Abstract
Line-active molecules (“linactants”) that bind to the boundary interface between different fluid lipid domains in membranes have a strong potential as regulators of the lateral heterogeneity that is important for many biological processes. Here, we use molecular dynamics simulations in combination with a coarse-grain model that retains near-atomic resolution to identify lipid species that can act as linactants in a model membrane that is segregated into two lipid domains of different fluidity. Our simulations predict that certain hybrid saturated/unsaturated chain lipids can bind to the interface and lower the line tension, whereas cone-shaped lysolipids have a less pronounced effect.
The lateral heterogeneity of biological membranes has important implications for the function of cells (1). Nevertheless, to study the organization of biological membranes remains a challenge, because it is inherently difficult to characterize fluctuating lipid assemblies in the membranes of living cells (2). Model membranes (3–6) and isolated plasma membranes (7–9) are more frequently studied, because large-scale phase separation can occur in these systems. In particular, ternary mixtures of saturated lipids, unsaturated lipids, and cholesterol can segregate into two coexisting fluid lipid domains, a liquid-ordered (Lo) and liquid-disordered (Ld) phase. Such domains have been widely studied, because they may be closely linked to lipid nanodomains in cell membranes (10). It is intriguing to devise molecules that specifically bind at the boundary interface between the different lipid domains, thereby modifying the boundary properties while leaving the bulk regions unaltered (11). As they are supposed to reduce the line tension (or energetic cost) of the one-dimensional boundary interface, such molecules can be called linactants, analogous to surfactants (which modify the surface tension at an oil/water interface). Possible line-active molecules could be, e.g., certain lipids, or lipid-anchored or transmembrane proteins.
In this work, our aim is to identify lipid species that can act as biological linactants. To that end, we inserted potentially line-active lipids into a lipid bilayer that consists of two coexisting fluid domains and studied their partitioning at the domain boundary during extensive coarse-grain (CG) molecular dynamics (MD) simulations. Two different types of lipids were chosen as potential candidates, hybrid saturated/unsaturated chain lipids, and a single-chain lysolipid. These different species were chosen to investigate two possible mechanisms: hybrid lipids might accumulate at the domain boundary due to their mixed hydrocarbon chains, whereas lysolipids are cone-shaped and may thus be attracted due to the (local) curvature at the domain boundary, which arises from the thickness mismatch between the domains (∼0.7 nm in our bilayer).
Fig. 1 shows the lipid bilayer studied, a ternary mixture of saturated diC16:0PC (dipalmitoyl-phosphatidylcholine, DPPC), doubly unsaturated diC18:2PC (dilinoleoyl-PC, DLiPC), and cholesterol (molar ratio 0.42:0.28:0.3). In a recent CG-MD study from our group, it was shown that this bilayer spontaneously segregates into two fluid domains at 295 K (12). DLiPC enhances the driving force for phase separation (13), while yielding domain properties similar to those observed in DOPC/DPPC/cholesterol bilayers (4). The liquid-ordered (Lo) domain mainly consists of DPPC and cholesterol, whereas the liquid-disordered (Ld) domain is enriched in DLiPC and contains less cholesterol. The domains are separated by a boundary interface that is ∼5 nm in width (14). Here, we added small amounts (40 molecules, 2 mol %) of the fourth component to this ternary mixture. The idea was to introduce enough molecules to obtain proper statistics during the MD simulations, while perturbing the phase diagram of the ternary system as weakly as possible. As hybrid lipids, C16:0C18:1PC (palmitoyl-oleoyl-PC, POPC) and C16:0C18:2PC (palmitoyl-linoleoyl-PC, PLiPC) were added; single-chain C16:0PC (palmitoyl-PC, LysoPC) was chosen as a cone-shaped lipid. As in the previous work (12), the GROMACS MD package (15) was used together with the MARTINI force field (16), a CG model that retains near-atomic resolution. For simulation parameters, see Risselada and Marrink (12) and Marrink et al. (16). Three MD simulations were carried out (with 2 mol % POPC, PLiPC, and LysoPC, respectively), each for 18 μs of simulation time. The last 8 μs of each simulation were analyzed (see the Supporting Material for details on equilibration times).
Figure 1.
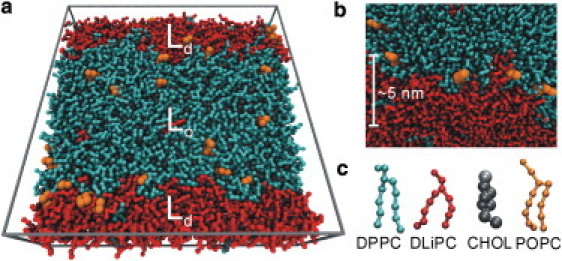
(a) The simulated lipid bilayer is segregated into an ordered (Lo) and a disordered (Ld) domain. The system comprises ∼2000 lipids, solvated by water (not shown). (b) Snapshot from simulation showing a part of the domain boundary interface. (c) MARTINI CG representations of DPPC (cyan), DLiPC (red), cholesterol (gray), and POPC (orange).
Fig. 2 shows the composition profiles obtained from the simulations. The POPC profile (Fig. 2 a, orange) has two pronounced peaks close to the Lo/Ld interface, which are slightly shifted toward the bulk of the Lo domain. The bulk domains have a reduced POPC content. Interestingly, the hybrid PLiPC does not show a clear preference for the interface (Fig. 2 b, violet), but colocalizes with the DLiPC lipids in the Ld domain. Obviously, two double bonds yield a chain that, in terms of the partitioning behavior, dominates over the saturated chain. LysoPC (Fig. 2 c, magenta) does accumulate at the domain boundary, albeit in a less pronounced manner as compared to POPC. Table 1 (also see (17)) summarizes the free energy differences of the fourth component between the bulk domains and the interface.
Figure 2.
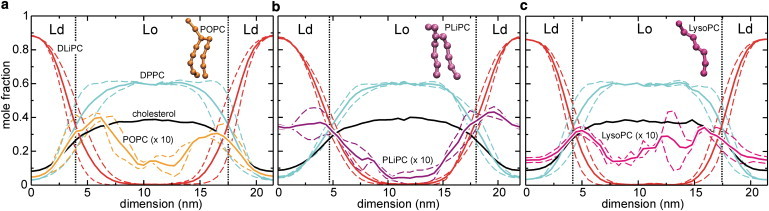
Lateral composition profiles along the dimension perpendicular to the domain boundaries (black dotted lines). The profiles obtained for the two bilayer leaflets are shown as dashed lines; solid lines represent the average. (a) POPC lipids (orange) accumulate close to the domain boundary. (b) PLiPC segregates into the Ld domain, whereas LysoPC (c) has a weak preference for the domain boundary interface.
Table 1.
Free energies and line tensions at the domain boundary interface (int)
| Fourth component (2 mol %) | ΔG(Lo/int) [kJ mol−1] | ΔG(Ld/int) [kJ mol−1] | σ [pN] (17) |
|---|---|---|---|
| POPC | −2.8 ± 0.5 | −6.0 ± 0.7 | 10.6 ± 1.2 |
| PLiPC | −4.9 ± 1.3 | −0.8 ± 0.5 | 14.8 ± 2.0 |
| LysoPC | −0.9 ± 0.5 | −2.9 ± 0.4 | 13.4 ± 2.6 |
Free energies ΔG was calculated according to ΔG = −RT ln (ρint/ρb), where ρb and ρint are the mass densities in the bulk and at the interface, respectively. Errors in ΔG were obtained from the difference between the two bilayer leaflets. Line tensions were σ = 14.4 ± 1.8 pN in the ternary mixture.
Among the three lipid species studied, only POPC clearly prefers the interface with respect to both the Lo and Ld domains. The free energy differences between the bulk of the Lo and Ld domains are rather small (2–4 kJ mol−1), in agreement with theoretical predictions (18). Although the POPC concentration of 2 mol % is too low to saturate the entire interface (see Fig. 1), it is sufficient to reduce the line tension, from σ = 14.4 ± 1.8 pN in the ternary mixture (19) to 10.6 ± 1.2 pN (see Table 1; for details, see the Supporting Material). By contrast, addition of PLiPC or LysoPC did not significantly reduce σ. The result that hybrid lipids can act as line-active compounds agrees with recent experiments on hydrocarbon/fluorocarbon monolayers (11) and theoretical findings based on a phenomenological thermodynamic model (20,21).
Furthermore, comparing the composition profiles of the two individual bilayer leaflets (dashed lines in Fig. 2) shows that the registration of the domains in the two leaflets is less pronounced in the POPC-containing bilayer as compared to, for example, the PLiPC-containing bilayer (domain registration mismatch ∼2 nm and 1 nm, respectively). The reduced domain registration with POPC is in line with increased fluctuations of the interfaces, which was hypothesized to lead to a stronger entropic repulsion between the opposing interfaces in the two leaflets (12).
The coarse-grain molecular dynamics simulations presented in this work yield detailed insights into the partitioning of different lipid species at a fluctuating domain boundary interface in a bilayer with coexisting fluid lipid domains, opening the way toward predicting linactant behavior through MD simulations. In this context, it is intriguing to also study line-active proteins and peptides. Together with their protein complements, we anticipate that linactant lipids might have a strong potential in regulating lateral heterogeneity, possibly also of biological membranes.
Acknowledgments
We thank H. J. Risselada for discussions and T. Baumgart for carefully reading the manuscript.
The Netherlands Organisation for Scientific Research supported this work through Top grant No. 700-57-303 (to S.J.M.), Veni grant No. 700-57-404 (to L.V.S.), and access to the National Supercomputing Facilities (NCF grant No. SH-148).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Contributor Information
Lars V. Schäfer, Email: l.schafer@rug.nl.
Siewert J. Marrink, Email: s.j.marrink@rug.nl.
Supporting Material
References and Footnotes
- 1.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Hancock J.F. Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahya N., Scherfeld D., Schwille P. Probing lipid mobility of raft-exhibiting model membranes by fluorescence correlation spectroscopy. J. Biol. Chem. 2003;278:28109–28115. doi: 10.1074/jbc.M302969200. [DOI] [PubMed] [Google Scholar]
- 4.Veatch S.L., Polozov I.V., Keller S.L. Liquid domains in vesicles investigated by NMR and fluorescence microscopy. Biophys. J. 2004;86:2910–2922. doi: 10.1016/S0006-3495(04)74342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgart T., Hess S.T., Webb W.W. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 6.Tian A., Johnson C., Baumgart T. Line tension at fluid membrane domain boundaries measured by micropipette aspiration. Phys. Rev. Lett. 2007;98:208102. doi: 10.1103/PhysRevLett.98.208102. [DOI] [PubMed] [Google Scholar]
- 7.Baumgart T., Hammond A.T., Webb W.W. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. USA. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaiser H.-J., Lingwood D., Simons K. Order of lipid phases in model and plasma membranes. Proc. Natl. Acad. Sci. USA. 2009;106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lingwood D., Ries J., Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc. Natl. Acad. Sci. USA. 2008;105:10005–10010. doi: 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edidin M. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 11.Trabelsi S., Zhang S., Schwartz D.K. Linactants: surfactant analogues in two dimensions. Phys. Rev. Lett. 2008;100:037802. doi: 10.1103/PhysRevLett.100.037802. [DOI] [PubMed] [Google Scholar]
- 12.Risselada H.J., Marrink S.J. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.In our MD simulations, the computational cost limits the lateral size of the simulation box to ∼22 × 22 nm. Thus, a strongly phase-separating system is needed, because otherwise large fluctuations would make the interface ill-defined on the time- and length-scale of the simulations.
- 14.Although the domain boundary interface is sharp on the molecular level (Fig. 1 b), its efficient width is ∼5 nm in our bilayer due to fluctuations.
- 15.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 16.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 17.Experiments report line tensions of approximately a few pN, depending on the composition and other parameters (5,6).
- 18.Uline M.J., Longo G.S., Szleifer I. Calculating partition coefficients of chain anchors in liquid-ordered and liquid-disordered phases. Biophys. J. 2010;98:1883–1892. doi: 10.1016/j.bpj.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.This line tension is higher than the 7 pN (3.5 pN per monolayer) reported in Risselada and Marrink (12). It was obtained by extending the previous simulation from Risselada and Marrink (12), in which the formation of the interface was not completely finished yet.
- 20.Brewster R., Pincus P.A., Safran S.A. Hybrid lipids as a biological surface-active component. Biophys. J. 2009;97:1087–1094. doi: 10.1016/j.bpj.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brewster R., Safran S.A. Line active hybrid lipids determine domain size in phase separation of saturated and unsaturated lipids. Biophys. J. 2010;98:L21–L23. doi: 10.1016/j.bpj.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


