Abstract
The hierarchical packaging of DNA into chromatin within a eukaryotic nucleus plays a pivotal role in both the accessibility of genomic information and the dynamics of replication. Our work addresses the role of nanoscale physical and geometric properties in determining the structure of chromatin at the mesoscale level. We study the packaging of DNA in chromatin fibers by optimization of regular helical morphologies, considering the elasticity of the linker DNA as well as steric packing of the nucleosomes and linkers. Our model predicts a broad range of preferred helix structures for a fixed linker length of DNA; changing the linker length alters the predicted ensemble. Specifically, we find that the twist registry of the nucleosomes, as set by the internucleosome repeat length, determines the preferred angle between the nucleosomes and the fiber axis. For moderate to long linker lengths, we find a number of energetically comparable configurations with different nucleosome-nucleosome interaction patterns, indicating a potential role for kinetic trapping in chromatin fiber formation. Our results highlight the key role played by DNA elasticity and local geometry in regulating the hierarchical packaging of the genome.
Introduction
In eukaryotic cells, a large amount of DNA must be packaged into a nucleus only a few microns in diameter. The organization of this packing plays a critical role in the function and regulation of cellular transcription, replication, and segregation machinery. The physical structure of chromatin is believed to regulate its dynamics and flexibility, which in turn dictate the accessibility of packaged DNA to regulatory factors. Understanding the structural properties of chromatin is thus fundamental to developing a quantitative picture of dynamic processes within the nucleus.
At the most basic level, chromatin is composed of nucleosomes—DNA-protein complexes containing 147 basepairs of DNA wrapped around a core of histone proteins (1). The nucleosomes and intervening linker DNA are further packaged into higher-order structures. The simplest higher-order structure observed in vitro is a fiber ∼30 nm in width (2,3), whose structure remains a source of much controversy. Electron microscopy (EM) studies (4) and force spectroscopy measurements (5) lend support to a one-start solenoidal model. However, the tetranucleosome crystal structure (6) indicates a crossed-linker two-start helical arrangement of the nucleosomes that is further supported by in vitro cross-linking studies (7,8) and in vivo analysis of chromatin fragmentation patterns (9). As the evidence for different fiber structures mounts, it is becoming increasingly clear that no single model is likely to encompass the structure of all 30-nm fibers.
Some of the contradictory results in the literature can be explained in part by the dependence of the structure on several physical and geometric factors (10). The feasibility of any structure is expected to depend on the geometry of the DNA attachment to the nucleosomes. Although this geometry can be extrapolated from the mononucleosome crystal structure (1), chromatin fibers in vivo have an additional linker histone that interacts with the DNA as it exits the nucleosome and helps stabilize compact configurations (11,12). Differences in fiber structure due to changes in entry/exit geometry may also arise in vivo through partial unwrapping of the nucleosomes, whether by acetylation of histone tails (13), the introduction of histone variants such as CENP-A (14) and H2A.BbD (15), or through the action of nucleosome remodeling complexes (16). Drastic alteration of nucleosome geometry might also occur if nucleosomes dissociate into hemisomes containing one copy of each histone protein as reported for Drosophila centromeric nucleosomes (17).
Furthermore, the length of the linker DNA separating the nucleosomes is likely to affect the stability of a compact fiber. Due to the inherent periodicity of the DNA double helix, the internucleosome separation length determines the twist registry of the nucleosomes. A repeat length (separation between nucleosome centers) that is an integer multiple of DNA pitch results in nucleosomes bound on the same side of the DNA, whereas one that is a half-integer multiple places the nucleosomes on opposite sides (18). Genomic studies of the preferred internucleosome separation lengths in several organisms indicate a corresponding 10–10.6 bp periodicity (19,20). The linker length also sets the total amount of DNA that must be sterically packed inside the fiber, an effect that is thought to be responsible for the discrete transition between small and large diameter fibers observed in EM studies (4,21).
Computational efforts at modeling the structure of compact chromatin fibers have focused primarily on the role of electrostatic internucleosomal interactions and the local geometry of the DNA. Sets of possible structures with straight linker DNA have been mapped out for a subset of nucleosomal geometries, emphasizing the importance of the twist angle between consecutive nucleosomes (22–24). Other studies have started with the assumption of maximally compact face-to-face packing of the nucleosomes and looked at the lengths of linker DNA that can sterically fit into such well-packed structures (21,25). Still other approaches involve large-scale Monte Carlo or Brownian dynamics simulations that model linker DNA elasticity and electrostatic interactions. Internucleosome interactions in such studies are treated with either simplified anisotropic potentials (22,26–30) or assemblies of charged patches derived from the crystal structure (8,31,32).
In this work, we choose to adopt a model that uses a minimal number of parameters (nucleosome geometry, DNA elastic moduli), all of which can be extracted from experimental data. We focus on the roles of linker elasticity, DNA geometry, and steric packing in determining compact structures that are energetically accessible for a given nucleosome array. By modeling the fiber in this way, we are able to estimate the nucleosome interaction energies that would be required to stabilize a fiber structure, without restriction to a given detailed potential. Our results thus underscore the importance of fundamental physical and geometric properties in limiting the ensemble of possible structures. Finally, we provide publicly available code for finding regular helical fiber structures with well-packed elastic linker DNA.
Methods
Regular helix coordinates
In this work, we focus on periodic chromatin fiber structures, which are defined by two consecutive nucleosomes and the path of the linker between them, repeated to create a helical array. We treat the nucleosomes as rigid bodies that fix the relative position and orientation of the DNA chain coming off either end. The geometry of the DNA end attachment is determined from the mononucleosome crystal structure (1) and is illustrated in Fig. 1 a. The geometric transformation from one nucleosome to the next can be expressed using screw coordinates (33) as a rotation around and translation along a single axis. This axis serves as the center of a helix formed by the nucleosome centers after repeated application of the same transformation to generate a long fiber. The position and orientation of the nucleosomes in the fiber can be fully defined by specifying six geometric parameters describing this helix. We choose as our helical coordinates (Fig. 1 b):
-
1.
Height (h) per nucleosome along the fiber axis.
-
2.
Angle (θ) per nucleosome around the fiber axis.
-
3.
Radius (R) of the helix.
-
4–6.
Euler angles (α, β, γ) defining the orientation of the first nucleosome relative to the fiber coordinate system.
Specifically, β is the angle between the symmetry axis of the nucleosomes (defined in Fig. 1 a) and the fiber axis. Note that h is inversely related to the linear compaction of the chromatin fiber (often reported as n nucleosomes per 11 nm) according to n = 11/h, and is measured between consecutive nucleosomes to bypass any dependence on the number of stacks in the structure. This choice of helical coordinates enables us to easily implement constraints on fiber geometry.
Figure 1.
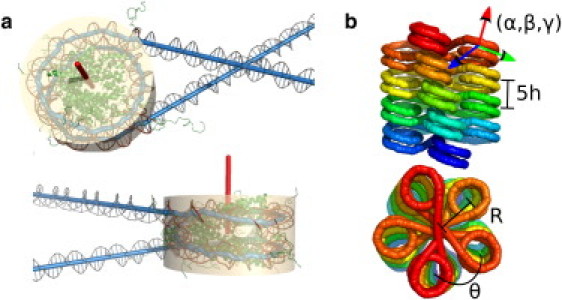
(a) Nucleosome geometry as obtained from crystallographic structure. (Beige cylinder) Shape of the nucleosome used for steric exclusion. (Red) Symmetry axis for the nucleosome. (b) The six coordinates used to specify a regular helix fiber structure. Because this is a five-stack structure, the height between each nucleosome and the one above it corresponds to 5h.
For a regular fiber structure, only the path of the first linker DNA must be defined. We approximate the linker DNA as a discrete elastic thread of N straight segments, defined by the positions of N–1 beads (), and bounded by the DNA attachment points on the nucleosomes. Because we are interested in the energetic effects of twisting as well as bending the linkers, each segment is assigned an associated coordinate system whose z axis is aligned along the segment. To represent the orientation of each segment we include another coordinate, ci = αi + γi, where (αi, βi, γi) are Euler angles associated with the ith segment coordinate system. In all calculations detailed below, we use N = 10 segments per linker. The configuration of the regular chromatin fiber is fully represented by the set of coordinates
Energetics
For each regular fiber configuration defined via the coordinate system described above, we assign a potential energy that includes the elasticity of the linker DNA and steric exclusion among nucleosomes and DNA segments. All energies are reported in units of kT per linker.
We treat the linker DNA as a discretized wormlike chain, with harmonic potentials for bending, twisting, and stretching deformations. Calculation of the energy requires finding the length of each segment
and its relative orientation with respect to the previous segment
Note that we define as the position and orientation of the DNA exiting the first nucleosome and as the position and orientation of DNA entering the second nucleosome.
The stretching energy is given by
| (1) |
where is the ground-state length of each segment, L is the linker length, and κs = 268 kT/nm is the stretch modulus for double-stranded DNA (34). If we express the relative rotation from one segment to the next using Euler angles the bend and twist elastic energies (in units of kT) are defined as
| (2) |
| (3) |
where lp = 50 nm and lt = 110 nm are the bend and twist persistence lengths of DNA, respectively, and τ = 2π/(10.46 bp) is the natural twist density (35,36).
To prevent overlap between nucleosomes and linker DNA segments, we introduce harmonic steric exclusion terms. The nucleosomes are treated as cylinders of radius 5.2 nm and height 5.5 nm (Fig. 1 a). The linker DNA segments are also treated as cylinders of length and radius 1 nm. We cut off interactions with nucleosomes and linkers more than Nr = 20 repeats away. We do not consider the unconstrained DNA tails on either end of the fiber. The steric interactions are quadratic, with the modulus κsteric = 103 kT taken sufficiently large that essentially no steric overlap occurs in all optimized structures. Details of the cylindrical sterics calculations are described in Appendix A in the Supporting Material.
Optimization and search
We employ the Broyden-Fletcher-Goldfarb-Shanno quasi-Newton optimization method (37) to find local minima in structure space using the energy function described above. Where indicated, some helix coordinates are held fixed whereas the optimization is carried out in the remaining free coordinates only. A high-dimensional energy landscape is expected to have a large number of local minima basins. We therefore use a global search technique called basin hopping to sample these different minima (38). The method consists of taking large Monte Carlo steps in configuration space, followed by local optimization, where the resulting locally minimized energy is used to determine whether the step is accepted. Details of our basin-hopping implementation are provided in Appendix B in the Supporting Material.
Results
Straight-linker structures from crystal geometry
In this work, we focus on regular fiber structures, where the geometric transformation between each pair of consecutive nucleosomes is constant throughout the fiber. The transformation between each nucleosome and the next is defined by the local geometry of the linker DNA coming on and off the nucleosome as well as the path taken by the linker. Due to the twist of the DNA chain, the linker length affects not only the relative position but also the orientation of consecutive nucleosomes. We begin by considering ground-state structures with straight linker DNA. The resulting families of superhelical fiber structures have previously been categorized for some sample nucleosome geometries (22–24). We note that, unlike Ben-Haïm et al. (23) and Schiessel et al. (24), we obtain the nucleosome entry and exit geometry directly from the mononucleosome crystal structure (1). The manipulations for obtaining the superhelix parameters (Fig. 1 b) from the nucleosome geometry and linker length are described in Appendix C in the Supporting Material.
Several superhelix parameters are plotted as a function of repeat length in Fig. 2. The twist registry of the nucleosomes along the DNA plays an important role in determining the straight-linker superhelical structure. In particular, the nucleosomes prefer to be oriented with their symmetry axes parallel to the fiber axis (β = 0) for repeat lengths that are integer multiples of the DNA pitch, and with perpendicular axes for half-integer multiples of the pitch. Note that bound DNA on the nucleosome is slightly overwound (1), so the repeat lengths yielding values of (β = 0) are in fact ∼1-bp shorter than integer multiples of the naked DNA pitch of 10.46 bp. Extended structures are formed for repeat lengths close to an integer multiple of pitch, with much shorter fibers near half-integer multiples. As has been noted previously (23), some linker lengths lead to a collapse of the superhelix into a planar structure. Such structures are unphysical, because they would involve overlap of the nucleosomes themselves. In Fig. 2, shaded regions indicate straight-linker structures with steric clashes. For the shaded repeat lengths, the ground-state structure requires deformation of the linker DNA to reposition the nucleosomes so as to avoid steric overlap.
Figure 2.
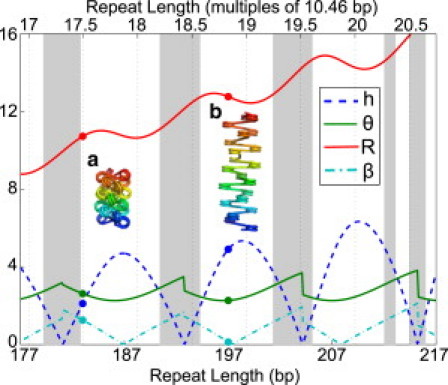
Superhelical parameters for straight-linker structures with different linker lengths. (Blue dashed line) Height per nucleosome (h) in nm. (Green solid line) Angle per nucleosome (θ) in radians. (Red solid line) Radius (R) in nm. (Cyan dash-dotted line) Angle of nucleosome relative to fiber axis (β) in radians. (Gray regions) Sterically excluded. Example structures are shown for (a) 36-bp and (b) 50-bp linker lengths.
Profiling regular helix structures
To fully characterize the effect of DNA elasticity on the set of possible compact structures, we generate a profile of optimized regular structures over a grid of helix height and angle values. For each pair (h,θ) of height and angle per nucleosome, the energy minimized over all other helix coordinates is plotted in Fig. 3. Values of θ close to 0 and 2π correspond to solenoidal structures while intermediate values yield crossed-linker structures.
Figure 3.
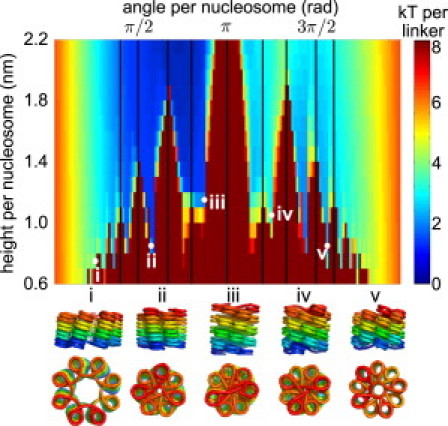
Profile of minimal energies for fiber structures with 50-bp linkers, as a function of height and angle per nucleosome. (Vertical lines) Angles that are small rational multiples of 2π and thus lead to steric clashes between nucleosomes. (Bottom) Example structures.
The profile can be thought of as an underlying smooth surface stemming from the elastic energy of linker deformation, overlaid with a series of excluded regions that arise from steric clashes between nucleosomes and/or linker DNA. The shape of the elastic energy surface depends upon both the twist registry of the nucleosomes, which sets the preferred helix coordinates, and the total linker length, which affects the flexibility of the fiber by setting the depth of the low-energy basins. Similar profiles for other linker lengths are shown in Fig. S2 in the Supporting Material.
It is interesting to note that in the illustrated profile there are elastically preferred local minima at low values of h where the linkers are crossed over so as to resist stretching of the fiber (e.g., Fig. 3 v). These structures are obtained by flipping the nucleosomes relative to the fiber axis, in a manner analogous to hockling, or formation of tension-resisting loops, in twisted cables (39). Nucleosome arrays folded into such structures would be particularly stable, as the elastic resistance of the linker DNA combines with any favorable internucleosome interactions to prevent the fiber from unraveling.
Steric clashes between nucleosomes and linker DNA completely exclude some combinations of height and angle values, leading to jagged ridges overlaid on the elastic energy landscape. In particular, when θ = 2π p/q for integer p and q, then every qth nucleosome lines up directly on top of the first, leading to steric clashes when qh is smaller than the size of the nucleosome (black lines in Fig. 3). In addition, the introduction of sterics raises the energy in other regions of the profile, as the nucleosomes are forced to reorient to avoid steric clashes, thereby increasing the elastic energy of the linker DNA.
Our focus on regular helical structures allows us to explore the full energy landscape of possible conformations, similar to another recent phase-diagram study (40). However, our explicit treatment of linker elastic deformation allows us to more accurately track the associated energetic cost of forming specific configurations. In addition, our choice of regular helix coordinates helps us to elucidate the geometric features of permitted structural regions. With 50-bp linkers, for instance, Fig. 3 shows a number of discrete rifts in the energy landscape that correspond to energetically feasible compact fibers. These valleys correspond to different ranges of θ and thus different numbers of helix starts in the superhelical structure.
Catalogue of potential structures
The energy profile illustrated in Fig. 3 indicates that, for a given linker length, compact fiber structures can fall into a number of local basins such that the fiber must partially unfold to interconvert between them. We employ a basin-hopping approach to pick out distinct low-energy compact structures. We begin by finding all local minima at each linker length that are stable under elastic and steric energies alone and that have a nucleosome line density within the experimentally measured range for compact fibers (>5 nucleosomes per 11 nm). Of particular interest are those stable structures where the elastic energy of deforming the linker DNA is sufficiently low that it can be compensated by favorable electrostatic interactions between the nucleosomes. Single-molecule force-extension experiments measured the magnitude of such internucleosomal interactions at ∼3.4 kT (41). A more recent study indicated that in the presence of Mg2+, the internucleosome stacking energy may be as high as 13 kT (5). Due to space constraints, we consider only those candidate structures that fall below the lower energy cutoff. Low-energy stable structures for 45- and 50-bp linkers are illustrated in Fig. S3. We find these linker lengths allow for a variety of locally optimized structures, ranging from two to nine nucleosome stacks in the fiber.
These locally stable fibers do not provide a comprehensive set of potential candidate structures. In particular, there are low-energy regions of structure space that could be stabilized by internucleosomal interactions but that unfold to more extended local minima under the influence of DNA elasticity and sterics alone. To generate a wider array of potential structure candidates, we search for fiber geometries that are local minima in all coordinates other than the height per nucleosome, while the height coordinate is fixed to discrete values within the experimentally measured range (5–16 nucleosomes per 11 nm). We then filter out structures that differ in the height coordinate but have very similar arrangements of the nucleosomes, as described in Appendix B in the Supporting Material.
The resulting sets of potential structures for two different linker lengths are shown in Fig. 4 a. These results emphasize the fact that linker DNA energetics and steric interactions allow for a wide variety of potential low-energy structures for a compact chromatin fiber.
Figure 4.
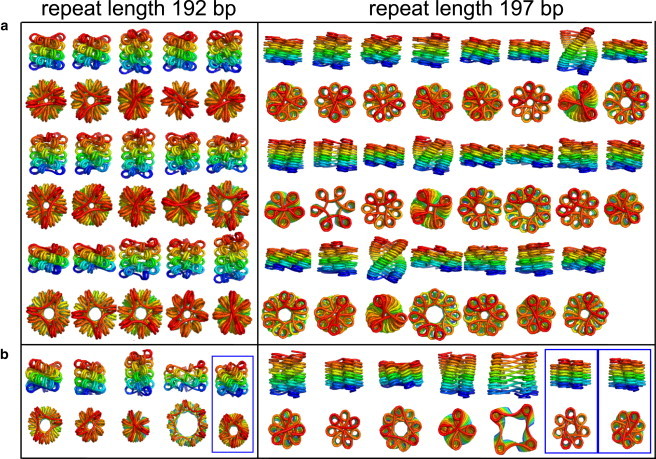
(a) Side and top views of all candidate structures for compact fibers with crystallographic nucleosome geometry, 192-bp and 197-bp repeat lengths (45-bp and 50-bp linkers, respectively). The structures shown are local minima in all coordinates other than the height per nucleosome. Only structures with height per nucleosome below 2.2 nm and energy below 3.4 kT are shown. Parameters are listed in Table S1 in the Supporting Material. (b) Example candidate structures for nucleosomes with 10 bp unwrapped at each end. Boxed structures have topologically entwined linkers.
Alterations in nucleosome geometry
Heretofore, our analysis of possible regular fiber structures has rested on the assumption that the geometry of the DNA coming on and off the nucleosome is rigidly fixed to that observed in the crystal structure. However, it has previously been shown that edges of the DNA chain can be unpeeled from the histone core with an energy penalty of ∼0.6 kT/bp (42). Certain modifications to the histones themselves, such as tail acetylation, are also believed to facilitate opening of the DNA wrapped around the nucleosome (13). Furthermore, recent experimental evidence showed that the replacement of histone H3 with the CENP-A analog in centromeric chromatin stabilizes a partially open nucleosome structure with ∼21 bp of DNA unwrapped (14).
To study possible structural effects of partially opened nucleosomes, we repeat our search for compact fiber structures with an altered nucleosome geometry obtained by unwrapping 10 bp at each end of the crystal structure. A few of the resulting structures are shown in Fig. 4 b, with the complete set of structures given in Fig. S4. Note that the relevant comparison is between wrapped and partially open structures with the same repeat length, because this parameter sets the relative twist registry of the nucleosomes on DNA.
We find that the altered nucleosome geometry produces many similar low-energy structures to those shown in Fig. 4 a, as well as several new structures that are stabilized by the partial unwrapping of the nucleosomes. The increased length of the flexible linkers due to nucleosome unwrapping allows for a number of more open structures of larger radius. The increased flexibility of longer linkers also makes structures with interwoven linkers energetically feasible. We thus observe several topologically knotted local minima with the open nucleosomes that were not present in Fig. 4 a. Although these structures would be inaccessible in vitro, they could conceivably appear in vivo through the action of type II topoisomerases, which are capable of passing segments of double-stranded DNA through one another.
Nucleosome angle determined by repeat length
The elasticity of the linker DNA allows for dozens of distinct candidate structures of a compact chromatin fiber. However, we find that one characteristic shared by all potential structures at a given linker length is the angle between the nucleosomes and the fiber axis. Furthermore, this characteristic is unaltered by unwrapping 20 bp of DNA to form an open nucleosome structure. Although the dependence of the nucleosome angle on the twist registry has previously been noted for straight-linker structures (23), our results demonstrate that this property is robust to deformation of the linkers through fiber compaction or nucleosome opening.
Fig. 5 a shows the angle (β) between the nucleosome symmetry axis and the fiber axis for all candidate structures discussed in the previous two sections. We find that nucleosomes separated by integer multiples of the DNA pitch prefer to pack with their symmetry axis aligned along the fiber axis (β ≈ 0, π), whereas nucleosomes separated by half-integer multiples of the pitch prefer to align perpendicular to the fiber axis (β ≈ π /2). A nontrivial cost in energy per nucleosome must be paid to rotate the nucleosomes away from their preferred angle to the axis. Shown in Fig. 5 b is a profile of the minimal energy per nucleosome necessary to rotate the nucleosomes in the regular fiber to different values of β for each linker length. This profile uses the crystal nucleosomal geometry, and the energy shown is the smallest among all local minima obtained by basin-hopping calculations with a fixed value of β. Our calculations show that over 10 kT elastic energy per nucleosome must be overcome to rotate the nucleosomes for short linker lengths, with lower energy penalties for longer linkers. Again, this result illustrates the greater flexibility with respect to permitted fiber structures that is inherent in longer separations between the nucleosomes.
Figure 5.
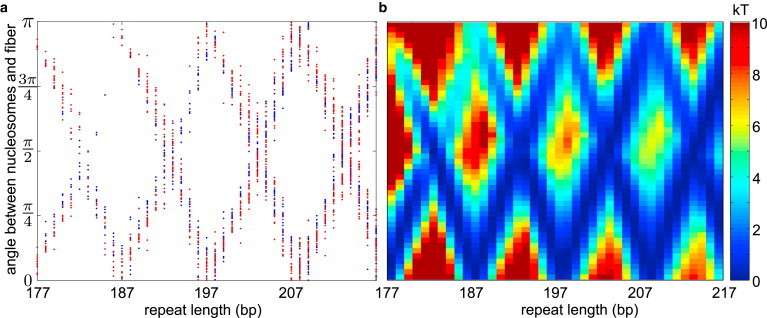
(a) Angle (β) between nucleosomes and fiber axis for all candidate structures at all repeat lengths. (Blue) Crystallographic geometry. (Red) Ten basepairs unwrapped at each edge. (b) Profile of minimal energies to form a structure with the given β-angle for different repeat lengths.
Structures with experimental constraints
The methods described in this article can be used to construct candidate fiber structures that match experimentally measured values of geometric parameters. For instance, the basin-hopping search can be carried out while holding the height per nucleosome and fiber diameter explicitly fixed to any given value. We treat the measured diameter as the distance from the fiber axis to the outermost point on the nucleosome cylinders (see Appendix A in the Supporting Material). For a fixed diameter, the radius (R) of the nucleosome centers can be set as a function of the remaining helical coordinates.
We use this approach to find locally optimized structures for fibers whose diameter and nucleosome line density have been measured via electron microscopy (4). Although several structures have previously been proposed to explain this data (4,25), our calculations have the advantage of explicitly considering the elastic and steric constraints on the packing of linker DNA inside the fiber. For each linker length between 30 and 80 bp, we run a basin-hopping calculation with the height and diameter fixed to the values reported by Robinson et al. (4). To avoid exclusion of densely packed structures, we use a steric height of 4.5 nm for the nucleosomes in these calculations. In Fig. 6, we show the three best local minima for each linker length, together with the elastic energy per linker.
Figure 6.
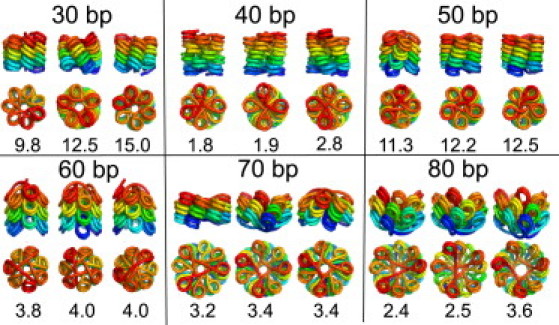
Top three structures for fibers of different linker lengths (30–90 bp) with height per nucleosome and fiber radius constrained to values measured by electron microscopy (4). The energy for each locally optimized structure is given in kT per linker.
For the shorter linker lengths, the lowest energy structures have the nucleosomes lined up with their symmetry axes oriented along the fiber axis, in contrast to previously constructed models (4). This is in keeping with the orientational preference discussed in the previous section. The fibers with 30-bp linkers assume solenoidal structures, because the linkers are not sufficiently long to stretch across a fiber of the experimentally measured diameter. This requires bending of the linker DNA, as manifested in the relatively high energies for these fibers. It must be noted that the experiments were carried out in the presence of linker histones, which are not currently included in our model. The favorable binding energy of the linker histones with the DNA could help compensate for the relatively high elastic energy required to form such fibers.
We find that fibers with longer linkers prefer to form crossed-linker structures when constrained to the experimentally measured geometries. The fact that fiber diameters do not scale linearly with linker length, but instead remain approximately constant in two structural classes, was previously cited as evidence for a solenoidal arrangement of nucleosomes (4). However, we show that the extra length of the linkers can be accommodated in crossed-linker structures without significantly changing the fiber diameter by tilting the nucleosomes, as shown in Fig. 6. The bending of the linker DNA in such fibers is a direct consequence of constraining the total fiber diameter to experimentally measured values. Although it is not clear whether there are internucleosomal forces that would be likely to stabilize such bent fibers in vitro, it is instructive to note that these structures nonetheless require less elastic deformation of the long linker DNA than the corresponding solenoids while still satisfying experimental constraints.
Discussion
Our results show that linker DNA length and local nucleosome geometry limit some key aspects of compact fiber structure, while allowing a variety of configurations to remain energetically feasible for any given fiber. In particular, we find that the twist registry of consecutive nucleosomes along the DNA has a notable effect on the preferred geometries of potential compact structures. The importance of twist registry for chromatin fiber compaction has previously been proposed as an explanation for the observed 10-bp quantization of linker lengths in vivo (19).
A recent study of nucleosome positioning on yeast DNA in vivo indicated that this quantization is not primarily dictated by DNA sequence preferences encoded in the genome (43,44). Alteration of the linker DNA twist by ethidium intercalation is known to cause dinucleosomes to transition from a compact to an unfolded state (45), providing direct evidence of the connection between twist registry and structure. Our results lend further support to previously proposed hypotheses that linker length quantization arises from structural considerations (20,22,25). If there is a preference for a particular family of chromatin structures, due either to their packing properties or the accessibility of the compacted DNA, the linker lengths separating the nucleosomes would have to be compatible with the necessary twist registry for such structures. The required quantization could then be maintained either by internucleosome interaction during the process of compaction or through active positioning by nucleosome remodeling factors.
Previous computational studies have pointed out that variation of the linker length has a strong effect on the stability of periodic fiber structures, both through the twist registry of consecutive nucleosomes and the total amount of DNA that must sterically fit within the compact fiber (22,25,46). Our simulations explore the full space of regular helix structures to more explicitly map out the structural preferences for fibers with different internucleosome separations. In particular, we find that the angle of the nucleosomes relative to the fiber axis is robustly determined by the twist registry of consecutive nucleosomes along the DNA, as set by the repeat length between nucleosome centers. Fibers where the nucleosomes are bound symmetrically on the same side of the DNA strand exhibit a strong energetic preference for configurations where the nucleosome symmetry axis is lined up along the fiber axis, whereas those with nucleosomes bound antisymmetrically prefer structures with the nucleosome axis oriented perpendicular to the fiber.
It is interesting to note that these two classes of fibers are expected to respond differently to perturbations of the fiber structure. Specifically, for fibers with nucleosome axes perpendicular to the helix, twisting portions of the fiber would require breaking face-to-face histone contacts, whereas linear extension of the fiber would only involve deforming the linker DNA. For fibers where the nucleosome axes are lined up with the helix axis, the response would be reversed, with linear stretching requiring separation of histone faces whereas twist is resisted by DNA elasticity alone. Because such opening of the compact fiber structures is required to access the buried linker DNA, our findings imply that linker length may set the preferred deformation mode for transcription and regulatory access to compacted DNA in vivo.
Quantification of this effect computationally would require introduction of a detailed model for internucleosomal interactions. However, it is possible that the two different types of structures could be distinguished experimentally in vitro by measuring the response of fiber elastic moduli to changing salt concentrations. Specifically, changes in the ionic strength should alter the relative importance of electrostatic interactions between nucleosome faces versus elastic resistance of linker DNA. We would thus expect the stretch modulus of fibers with repeat lengths that are integer multiples of DNA pitch to be more sensitive than the twist modulus to salt concentrations, whereas the opposite relationship would hold for fibers with nucleosomes separated by half-integer multiples of the pitch.
Most in vitro experimental studies of well-defined nucleosome arrays folded into compact fibers have focused on repeat lengths that are approximately integer multiples of the DNA pitch (4–8). The models constructed to explain the results of these studies have assumed perpendicular alignment of the nucleosomes to the fiber axis (4,7), in contrast to our predictions that fibers with these linker lengths prefer structures where the nucleosome axes point approximately along the helix. Models that assume compact face-to-face packing of the nucleosomes (21,25), on the other hand, have a much smaller tilt of the nucleosomes relative to the fiber. An all-atom model constructed by fitting linker DNA into several such compactly packed structures (25) showed that the linker lengths used are compatible with the fiber dimensions observed by EM measurements (4). Our models of specific fiber structures that fit this experimental data show similar low tilt angles of the nucleosomes for short linker lengths, but different packing arrangements and different sets of internucleosome contacts.
We emphasize that the primary thrust of our calculations is an extensive search for as many low-energy structures as are compatible with experimentally measured criteria. We intentionally keep our model as simple as possible, including elastic energies and cylindrical steric exclusion only, while avoiding the introduction of variables for which there is as yet no quantitative experimental consensus. In contrast to, for example, Grigoryev et al. (8) and Wong et al. (25), we do not include linker histones, modification of DNA elastic parameters, internucleosomal electrostatic interactions, or direct enforcement of compact space-filling nucleosome packing. Our work thus helps elucidate which aspects of chromatin fiber structure are determined by local geometric parameters alone, and which will vary when additional details are introduced.
For instance, the robustness of the β-angle preference to DNA unwrapping implies that merely sequestering more of the DNA by linker histone binding is unlikely to alter the preferred angle between the nucleosome and fiber axis. For short linkers of 30 or 40 bp, stabilizing previously proposed models with nucleosomes perpendicular to the axis would require the linker histone to directly impart a twist (or writhe) to the DNA configuration.
In contrast, we find that the internucleosome stacking pattern is not robustly determined by DNA elasticity and sterics alone. Even for reasonably short linkers there are a number of configurations with comparable elastic energies. Our catalog of low-energy compact structures expands upon the previously noted polymorphism of chromatin fibers (21,25) by providing a wide array of potential structures that are sterically feasible yet do not require energetically expensive deformation of the linker DNA. The preference for specific configurations is dependent on the exact local geometry of the nucleosome, so that unwrapping even a few of the bound basepairs in the crystal structure can change the energetic preference for the number of nucleosome stacks in the fiber.
Furthermore, minimization of our structural candidates with the inclusion of previously published internucleosome potentials again yielded structures with multiple different stacking patterns but comparable energies. Specifically, we minimized the candidate structures shown in Fig. 4 while applying both an anisotropic Lennard-Jones potential (30) and a much more detailed potential with flexible histone tails and 300 charges representing core nucleosome electrostatics (47) (results not shown). We found that both types of internucleosomal potentials were insufficient to establish a strong energetic preference for specific fiber packings. In fact, because very similar face-to-face contacts can occur in fibers with different numbers of nucleosome stacks, it seems likely that any interaction potential that depends on the nucleosome geometry alone would be insufficient to provide a large energetic distinction between these different structures.
Our findings imply that a specific fiber structure is unlikely to be thermodynamically determined. However, steric exclusion of nucleosomes in a densely packed fiber means that interconversion between different packing patterns requires large-scale unraveling of the fiber structure. The structures observed in experimental studies may thus be manifestations of the resultant kinetic traps. If kinetic trapping does play an integral role in compact fiber formation, the precise chromatin structure formed would then be quite sensitive to the details of the reconstitution procedure, possibly explaining some of the contradictory results in the literature concerning the structure of the 30-nm fiber (10).
A primary goal of our study was to elucidate the role of DNA elasticity and local nucleosome geometry in stabilizing chromatin fiber structure. We found three classes of compact structures where the linker DNA played an important role in maintaining fiber stability against fluctuations.
First, for nucleosomes separated by half-integer multiples of the DNA pitch, the preferred straight-linker conformations have a high nucleosome linear density. In such cases, the height of the fiber is set by the compact steric-limited packing of the nucleosomes, and the linker DNA elasticity resists unraveling.
Although most in vitro experiments have focused on integer twist repeat lengths, recent genomic evidence suggests a preference for repeat lengths in yeast (19), which would lead to an elastic driving force for fiber compaction.
Second, we find that even for repeat lengths where the straight-linker helix is highly extended, locally stable compact structures can be formed with a fairly low elastic energy cost per linker. These structures have the nucleosomes flipped upside down relative to the fiber axis, so that extension of the fiber would require deformation or crossing of the linkers. Such structures could be made even more favorable by interactions of the linker histone or core histone tails with the DNA at the entry/exit site in such a way as to promote bending of the entering or exiting DNA toward the opposite face of the nucleosome.
Third, there are a number of possible fiber structures where the DNA linkers are topologically entangled. Although these structures are not relevant for fibers reconstituted in vitro, they are of potential interest when considering in vivo chromatin packing. In the nucleus of eukaryotic cells, topoisomerase proteins capable of passing double-stranded DNA helices through one another play an important role in maintaining genome organization, with topoisomerase II involved in distentangling chromatin strands during DNA replication (48). It is thus possible that short stretches of the interwoven chromatin fibers could be formed in vivo, thereby helping to stabilize higher order chromatin structures through topological entanglement.
It must be acknowledged that there is currently no experimental evidence of an in vivo role for many of the structures presented in this article, including topologically knotted structures in Fig. 4 b and bent structures in Fig. 6. However, our calculations indicate that such structures can be achieved with a lower elastic energy than many commonly proposed structures (e.g., solenoids). These results imply that such packing arrangements could at least serve as transient, localized intermediates in the formation of chromatin fibers. Our primary conclusion thus remains the existence of a diverse set of low-energy configurations available to fibers undergoing compaction.
Summary
By exploring the landscape of compact helical chromatin fibers, we show that linker length and nucleosome geometry play an important role in setting preferences for the formation of different classes of fiber structures. Specifically, we show that the tilt of the nucleosomes relative to the fiber axis is robustly set by the repeat length modulo the DNA pitch. However, the many energetically comparable structures found for any given linker length underscore the fact that geometry and elasticity alone are insufficient to fix any fiber configuration as the single preferred structure.
Furthermore, multiple regular structures exhibit similar face-to-face interactions between nucleosomes arranged in different numbers of helical stacks, thereby implying that internucleosomal interactions also are unlikely to robustly distinguish between the different structures. Our findings thus lend support to the idea that there is no one single preferred structure for the 30-nm fiber and that kinetic trapping modulated by details of the folding process may be responsible for the actual structures seen in vitro and in vivo.
To aid others studying the role of DNA elasticity in chromatin fiber models, we provide publicly available code for searching through regular helical fiber structures, optimizing linker DNA packing, and calculating the elastic energies associated with different fiber arrangements. Code and documentation is available at http://www.stanford.edu/∼ajspakow/.
Acknowledgments
This research was carried out in part with TeraGrid resources provided by the Purdue Condor pool.
We acknowledge funding from the National Science Foundation Graduate Student Fellowship, the Hertz Foundation, and the National Science Foundation CAREER award.
Supporting Material
References
- 1.Richmond T.J., Davey C.A. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 2.Huynh V.A., Robinson P.J., Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fiber containing stoichiometric amounts of the linker histone. J. Mol. Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 3.Langmore J.P., Schutt C. The higher order structure of chicken erythrocyte chromosomes in vivo. Nature. 1980;288:620–622. doi: 10.1038/288620a0. [DOI] [PubMed] [Google Scholar]
- 4.Robinson P.J.J., Fairall L., Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl. Acad. Sci. USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruithof M., Chien F.T., van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 6.Schalch T., Duda S., Richmond T.J. X-ray structure of a tetranucleosome and its implications for the chromatin fiber. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 7.Dorigo B., Schalch T., Richmond T.J. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 8.Grigoryev S.A., Arya G., Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rydberg B., Holley W.R., Chatterjee A. Chromatin conformation in living cells: support for a zig-zag model of the 30 nm chromatin fiber. J. Mol. Biol. 1998;284:71–84. doi: 10.1006/jmbi.1998.2150. [DOI] [PubMed] [Google Scholar]
- 10.van Holde K., Zlatanova J. Seminars in Cell & Developmental Biology. Vol. 18. Elsevier; Dordrecht, The Netherlands: 2007. Chromatin fiber structure: where is the problem now? pp. 651–658. [DOI] [PubMed] [Google Scholar]
- 11.Bednar J., Horowitz R.A., Woodcock C.L. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson R.T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 13.Manohar M., Mooney A.M., Ottesen J.J. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J. Biol. Chem. 2009;284:23312–23321. doi: 10.1074/jbc.M109.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conde e Silva N., Black B.E., Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 15.Doyen C.M., Montel F., Dimitrov S. Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome. EMBO J. 2006;25:4234–4244. doi: 10.1038/sj.emboj.7601310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kassabov S.R., Zhang B., Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 17.Dalal Y., Wang H., Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koslover E.F., Spakowitz A.J. Twist- and tension-mediated elastic coupling between DNA-binding proteins. Phys. Rev. Lett. 2009;102:178102. doi: 10.1103/PhysRevLett.102.178102. [DOI] [PubMed] [Google Scholar]
- 19.Wang J.P., Fondufe-Mittendorf Y., Widom J. Preferentially quantized linker DNA lengths in Saccharomyces cerevisiae. PLOS Comput. Biol. 2008;4:e1000175. doi: 10.1371/journal.pcbi.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc. Natl. Acad. Sci. USA. 1992;89:1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Depken M., Schiessel H. Nucleosome shape dictates chromatin fiber structure. Biophys. J. 2009;96:777–784. doi: 10.1016/j.bpj.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aumann F., Suhnel J., Diekmann S. Rigid assembly and Monte Carlo models of stable and unstable chromatin structures: the effect of nucleosomal spacing. Theor. Chim. Acta Theory Comp. Modeling. 2010;125:217–231. [Google Scholar]
- 23.Ben-Haïm E., Lesne A., Victor J.M. Chromatin: a tunable spring at work inside chromosomes. Phys. Rev. E. 2001;64:051921. doi: 10.1103/PhysRevE.64.051921. [DOI] [PubMed] [Google Scholar]
- 24.Schiessel H., Gelbart W.M., Bruinsma R. DNA folding: structural and mechanical properties of the two-angle model for chromatin. Biophys. J. 2001;80:1940–1956. doi: 10.1016/S0006-3495(01)76164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong H., Victor J.M., Mozziconacci J. An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length. PLoS ONE. 2007;2:e877. doi: 10.1371/journal.pone.0000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cinacchi G., La Penna G., Perico A. Anisotropic internucleosome interactions and geometrical constraints in the organization of chromatin. Macromolecules. 2007;40:9603–9613. [Google Scholar]
- 27.Kepper N., Foethke D., Rippe K. Nucleosome geometry and internucleosomal interactions control the chromatin fiber conformation. Biophys. J. 2008;95:3692–3705. doi: 10.1529/biophysj.107.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langowski J. Polymer chain models of DNA and chromatin. Eur .Phys. J. E Soft Matter. 2006;19:241–249. doi: 10.1140/epje/i2005-10067-9. [DOI] [PubMed] [Google Scholar]
- 29.Stehr R., Kepper N., Wedemann G. The effect of internucleosomal interaction on folding of the chromatin fiber. Biophys. J. 2008;95:3677–3691. doi: 10.1529/biophysj.107.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wedemann G., Langowski J. Computer simulation of the 30-nanometer chromatin fiber. Biophys. J. 2002;82:2847–2859. doi: 10.1016/S0006-3495(02)75627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arya G., Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun J., Zhang Q., Schlick T. Electrostatic mechanism of nucleosomal array folding revealed by computer simulation. Proc. Natl. Acad. Sci. USA. 2005;102:8180–8185. doi: 10.1073/pnas.0408867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray R., Li Z., Sastry S. CRC Press; Boca Raton, FL: 1994. A Mathematical Introduction to Robotic Manipulation. [Google Scholar]
- 34.Smith S.B., Cui Y., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 35.Bryant Z., Stone M.D., Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 36.Smith S.B., Finzi L., Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 37.Nocedal J., Wright S. Springer; New York: 1999. Numerical Optimization. Springer Series in Operations Research. [Google Scholar]
- 38.Wales D.J. Cambridge University Press; Cambridge, UK: 2003. Energy Landscapes. [Google Scholar]
- 39.Coyne J. Analysis of the formation and elimination of loops in twisted cable. IEEE J. Oceanic Eng. 1990;15:72–83. [Google Scholar]
- 40.Stehr R., Schöpflin R., Wedemann G. Exploring the conformational space of chromatin fibers and their stability by numerical dynamic phase diagrams. Biophys. J. 2010;98:1028–1037. doi: 10.1016/j.bpj.2009.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui Y., Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl. Acad. Sci. USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mihardja S., Spakowitz A.J., Bustamante C. Effect of force on mononucleosomal dynamics. Proc. Natl. Acad. Sci. USA. 2006;103:15871–15876. doi: 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein A., Takasuka T., Collings C. Are nucleosome positions in vivo primarily determined by histone-DNA sequence preferences? Nucl. Acids Res. 2009;38:709–719. doi: 10.1093/nar/gkp1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Moqtaderi Z., Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao J., Lowary P.T., Widom J. Twist constraints on linker DNA in the 30-nm chromatin fiber: implications for nucleosome phasing. Proc. Natl. Acad. Sci. USA. 1993;90:9364–9368. doi: 10.1073/pnas.90.20.9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staynov D.Z., Proykova Y.G. Topological constraints on the possible structures of the 30 nm chromatin fiber. Chromosoma. 2008;117:67–76. doi: 10.1007/s00412-007-0127-3. [DOI] [PubMed] [Google Scholar]
- 47.Arya G., Zhang Q., Schlick T. Flexible histone tails in a new mesoscopic oligonucleosome model. Biophys. J. 2006;91:133–150. doi: 10.1529/biophysj.106.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hardy C.D., Crisona N.J., Cozzarelli N.R. Disentangling DNA during replication: a tale of two strands. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:39–47. doi: 10.1098/rstb.2003.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


