is to employ protease biosensors in FRET-based screens for inhibitors of clinically important enzymes such as poliovirus 2Apro protease.31
Following closely on the heels of the first demonstrations of inter-FP FRET came the ‘cameleon’ class of FRET-based biosensors for Ca2+.5,32 This breakthrough clearly demonstrated the potential of FRET-based biosensors to be applied to a wide variety of intracellular imaging applications. Cameleon-type biosensors are assembled by genetic fusion of four components: a donor FP, calmodulin (which binds to Ca2+), a peptide (such as M13) that binds to the Ca2+-bound form of calmodulin as an actuator, and an acceptor FP. An increase in Ca2+ concentration results in binding of the ion to calmodulin, which in turn binds to M13. The overall result is a conformational change that brings the donor and acceptor FP into closer proximity and thus increases FRET efficiency and, thus, the Ia/Id emission ratio. The original calcium biosensors had a relatively low signal-to-noise ratio, therefore efforts to further improve cameleon-type biosensors continue to be an active area of research.33
Other promising classes of FRET-based biosensors to emerge in recent years are those that are suitable for the real-time imaging of kinase activity.34 These biosensors tend to follow the basic design of having a sensing domain composed of a peptide substrate for the kinase of interest linked to a protein domain that binds only to the phosphorylated form of the substrate. As is typical for FRET-based biosensors, a sensing domain is flanked by a donor and acceptor FP fused at either end. In the absence of the kinase activity of interest, the sensing domain is in an extended conformation and the FPs are relatively far from each other. If the active kinase is present and has phosphorylated the substrate, the peptide will bind to the linked domain and the sensing domain will assume a more compact conformation. As with the cameleon-type indicators described above, this change from an extended to a compact conformation for the sensing domain changes the distance between the linked FPs and the Ia/Id emission ratio increases (typically) due to the higher FRET efficiency. This design has now been applied to a growing number of kinases, many of which are listed in Supplementary Table 1.‡ An important application of FRET-based kinase biosensors is in the development of high throughput assays for the discovery of specific kinase inhibitors.35
Consideration of the large number of biosensor variants in which linker regions have been mutated leads to the conclusion that it is generally difficult to predict whether the design of a FRET biosensor will be successful and whether the ratio change will be positive or negative. The challenge of predicting the response of a given biosensor variant is exemplified by the fact that effective biosensors based on periplasmic binding proteins could be engineered using a design that does not directly exploit the large conformational changes occurring between the two lobes of the protein (Fig. 4d). For example, researchers have created a glucose biosensor with very high FRET change in which both FPs are fused to the same lobe of the protein.16 It is likely that ligand binding induces changes in surface properties of the recognition element and that these changes lead to a change in the relative distance and orientation of the average position of the FPs.
Despite the lack of predictability in terms of biosensor responses, the success rate for development of new FRET-based biosensors has been surprisingly high, to the extent that it is worthwhile to invest a couple of weeks to design a novel biosensor. Design principles are basically empirical and include the use of as many scaffolds as possible for the recognition element and variations in the linkers used to attach the fluorophores. A recent successful example is a FRET-based biosensor of the serine/threonine phosphatase calcineurin activity that was developed by fusing a FRET pair of FPs to the nuclear factor of activated T-cells (NFAT).36 No structure was available for the conformationally mobile NFAT domain. By exploring a variety of NFAT1 truncations, the authors found a construct with a ratio change sufficient for imaging of phosphatase activity in live cells. In another example, a voltage biosensor was constructed by linking donor and acceptor FPs (in tandem) to a membrane associated voltage sensitive domain (VSD).37,38 Apparently, voltage induced conformational changes in the VSD alter the relative orientations of the FPs. Through systematic variation of the VSD to FP and FP to FP linkers, ratio changes as high as 40% have been achieved.38 In yet another example, researchers made more than 50 different constructs with various homologues of the sensing domain and various linker lengths in an effort to identify cGMP biosensors with large FRET changes.39 Similarly, through a combination of linker variation and insertion of the fluorophore into the backbone of periplasmic binding proteins, high signal-to-noise FRET biosensors for a wide spectrum of metabolites including glucose and glutamate have been developed.40 Linker deletion of insertion of the fluorophore into the backbone of the recognition element as well as the use of circular permutated fluorophores are means for optimizing the sensors by changing the orientation or distance factors of the transition dipole moments.
Although this review focuses on biosensors that consist of a single polypeptide chain, it is important to note that similar design strategies can be used for the construction of ‘split’ biosensors in which two different polypeptide chains, each fused to one member of the FRET pair, interact in a target analyte- or activity-specific manner. The advantage of split biosensors is that they can potentially provide larger FRET efficiency changes due to the fact that the two FPs are effectively infinitely far apart in the dissociated state. The classic example of such a split FRET-biosensor is the fusion of FP variants to the catalytic and regulatory subunits of PKA for detection of cAMP.41
4.2.2 Single FP-based biosensors
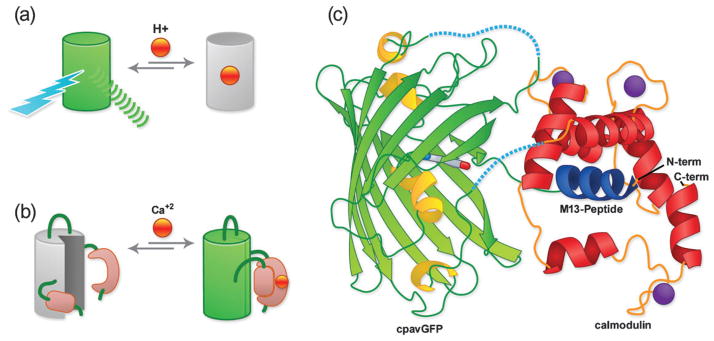
As their name implies, single FP-based biosensors contain only one engineered FP domain. The key to creation of such a biosensor is to engineer this single FP to undergo a change in fluorescent intensity, or excitation profile, or emission profile, in response to the biochemical stimulus of interest. The advantage of this approach is that the fluorescence intensity changes at a particular wavelength tend to be greater in magnitude for a single FP-based biosensor compared to a FRET-based biosensor. On the other hand, single FP-based biosensors may not have a ratiometric response (although in some cases they do) that is advantageous with respect to calibration and quantitative imaging applications. A ratiometric response is inherent in the design of FRET-based biosensors. One way to create a single FP-based biosensor is to take advantage of intrinsic sensitivities of certain FP variants (Fig. 5a). The classic examples of such a biosensor are ones that take advantage of the fact that the fluorescence of all FPs exhibits a pH dependence, and some variants happen to have apparent pKas (pK′a) that are close to physiologically relevant pH values. Among the many examples of such pH biosensors are the pHlourin variants of avGFP.42 In at least one case it has proven possible to rationally engineer an intrinsic sensitivity, namely a fluorescent response to redox potential, into a FP.43
Fig. 5.
Single FP-based biosensors. (a) Single FP biosensor based on intrinsic (i.e., pH) sensitivity. (b) Single FP biosensor based on the extrinsic sensitivity (i.e., Ca2+) of a genetically fused domain (i.e., calmodulin). (c) GCaMP X-ray crystal structure.45 Linker regions that were not visible in the crystal structure are represented with dashed lines.
Relative to these intrinsic FP biosensors, a wider range of target specificities is accessible to single FP-based biosensors that are engineered to respond to an extrinsic sensing event mediated through a fused protein domain (Fig. 5b). The strategy for making such extrinsic single FP-type biosensors is to insert a protein domain that undergoes a conformational change in response to binding of a target of interest (i.e., its ligand) into a FP such that ligand binding is ‘communicated’ to the FP as a local distortion in its structure.6 This distortion can quench the fluorescence due to an increase in chromophore solvent accessibility or alter the equilibrium between the protonated and unprotonated forms of the chromophore (refer to Fig. 1). The main challenge in such a design is how to circumvent the ‘protective shell’ of the beta-barrel that surrounds the chromophore. Fortunately, researchers have developed a series of topological variants of avGFP, which have been circularly permuted (cp) such that their new N- and C-termini are in very close proximity to the chromophore. These cpavGFP variants are the starting point of choice for creation of single FP-based biosensors. As with the development of FRET-based biosensors, progress in the area of single- FP-based biosensors has been driven by the development of Ca2+ biosensors.6 Similar designs have recently proven useful for imaging of a variety of other analytes, with one recent example being a new biosensor for ATP : ADP ratio.44
In the push for ever more effective Ca2+ biosensors, the X-ray crystal structure of the single FP-biosensor known as GCaMP was recently determined (Fig. 5c).45 This structure reveals that the ‘hole’ in the cpGFP is quite dramatic and perhaps larger than expected. This structure has already provided valuable insight that has allowed the rational engineering of an improved GCaMP variant.
Many of the sensors shown here were generated to detect small molecules, but apparently, the same concept can be used to detect interactions with macromolecules, e.g. to identify protein interactions.46
5. Conclusions and outlook
One of the most interesting new trends in the area of FP-based biosensors are published reports of rigorous head-to-head comparisons of various biosensors from within a given target class. The very fact that such comparisons are necessary (and feasible) is reflective of the high amount of effort currently being expended on the development of new and improved biosensors. Such analyses serve as a valuable benchmark for different groups working on development of different versions of biosensors and help to focus their efforts on the most critical shortcomings of their particular design. We are encouraged by this trend and expect particular research groups will emerge as ‘specialists’ in the application of particular classes of biosensors and serve as a community resource for validation and testing of new designs and improved variations.
Another exciting trend in the FP-biosensor field is the application of these biosensors in tissues and transgenic animals. For example, various research groups have recently reported imaging of glutamate, protein kinase A activity, Ca2+, and cyclic nucleotides, in prepared tissues or transgenic animals as well as glucose flux in roots of intact plants. The biosensors that have proven most effective for true in vivo imaging are those that have been highly optimized for expression and FRET response. The current challenge facing protein engineers will be to optimize biosensors with a variety of different specificities to the point where they can also be useful in vivo.
Yet another area where we expect to see exciting advances in the years to come is in the use of FP-based biosensors for multiparameter imaging. Despite the impressive selection of new colours and new FRET pairs, their broad spectral profiles have hampered the development of spectrally orthogonal FRET pairs that can be individually and ratiometrically imaged in a single cell. Only recently have researchers reported the first applications of spectrally orthogonal FRET pairs.47 An analogous breakthrough in the area of single FP-based biosensors, probably through engineering of a red homologue of the avGFP-derived biosensors, is a highly anticipated advance that would greatly facilitate multi-parameter biosensor imaging.
Supplementary Material
Acknowledgments
WBF is supported by grants from the National Institute of Health (NIDDK 1RO1DK079109-01) and the Department of Energy (DE-FG02-04ER15542). REC acknowledges the support of the Canada Research Chairs program, NSERC, and Alberta Ingenuity and thanks Haley Carlson for assistance with the literature review.
Abbreviations and common names used
- avGFP
Aequorea victoria GFP
- BFP
blue FP
- CFP
cyan FP
- Citrine
a YFP
- cpXFP
circularly permuted FP
- EGFP
enhanced avGFP
- FP
fluorescent protein
- FRET
Förster resonance energy transfer
- GFP
green FP
- mXFP
a monomeric FP
- RFP
red FP
- YFP
yellow FP
Biographies
 Wolf B. Frommer is Director of the Department of Plant Biology, Carnegie Institution for Science, Stanford, CA. He obtained his PhD with Peter Starlinger, University of Köln, did a postdoc with Lothar Willmitzer, IGF Berlin, and was awarded a BMBF young investigator fellowship from the German Ministry for Education and Research to head a group at the IGF. He moved to Tübingen University as Chair of Plant Physiology. He is a recipient of the G. W. Leibniz award of the German Research Foundation and the European Science Award of the Körber Foundation and is a fellow of AAAS.
Wolf B. Frommer is Director of the Department of Plant Biology, Carnegie Institution for Science, Stanford, CA. He obtained his PhD with Peter Starlinger, University of Köln, did a postdoc with Lothar Willmitzer, IGF Berlin, and was awarded a BMBF young investigator fellowship from the German Ministry for Education and Research to head a group at the IGF. He moved to Tübingen University as Chair of Plant Physiology. He is a recipient of the G. W. Leibniz award of the German Research Foundation and the European Science Award of the Körber Foundation and is a fellow of AAAS.
 Michael W. Davidson is an Associate in Research affiliated with the Department of Biological Science and the National High Magnetic Field Laboratory at the Florida State University. Davidson is involved in development of educational websites that address all phases of optical microscopy, including bright-field, phase contrast, DIC, fluorescence, confocal, TIRF, and multiphoton. Davidson is also involved in the performance of traditional fluorescent proteins and optical highlighters in fusions for targeting and dynamics studies in live cells. Davidson’s digital images and photomicrographs have graced the covers of over 2000 publications in the past two decades and he has licensed images to numerous commercial partners.
Michael W. Davidson is an Associate in Research affiliated with the Department of Biological Science and the National High Magnetic Field Laboratory at the Florida State University. Davidson is involved in development of educational websites that address all phases of optical microscopy, including bright-field, phase contrast, DIC, fluorescence, confocal, TIRF, and multiphoton. Davidson is also involved in the performance of traditional fluorescent proteins and optical highlighters in fusions for targeting and dynamics studies in live cells. Davidson’s digital images and photomicrographs have graced the covers of over 2000 publications in the past two decades and he has licensed images to numerous commercial partners.
 Robert E. Campbell is an associate professor and Canada Research Chair in Bioanalytical Chemistry in the Department of Chemistry of the University of Alberta. He did his PhD research with Martin E. Tanner at the University of British Columbia and undertook a CIHR-funded postdoctoral fellowship with Roger Y. Tsien at the University of California, San Diego. Research in his laboratory is focused on protein engineering and the development of new fluorescent protein variants for construction of FRET-based biosensors.
Robert E. Campbell is an associate professor and Canada Research Chair in Bioanalytical Chemistry in the Department of Chemistry of the University of Alberta. He did his PhD research with Martin E. Tanner at the University of British Columbia and undertook a CIHR-funded postdoctoral fellowship with Roger Y. Tsien at the University of California, San Diego. Research in his laboratory is focused on protein engineering and the development of new fluorescent protein variants for construction of FRET-based biosensors.
Footnotes
Part of a themed issue on the topic of green fluorescent protein (GFP) in honour of the 2008 Nobel Prize winners in Chemistry, Professors Osamu Shimomura, Martin Chalfie and Roger Y. Tsien.
Electronic supplementary information (ESI) available: A list of published biosensor constructs. See DOI: 10.1039/b907749a
Contributor Information
Wolf B. Frommer, Email: wfrommer@stanford.edu.
Michael W. Davidson, Email: davidson@magnet.fsu.edu.
Robert E. Campbell, Email: robert.e.campbell@ualberta.ca.
References
- 1.Shimomura O, Johnson FH, Saiga Y. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 2.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 3.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Tsien RY. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 5.Miyawaki A, Llopis J, Heim R, McCaffery JM, et al. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 6.Baird GS, Zacharias DA, Tsien RY. Proc Natl Acad Sci U S A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ormo M, Cubitt AB, Kallio K, Gross LA, et al. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 8.Heim R, Prasher DC, Tsien RY. Proc Natl Acad Sci U S A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shagin DA, Barsova EV, Yanushevich YG, Fradkov AF, et al. Mol Biol Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- 10.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, et al. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 11.Hell SW. Nat Methods. 2009;6:24–32. doi: 10.1038/nmeth.1291. [DOI] [PubMed] [Google Scholar]
- 12.Matz MV, Fradkov AF, Labas YA, Savitsky AP, et al. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 13.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. Proc Natl Acad Sci U S A. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell RE, Tour O, Palmer AE, Steinbach PA, et al. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaner NC, Patterson GH, Davidson MW. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 16.Deuschle K, Okumoto S, Fehr M, Looger LL, et al. Protein Sci. 2005;14:2304–2314. doi: 10.1110/ps.051508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH. Nat Cell Biol. 2003;(Suppl):S7–14. [PubMed] [Google Scholar]
- 18.Kogure T, Karasawa S, Araki T, Saito K, et al. Nat Biotechnol. 2006;24:577–581. doi: 10.1038/nbt1207. [DOI] [PubMed] [Google Scholar]
- 19.Huh WK, Falvo JV, Gerke LC, Carroll AS, et al. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 20.Meyer T, Teruel MN. Trends Cell Biol. 2003;13:101–106. doi: 10.1016/s0962-8924(02)00040-5. [DOI] [PubMed] [Google Scholar]
- 21.Violin JD, Zhang J, Tsien RY, Newton AC. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabantous S, Terwilliger TC, Waldo GS. Nat Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 23.Forster T. Discuss Faraday Soc. 1959;27:7–17. [Google Scholar]
- 24.Chen H, Puhl HL, Ikeda SR. J Biomed Opt. 2007;12:054011. doi: 10.1117/1.2799171. [DOI] [PubMed] [Google Scholar]
- 25.Kwok S, Lee C, Sánchez SA, Hazlett TL, et al. Biochem Biophys Res Commun. 2008;369:519–525. doi: 10.1016/j.bbrc.2008.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ai H, Hazelwood KL, Davidson MW, Campbell RE. Nat Methods. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 27.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, et al. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lalonde S, Ehrhardt DW, Frommer WB. Curr Opin Plant Biol. 2005;8:574–581. doi: 10.1016/j.pbi.2005.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, et al. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 30.Heim R, Tsien RY. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 31.Hwang YC, Chu JJ, Yang PL, Chen W, Yates MV. Antiviral Res. 2008;77:232–236. doi: 10.1016/j.antiviral.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romoser VA, Hinkle PM, Persechini A. J Biol Chem. 1997;272:13270–13274. doi: 10.1074/jbc.272.20.13270. [DOI] [PubMed] [Google Scholar]
- 33.Mank M, Griesbeck O. Chem Rev. 2008;108:1550–1564. doi: 10.1021/cr078213v. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Allen MD. Mol BioSyst. 2007;3:759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 35.Allen MD, DiPilato LM, Rahdar M, Ren YR, et al. ACS Chem Biol. 2006;1:371–376. doi: 10.1021/cb600202f. [DOI] [PubMed] [Google Scholar]
- 36.Newman RH, Zhang J. Mol BioSyst. 2008;4:496–501. doi: 10.1039/b720034j. [DOI] [PubMed] [Google Scholar]
- 37.Dimitrov D, He Y, Mutoh H, Baker BJ, et al. PLoS ONE. 2007;2:e440. doi: 10.1371/journal.pone.0000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Nat Methods. 2008;5:683–685. doi: 10.1038/nmeth.1235. [DOI] [PubMed] [Google Scholar]
- 39.Russwurm M, Mullershausen F, Friebe A, Jäger R, et al. Biochem J. 2007;407:69–77. doi: 10.1042/BJ20070348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okumoto S, Takanaga H, Frommer WB. New Phytol. 2008;180:271–295. doi: 10.1111/j.1469-8137.2008.02611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaccolo M, De Giorgi F, Cho CY, Feng L, et al. Nat Cell Biol. 2000;2:25–29. doi: 10.1038/71345. [DOI] [PubMed] [Google Scholar]
- 42.Miesenböck G, De Angelis DA, Rothman JE. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 43.Dooley CT, Dore TM, Hanson GT, Jackson WC, et al. J Biol Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 44.Berg J, Hung YP, Yellen G. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akerboom J, Rivera JD, Guilbe MM, Malavé EC, et al. J Biol Chem. 2009;284:6455–6464. doi: 10.1074/jbc.M807657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lalonde S, Ehrhardt DW, Loqué D, Chen J, et al. Plant J. 2008;53:610–635. doi: 10.1111/j.1365-313X.2007.03332.x. [DOI] [PubMed] [Google Scholar]
- 47.Carlson HJ, Campbell RE. Curr Opin Biotechnol. 2009;20:19–27. doi: 10.1016/j.copbio.2009.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.