Abstract
In the nucleus of eukaryotic cells, nascent transcripts are associated with heterogeneous nuclear ribonucleoprotein (hnRNP) particles that are nucleated by hnRNP C. Despite their abundance however, it remained unclear whether these particles control pre-mRNA processing. Here, we developed individual-nucleotide resolution UV-cross-linking and immunoprecipitation (iCLIP) to study the role of hnRNP C in splicing regulation. iCLIP data demonstrate that hnRNP C recognizes uridine tracts with a defined long-range spacing consistent with hnRNP particle organization. hnRNP particles assemble on both introns and exons, but remain generally excluded from splice sites. Integration of transcriptome-wide iCLIP data and alternative splicing profiles into an ‘RNA map’ indicates how the positioning of hnRNP particles determines their effect on inclusion of alternative exons. The ability of high-resolution iCLIP data to provide insights into the mechanism of this regulation holds promise for studies of other higher-order ribonucleoprotein complexes.
A major source of proteomic diversity in multicellular eukaryotes is the production of multiple mRNA isoforms. In humans, it was recently estimated that 95-100% of all multi-exon transcripts undergo alternative splicing1. Splice-site selection is primarily mediated by RNA-binding proteins that bind regulatory elements within nascent transcripts2,3. Heterogeneous nuclear ribonucleoprotein C1/C2 (hnRNP C) was identified over 30 years ago as a core component of hnRNP particles that form on all nascent transcripts4. However, although hnRNP C is one of the most abundant proteins in the nucleus, its role in splicing regulation remained unresolved. Whereas some studies suggested that hnRNP particles generally facilitate splicing5,6, individual hnRNP proteins were thought to function as splicing silencers7,8. Resolving these seemingly contradictory observations was hindered by the inability to locate precisely hnRNP particles on nascent transcripts in vivo. In particular, genome-wide mapping of hnRNP C positioning would provide critical information on how hnRNP particles control splicing. Since these highly abundant particles are likely to constitute a general platform for other splicing regulators, deciphering their function would greatly advance our understanding of splicing regulation.
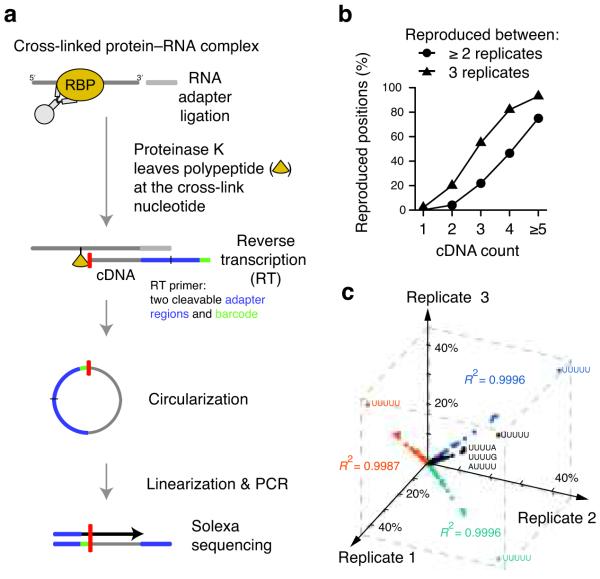
UV-cross-linking and immunoprecipitation (CLIP) combined with high-throughput sequencing was previously used to generate transcriptome-wide binding maps of several RNA-binding proteins9-12. However, since identification of binding sites relied on the analysis of overlapping sequence clusters, distances of less than 30 nucleotides were not resolved. An additional disadvantage of CLIP is the requirement of reverse transcription to pass over residual amino acids that remain covalently attached to the RNA at the cross-link site. Primer extension assays have shown that the vast majority of cDNAs prematurely truncate immediately before the ‘cross-link nucleotide’13. Here, we exploited this apparent limitation to achieve single nucleotide resolution by capturing these truncated cDNAs through the introduction of a second adapter after reverse transcription via self-circularization (Fig. 1). In order to quantify cDNA molecules that truncate at the same nucleotide, we added a random barcode to the DNA adapter. This allowed us to discriminate between unique cDNA products and PCR duplicates. We successfully applied individual-nucleotide resolution CLIP (iCLIP) to study hnRNP C-dependent splicing regulation in human cells. Taken together, iCLIP enables precise mapping of protein–RNA interactions in intact cells.
Figure 1.
iCLIP identifies hnRNP C cross-link nucleotides on RNAs. (a) Schematic representation of the iCLIP protocol. After UV irradiation, the covalently linked RNA is co-immunoprecipitated with the RNA-binding protein (RBP) and ligated to an RNA adapter at the 3′ end. Proteinase K digestion leaves a covalently bound polypeptide fragment on the RNA that causes premature truncation of reverse transcription (RT) at the cross-link site. The red bar indicates the last nucleotide added during reverse transcription. Resulting cDNA molecules are circularized, linearized, PCR-amplified and subjected to high-throughput sequencing. The first nucleotides of each sequence contain the barcode followed by the nucleotide where cDNAs truncated during reverse transcription. (b) Reproducibility of cross-link nucleotide positions. Percentage of cross-link nucleotides with a given cDNA count that were identified in at least two (circles) or all three experiments (triangles) are shown. The percentage of reproduced cross-link nucleotides increased with the incidence of hnRNP C cross-linking (cDNA count). (c) Reproducibility of sequence composition at cross-link nucleotides. Frequencies of pentanucleotides overlapping with cross-link nucleotides are shown for the three replicate experiments (R2 = 0.9996, R2 = 0.9987 and R2 = 0.9996) with the sequence shown for the four most highly enriched pentanucleotides. 42% of cross-link nucleotides overlap with UUUUU in all three replicate experiments.
Results
iCLIP maps hnRNP C binding to pre-mRNAs at nucleotide resolution
We employed iCLIP to examine the positioning of hnRNP C on pre-mRNAs in vivo. Three replicate iCLIP experiments were performed using an hnRNP C antibody on human HeLa cell lysates. The purified protein–RNA complex was absent when omitting UV-cross-linking or the use of hnRNP C antibody, and was diminished when hnRNP C knockdown cells were used (Supplementary Fig. 1a). Cross-linked RNA was reverse transcribed and PCR amplified, controlling PCR specificity with an experiment that lacked the antibody during purification (Supplementary Fig. 1b). High-throughput sequencing using Illumina GA2 generated a total of 6.5 million sequence reads (Supplementary Table 1). 4.2 million sequence reads aligned to the human genome by allowing only single genomic hits and one nucleotide mismatch. Next, we eliminated PCR amplification artifacts by removing sequences that truncated at the same nucleotide in the genome and shared the same random barcode. This identified 641,350 reads in total for the three replicate experiments, each representing a uniquely cross-linked RNA molecule. Finally, we summarized the number of sequences at each cross-link nucleotide into a ‘cDNA count’, representing a quantitative measure of the amount of hnRNP C cross-linking to each position (Fig. 2a). For the analyses of three independent no-antibody control samples we generated a total of 18 million sequence reads. After elimination of PCR amplification artifacts only 1,780 unique cDNAs remained (Supplementary Table 1), reflecting the high quality of purification and library preparation steps.
Figure 2.
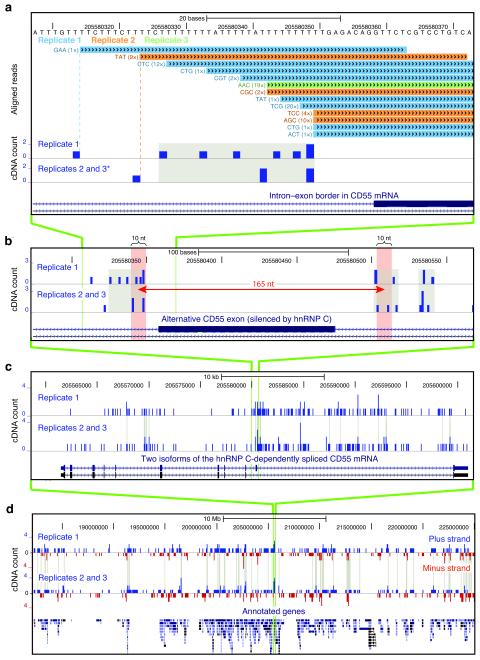
The genomic location of hnRNP C cross-link nucleotides. (a) Conversion of mapped iCLIP sequence reads into cDNA count values. Genomic sequence is shown above the color-coded positions of cDNA sequences from replicate experiments, preceded by the associated random barcode and the number of sequenced PCR duplicates (given in brackets). In the lower panel, a ‘cDNA count’ was assigned to the upstream ‘cross-link nucleotide’. Cross-link nucleotides within filtered clusters are highlighted in grey. The position of an alternative exon in CD55 mRNA is shown at the bottom. Modified image of the UCSC genome browser (human genome, version hg18, chromosome 1, nucleotides 205,580,308 to 205,580,373). * Due to space limitations, replicates 2 and 3 were merged into one lane. (b) Long-range spaced cross-link nucleotides flank the alternative exon in CD55 pre-mRNA. A distance of 165 nucleotides is marked by a red arrow with red shaded bars on either side representing ten nucleotide surrounding intervals. (c) Cross-link nucleotides are present along the entire length of CD55 pre-mRNA and accumulate around the alternative exon. Clustered cross-link nucleotides are indicated with grey lines. Annotation below shows position of exons in two alternative transcripts. (d) Global view of cross-link nucleotides on chromosome 11 (nucleotides 182,200,000 to 225,000,000). cDNA counts corresponding to positions in plus and minus strand transcripts are shown in blue and red, respectively. Gene annotations are given below. Cross-linking to individual genes and strand specificity are reproduced between replicates.
The iCLIP data were of high positional precision. Reproducibility of iCLIP data was demonstrated by the observation that 12,790 cross-link nucleotides were identified in at least two independent experiments (Fig. 1b). 75% of cross-link nucleotides with a cDNA count of five or more were seen in all three experiments showing that the strongest cross-link sites of hnRNP C are the most reproducible (Fig. 1b). Furthermore, there was an enrichment of cross-link nucleotides with an offset of one or two nucleotides (Supplementary Fig. 2). This observation may arise from protein contacts to more than one nucleotide of the RNA. In addition, the steric hindrance of the peptide fragment remaining on RNA may cause reverse transcription to terminate more than one nucleotide upstream of the cross-link site. As an independent measure of reproducibility we compared the occurrence of pentanucleotides overlapping the cross-link nucleotides; we found a high correlation between the three experiments (Fig. 1c), underlining the high precision of iCLIP in capturing protein–RNA interactions.
iCLIP identified large-scale binding of hnRNP C across the whole transcriptome. Although only a few direct targets were known prior to this study, we found hnRNP C cross-linking to transcripts from 55% of all annotated protein-coding genes (Supplementary Fig. 3, Fig. 2). This places hnRNP C as a major post-transcriptional regulator of similar importance as, for example, the poly-pyrimidine tract-binding protein (PTB) that was shown to bind transcripts of 43% of annotated human genes14. Among previously described hnRNP C targets, we observed binding to the regulatory element that determines start codon selection within the c-myc mRNA and to the 3′ untranslated region of the APP mRNA15,16 (Supplementary Fig. 4). 79% of cDNAs mapped in a sense orientation relative to introns, 9% to exons and 1% to non-coding RNAs. 11% mapped to intergenic regions, indicating that these harbor previously undescribed transcribed regions. Only 2% mapped in an antisense orientation relative to annotated genes, confirming that iCLIP generates strand-specific information on RNA binding (Supplementary Fig. 3, Fig. 2d). In summary, our data demonstrate that hnRNP C has a central role as a regulator of nascent transcripts.
In order to reduce false positive hits and to increase the resolution of the data, previous CLIP studies have applied filtering algorithms to identify clusters of CLIP cDNAs. Applying this approach to the hnRNP C dataset, we identified 33,991 clustered cross-link nucleotides (FDR < 0.05)12. This filtering removed 94% of all cross-link nucleotides, which most likely included true binding sites. Since the iCLIP libraries prepared during this study are not fully saturated – a limitation that currently applies to all CLIP methods – many real binding sites are represented by only few cDNAs. This view was supported by the observation that 6,367 out of 12,790 reproduced cross-link nucleotides were removed during the filtering process. Therefore, we performed all the analyses described below on the complete and the filtered datasets; as shown in Supplementary Figure 5, the results are quantitatively and qualitatively consistent, indicating that both sets are of high quality. Therefore in order to minimize loss of information, we describe findings for the complete dataset in the remainder of this work.
hnRNP C cross-links to uridine tracts
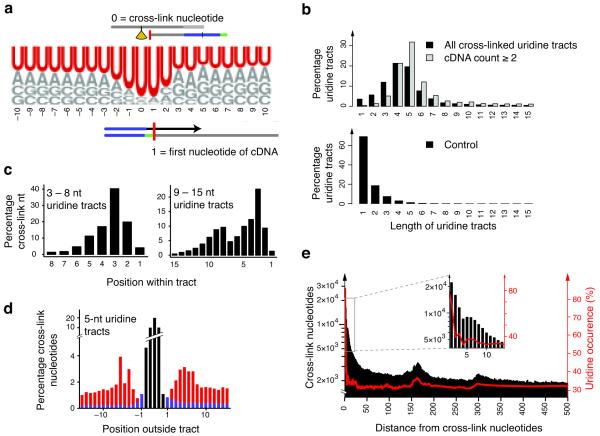
The high resolution of iCLIP data allowed us to assess the sequence specificity of hnRNP C binding. Strikingly, uridine represented 85% of cross-link nucleotides (p-value < 0.001 by hypergeometric distribution for enrichment relative to background base frequencies; Fig. 3a, Supplementary Fig. 5a). Surrounding positions were also strongly enriched for uridines, such that 65% of cross-link nucleotides were part of a contiguous tract of four or more uridines (Fig. 3b, Supplementary Fig. 5b). These results agree with the in vitro observation that the RRM domains of hnRNP C bind to uridine tracts17-19, suggesting that cross-link nucleotides reflect the positions where the RRM domains contact RNA in vivo. In comparison, only 15 – 24% of cross-link nucleotides from the no-antibody control experiments were located in a tract of four or more uridines, demonstrating a significant enrichment of uridine tract binding in the hnRNP C iCLIP data (p value < 0.01 by Student’s t-test). We note that the control displays a bias to bind uridine tracts compared with the expected 5% from the background distribution in transcribed regions. However, this is in line with previous studies on single-stranded DNA-binding proteins that show preferential cross-linking to thymidine residues20,21. Nonetheless, the small number of sequence reads and the low cross-linking bias in the control data contrast the strong preference for uridine by hnRNP C, indicating that the vast majority of iCLIP sequence reads reflect real hnRNP C binding events. Furthermore, the ability of iCLIP to quantify the number of cDNAs mapping to each cross-link nucleotide allowed us to analyze the affinity of hnRNP C to uridine tracts of different lengths. We found that cDNA counts increased with the number of uridines in the tract, suggesting that hnRNP C binds longer tracts with higher affinity (Fig. 3b, Supplementary Fig. 5b, 6a).
Figure 3.
hnRNP C binds uridine tracts with a defined spacing. (a) Weblogo showing base frequencies of cross-link nucleotides and 20 nucleotides of surrounding genomic sequence. Positions 0 and 1 correspond to cross-link nucleotide and first position of cDNA sequence, respectively. For comparison, the background distribution of bases within transcribed regions is: U, 30.3%, A, 27.7%, G, 21.4% and C, 20.6%. (b) Length distribution of uridine tracts harboring cross-link nucleotides. The percentage of tracts of a certain length is given relative to all bound tracts. Panels compare all cross-link nucleotides (black) to those with a cDNA count of 2 or higher (grey, top), and length distribution of tracts within the transcriptome as control (bottom). (c) Positioning of cross-link nucleotides within uridine tracts. Positions were summarized over shorter (3 – 8 uridines) and longer tracts (9 – 15 uridines) aligned at their 3′ ends. Longer tracts contain two peaks at a defined spacing of 5 – 6 nucleotides (Supplementary Fig. 6b). (d) Binding neighborhood of five-nucleotide uridine tracts (black). Occurrence of cross-link nucleotides at a given position is given as a fraction of all positions. Cross-link nucleotides within flanking uridine tracts of at least three uridines are shown in red, and those remaining in blue. (e) Long-range spacing of cross-link nucleotides. Distances to all downstream cross-link nucleotides were summarized (black). Uridine densities at the same distances are superimposed (red). Inlay shows an enlarged region of the graph. Increased occurrence of cross-link nucleotides coincided with peaks in uridine density at 165 and 300 nucleotides distance.
The spacing of cross-link sites reflects hnRNP particle formation
iCLIP allowed us to resolve adjacent binding sites within uridine tracts. We found that regardless of the length of the uridine tract, hnRNP C primarily cross-linked to the third uridine from the 3′ end (Fig. 3c, Supplementary Fig. 5c, 6b). In addition, we identified a second peak of hnRNP C cross-linking positioned five or six nucleotides upstream on tracts longer than nine uridines. Consistently, such dual binding also occurred on shorter tracts when flanked by neighboring uridine tracts (Fig. 3d, Supplementary Fig. 5d). Since the hnRNP C tetramer binds RNA with two RRM domains positioned proximally to each other6,22, the dual cross-linking pattern could result from adjacent binding by the two RRM domains. These results show that the high resolution of iCLIP can elucidate combinatorial binding by multiple RNA-binding domains to proximal RNA binding sites, which would otherwise remain unresolved.
In addition to the short-range spacing within uridine tracts, iCLIP also identified a pattern of long-range spacing of cross-link nucleotides. We found peaks at distances of 165 and 300 nucleotides (Fig. 3e, Supplementary Fig. 5e). Strikingly, the uridine density also peaked at the same positions (Fig. 3e, Supplementary Fig. 5e). The defined spacing between cross-link nucleotides suggests that the intervening RNA is incorporated into the hnRNP particles. This model agrees with the organization of hnRNP particles as proposed by previous studies6,23,24. Taken together, the precise mapping of hnRNP C cross-link sites provides insights into the structure of hnRNP particles.
The positioning of hnRNP particles determines the splicing outcome
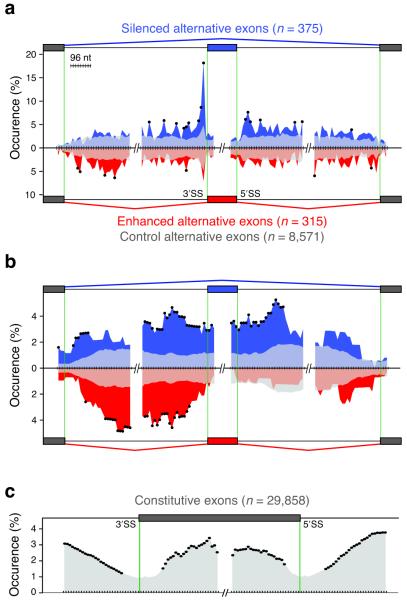
iCLIP allowed us to assess precisely the positioning of hnRNP C on alternatively spliced pre-mRNAs. Comparing transcript abundance in hnRNP C knockdown and control HeLa cells using high-resolution splice-junction microarrays, we detected significant increases and decreases by a factor of at least 2 for 47 and 115 transcripts, respectively (p-value < 0.01 by Student’s t-test). Transcript changes showed no apparent correlation with the amount of hnRNP C cross-linking (Supplementary Fig. 7). By far the strongest change was seen for the hnRNP C transcript (decreased by a factor of 10.6), underlining the efficiency and specificity of the knockdown. This was also verified by Western blot analysis (Supplementary Fig. 8). Using the ASPIRE3 algorithm, we detected changes in splicing at 1,340 alternative exons. Transcripts harboring at least one alternatively spliced exon were significantly overrepresented among the differentially expressed transcripts and vice versa (Supplementary Fig. 7b), indicating a relation between alternative splicing and transcript abundance. We observed a similar incidence of increased or decreased exon inclusion in hnRNP C knockdown cells, indicating that hnRNP C can either silence or enhance exon inclusion, respectively. We validated changes at 26 exons by RT-PCR with a 92% success rate (Supplementary Table 2, Supplementary Fig. 9). In order to address the role of hnRNP C binding in these changes, iCLIP data and splicing profiles were integrated into an ‘RNA map’25. Increased density of cross-link nucleotides was seen at the splice sites of silenced alternative exons (Fig. 4, Supplementary Fig. 5f). At the 3′ splice site, hnRNP C predominantly cross-linked within the first 30 nucleotides that generally coincide with the poly-pyrimidine tracts, as seen in the CD55 pre-mRNA (Fig. 2a, Fig. 4a). This suggests that similar to PTB, hnRNP C can regulate alternative splicing by repressing specific 3′ splice sites26. In conclusion, the ability of iCLIP to map cross-link nucleotides to characterized RNA regulatory elements can indicate the function of protein–RNA interactions.
Figure 4.
The RNA map relates hnRNP particle positioning to splicing regulation. (a) The RNA map of cross-link sites within regulated pre-mRNAs. Positioning of cross-link nucleotides was assessed at exon–intron boundaries of alternative (375 silenced, blue; 315 enhanced, red; 8,571 control alternative exons, grey; regions of overlap are shown as lighter shades of blue/red) and flanking constitutive exons. “Occurrence (%)” indicates the percentage of exons that have at least one cross-link nucleotide within a given window. Black dots mark significant enrichment of regulated exons containing cross-link nucleotides within a given window relative to control alternative exons (p value < 0.01 by Fisher’s Exact test). Silenced alternative exons show strong enrichment of cross-link nucleotides proximal to the 3′ and the 5′ splice sites (3′SS and 5′SS). (b) The RNA map of hnRNP particles on regulated pre-mRNAs. Positioning of regions intervening cross-link nucleotides with defined 160 – 170 nucleotide spacing was analyzed as in Fig. 4a. Silenced alternative exons show incorporation of the entire regulated exon into hnRNP particles, whereas particle incorporation is confined to the preceding intron at enhanced alternative exons. (c) The RNA map of hnRNP particles at constitutive exons. Positioning of regions intervening the cross-link nucleotides with a spacing of 160 – 170 nucleotides was assessed at exon–intron boundaries of constitutive exons (29,858 exons analyzed as in Fig. 4a). Splice sites show decreased incorporation into hnRNP particles.
In order to understand the impact of higher-order hnRNP particles on the observed splicing changes, we restricted the analysis to the cross-link sites displaying long-range spacing indicative of particle formation. We considered the regions between these cross-link sites as being incorporated into the particles. Due to the limited complexity of the clustered dataset, we restricted this analysis to the complete dataset. We found that silenced exons and proximal intronic regions showed increased incorporation into hnRNP particles (Fig. 4b). Long-range spaced binding across an exon, as seen in CD55 pre-mRNA (Fig. 2b), might silence splicing by incorporating the exon into the hnRNP particle. A related hypothesis proposed that binding of PTB via its four RRM domains to sites flanking an exon silences splicing by looping out the exon14,27,28. In addition, we found that hnRNP particles enhance splicing when binding within the intron preceding the alternative exon (Fig. 4b). Thus, by incorporating long regions of RNA, hnRNP particles can play a dual role in splicing regulation. Importantly, the outcome of this regulation depends on the positioning of hnRNP particles on pre-mRNAs.
The RNA map of hnRNP C regulation described that silenced exons are flanked by precisely spaced cross-link nucleotides. In order to assess whether hnRNP C binding could predict silenced exons, we used the iCLIP data to search the transcriptome for exons that are flanked by hnRNP C cross-link nucleotides with a defined spacing of 160 – 170 nucleotides. We then chose nine alternatively spliced exons that had not shown hnRNP C-dependent regulation in our microarray analyses, and quantified their splicing behavior using RT-PCR. Strikingly, five of these (56%) showed significantly increased inclusion in hnRNP C knockdown cells (p value < 0.05 by Student’s t-test), while the others remained unchanged (Fig. 5, Supplementary Table 3). Thus, the hnRNP C binding patterns identified by the iCLIP data could predict exon silencing, further substantiating our model of position-dependent splicing regulation by hnRNP particles.
Figure 5.
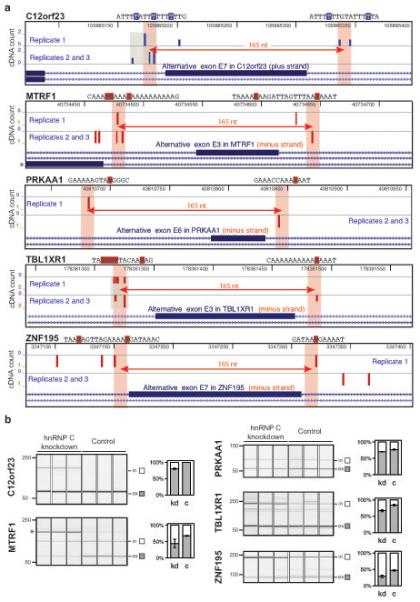
iCLIP data predict exons that are silenced by hnRNP C. (a) Genomic location of hnRNP C cross-link nucleotides surrounding silenced exons that were predicted from iCLIP data. Five exons that are flanked by cross-link nucleotides with defined spacing and showed a significant increase in inclusion in the hnRNP C knockdown cells are depicted. cDNA counts corresponding to positions in plus and minus strand transcripts are shown in blue and red, respectively. Gene names and genomic sequence around cross-link nucleotides (highlighted by blue or red boxes indicating plus-strand or minus-strand location) are given above each panel. A distance of 165 nucleotides is marked by a red arrow with shaded bars on either side representing ten nucleotide intervals. Clustered cross-link nucleotides are highlighted in grey. A mutual exclusive exon in MTRF1 pre-mRNA is indicated by an asterisk. Images are based on the UCSC genome browser (human genome, version hg18; C12orf23, chromosome 12, nucleotides 105,885,065 – 105,885,394; MTRF1, chromosome 13, nucleotides 40,734,402 – 40,734,731; PRKAA1, chromosome 5, nucleotides 40,810,631 – 40,810,960; TBL1XR1, chromosome 3, nucleotides 178,361,247 – 178,361,576; ZNF195, chromosome 11, nucleotides 3,347,071 – 3,347,400). (b) Quantification of splicing changes of the alternative exons depicted in (a). RNA from hnRNP C knockdown (kd) and control (c) HeLa cells was analyzed using RT-PCR and capillary electrophoresis. Capillary electrophoresis image and signal quantification are shown for each exon. Quantified transcripts including (in) or excluding (ex) the regulated alternative exon are marked on the right. Average quantification values of exon inclusion (white) and exclusion (grey) are given as a fraction of summed values. Error bars represent standard deviation of three replicates. Change in exon inclusion and p values are given in Supplementary Table 3. The asterisk indicates the PCR product for the RNA isoform of a mutually exclusive exon in MTRF1 pre-mRNA as depicted in (a). Its inclusion is strongly increased in hnRNP C knockdown cells consistent with our model that hnRNP C binding within the polypyrimidine tract leads to silencing of exons.
The broad distribution of hnRNP C cross-link sites over complete transcripts (Fig. 2c) suggested that the hnRNP C activity is not restricted to regulation of alternative splicing. Therefore, we analyzed hnRNP particle formation on constitutive exons and flanking intronic regions to find a similar coverage on exons and introns, as predicted by previous studies5. However, we found a decreased coverage at the splice sites, agreeing with the hypothesis that hnRNP particles need to be excluded from regions required for splicing7 (Fig. 4c). These results suggest that hnRNP particles maintain splicing fidelity by incorporating introns and exons, while leaving the splice sites free to interact with the splicing machinery.
Discussion
Global profiling of protein–RNA interactions has been successful in elucidating principles of post-transcriptional regulation. Over the past years, CLIP was proven as a powerful method to determine protein–RNA interactions in vivo on a global scale9-12. However, the resolution of this method is limited due to the inability to directly identify the cross-linked nucleotides. Moreover, CLIP suffers from the inherent problem that most cDNAs truncate at the cross-link site and are thus lost during the amplification process. Here, we developed iCLIP, which overcomes these obstacles and identifies the positions of cross-link sites at nucleotide resolution. iCLIP also introduces a random barcode to mark individual cDNA molecules, thereby solving an inherent problem of all current high-throughput sequencing methods that suffer from PCR artefacts. Therefore, exploiting the random barcode strongly improves the quality of quantitative information. Due to the low abundance of introns, the obtained sequence coverage is at present insufficient to quantitatively compare individual binding sites at single nucleotide resolution. However, the quantitative information could be exploited on a transcriptome-wide scale to show that hnRNP C binds longer uridine tracts with higher affinity, underlining the great potential of iCLIP’s quantitative nature. In order to identify clustered cross-link nucleotides, we applied a statistical algorithm to filter for enriched hnRNP C binding. Comparison of the clustered cross-link nucleotides with the complete dataset showed that both datasets generate consistent results, suggesting that real binding sites constitute a major proportion of both. This observation underlines the high quality of iCLIP data, achieved by high stringency of purification and library preparation. Thus, iCLIP allows the transcriptome-wide analysis of protein–RNA interactions at individual nucleotide resolution.
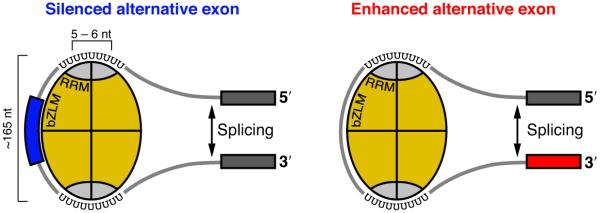
We used iCLIP to show that hnRNP C binds to uridine tracts in nascent transcripts with a defined spacing of 165 and 300 nucleotides. These data agree with past findings that the hnRNP C tetramer binds in repetitive units of approximately 150 – 300 nucleotides6,23,24. Whereas some studies suggested that this binding occurs in a sequence-independent manner6,23,24, other studies proposed that the sequence-specific RRM domains critically contribute to high-affinity RNA binding of the hnRNP C tetramer17-19. iCLIP data agree with the latter model that hnRNP C is positioned on pre-mRNAs via sequence-specific binding of its RRM domains (Fig. 6). In addition, the precise spacing between the hnRNP C cross-link sites suggests that in accordance with the former model the basic leucine zipper-like RNA-binding motif (bZLM) domains guide the intervening RNA along the axis of the hnRNP C tetramer via sequence-independent electrostatic interactions22,29. Thus, by measuring the spacing between distant binding sites, iCLIP can yield structural insights into ribonucleoprotein complexes.
Figure 6.
A model of hnRNP C tetramer binding at silenced and enhanced alternative exons. hnRNP C protein monomers are depicted in yellow with the RRM domains in grey. The schematic RNA molecule is shown to contact the RRM domains via uridine tracts and the bZLM domains via electrostatic interactions. Binding of the RRM domains on both sides of an alternative exon results in silencing of exon inclusion (blue), whereas tetramer binding to the preceding intron enhances exon inclusion (red).
Even though hnRNP particles were found to form on nuclear RNAs more than 30 years ago, their function in pre-mRNA processing remained unresolved4-8. Here, we present nucleotide-resolution mapping of in vivo hnRNP C cross-link sites, which reveals a role of hnRNP particles in splicing regulation. Importantly, we found that binding of hnRNP particles is guided by the pre-mRNA sequence to determine the splicing outcome in a position-dependent manner. In particular, alternative exons are silenced by incorporation into the hnRNP particles, whereas binding to the preceding intron enhances inclusion of alternative exons. Early studies had hypothesized that hnRNP particles might function to organize long introns for efficient splicing30. This was based on the observation that long pre-mRNAs are highly compacted in hnRNP particles. In accordance with this hypothesis, we propose that hnRNP particles might act as ‘RNA nucleosomes’ that bind long regions of pre-mRNA, but maintain the correct splice sites accessible to the splicing machinery. The ability of iCLIP to study protein–RNA interactions with high resolution and in a quantitative manner holds promise for future studies of the structure and function of ribonucleoprotein complexes.
Supplementary Material
Acknowledgements
The authors thank all members of the Ule laboratory for experimental assistance and discussion. We thank Aaron Klug, Kiyoshi Nagai, Madan Babu, Sebastian Eustermann, Nicholas McGlincy, Dalia Daujotyte and Oliver Rossbach for fruitful discussions and comments on the manuscript. We thank James Hadfield and Nik Matthews for high-throughput sequencing. We thank Brooke Rhead and Brian Raney for modifying the UCSC Genome Browser BedGraph format. This work was supported by the European Research Council grant 206726-CLIP to J.U. and a Long-term Human Frontiers Science Program fellowship to J.K.
Methods
iCLIP analyses
HeLa cells were irradiated with UV-C light to covalently cross-link proteins to nucleic acids in vivo. Upon cell lysis, RNA was partially fragmented using low concentrations of RNase I, and hnRNP C-RNA complexes were immuno-purified with the antibody immobilized on immunoglobulin G-coated magnetic beads. After stringent washing, RNAs were ligated at their 3′ends to an RNA adapter and radioactively labelled to allow visualization. Denaturing gel electrophoresis and transfer to a nitrocellulose membrane removed RNAs that were not covalently linked to the protein. Two size fractions of the RNA (Supplementary Fig. 1a) were recovered from the membrane by proteinase K digestion. The oligonucleotides for reverse transcription contained two inversely oriented adapter regions separated by a BamHI restriction site as well as a barcode region at their 5′ end containing a two nucleotide barcode to mark the experiment and a three nucleotide random barcode to mark individual cDNA molecules. cDNA molecules were size-purified using denaturing gel electrophoresis, circularized by single-stranded DNA ligase, annealed to an oligonucleotide complementary to the restriction site and cut between the two adapter regions by BamHI. Linearized cDNAs were then PCR-amplified using primers complementary to the adapter regions (Supplementary Fig. 1b) and subjected to high-throughput sequencing using Illumina GA2. A more detailed description of the iCLIP protocol is available in Supplementary Methods.
High-throughout sequencing and mapping
High-throughput sequencing of iCLIP cDNA libraries from three replicate experiments was performed on one lane of an Illumina GA2 flow cell with 54 nt run length. Mapping of sequence reads was performed against the human genome (version Hg18/NCBI36) using bowtie version 0.10.131 (Supplementary Methods). The 3-nt random barcode enabled us to discriminate PCR duplicates from sequences, which start at the same nucleotide, but derived from individual cDNA molecules. Random barcodes with more than one identical nucleotide were considered to be PCR duplicates, which were excluded from the data set (for more details, see Rot et al., manuscript in preparation). Following this strategy, a total of 3521462 sequences were removed from the analysis (85% of mapped reads), resulting in a final set of 309489, 216295 and 115566 sequences representing individual cDNA molecules from the three replicates. Last, the first nucleotide in the genome upstream of a mapping cDNA sequence was defined as ‘cross-link nucleotide’ and the total of corresponding cDNA sequences assigned as ‘cDNA count’ at this position. For subsequent analyses, replicates were merged into one iCLIP dataset by summing cDNA counts from all three replicates for each cross-link nucleotide.
Analyses of reproducibility and significance of cross-link nucleotides
Reproducibility was assessed by comparing cross-link nucleotide positions from three independent replicates (Supplementary Methods). The false-discovery rate (FDR) for each position was determined according to Yeo and coworkers12 (Supplementary Methods).
Analysis of sequence and positioning of cross-link nucleotides
All analyses of hnRNP C binding were based on sequences mapping to human nuclear chromosomes. In order to determine pentanucleotide frequencies at cross-link nucleotides (Fig. 1c), we assessed all pentanucleotides overlapping each cross-link nucleotide within the three replicate experiments. Multiple occurrences at the same cross-link nucleotide were counted only once. Frequencies were calculated as the number of cross-link nucleotides that are associated with a certain pentanucleotide.
For calculating base frequencies of iCLIP sequence reads (Fig. 3a), we extracted genomic sequence corresponding to the first 10 nucleotides of all reads plus 11 nucleotides of preceding sequence. Graphic representation was generated using Weblogo 332 (http://weblogo.berkeley.edu). Background distribution of bases was calculated using all transcribed regions annotated in the Ensembl database33 (release 54; http://www.ensembl.org/).
In order to determine the lengths distribution of uridine tract bound by hnRNP C (Fig. 3b), we extracted all uridine tracts in the genome that harbored at least one cross-link nucleotide. Distribution of uridine tracts within the transcriptome was calculated again based on all transcribed regions.
The percentage of cross-link nucleotides located within a tract of at least four uridines was calculated as a fraction of all identified cross-link nucleotides. The expected background was calculated upon randomization of cross-link nucleotide positions as described above. Finally, the expected value for background localization to tracts of at least four uridines was calculated as mean percentage from 100 random permutations.
In order to assess the spacing of cross-link nucleotides (Fig. 3e), we summarized the distances of all cross-link nucleotides to all downstream cross-link nucleotides within a window of 500 nt. In order to analyze the short-range binding patterns, we summarized all cross-link nucleotides on each position of uridine tracts of the same length (Fig. 3c, Supplementary Fig. 6b). For tracts of five uridines, we additionally assessed distribution of surrounding cross-link nucleotides (Fig. 3d), using only those tracts that displayed at least one additional cross-link nucleotide at a distance of no more than 15 nt to either side.
In order to examine the influence of uridine tract length on the occurrence of cross-linking (Supplementary Fig. 6a), the percentage of tracts with a cDNA count of at least two at the third position from the 3′ end was calculated relative to all tracts of the same length containing a cross-link site at this position.
Knockdown of hnRNP C
hnRNP C was depleted in HeLa cells using two different siRNAs (Supplementary Methods).
Splice-junction microarrays
Microarray analyses and PCR validations were performed as described in Supplementary Methods. The microarray data was analyzed using ASPIRE version 3 that was modified relative to previous versions11,34 by adding background subtraction and significance ranking of predicted splicing changes (Supplementary Methods). By analysing the signal of reciprocal probe sets, ASPIRE3 was able to monitor 53632 alternative splicing events. Applying a threshold of |ΔIrank| ≥ 1, we identified 1340 differentially spliced alternative exons, of which 662 and 678 were increased and decreased in the hnRNP C knockdown cells, respectively.
RNA map
In order to analyze the impact of hnRNP C positioning on splicing regulation, we assessed the positioning of hnRNP C cross-link sites at exon-intron boundaries of alternative exons and flanking constitutive exons (Supplementary Methods).
Manuscript preparation
All figures were prepared using R Lattice Graphics (http://r-forge.r-project.org/projects/lattice/) and Adobe Illustrator® CS4.
References
- 1.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyer AL, Christensen ME, Walker BW, LeStourgeon WM. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977;11:127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- 5.Steitz JA, Kamen R. Arrangement of 30S heterogeneous nuclear ribonucleoprotein on polyoma virus late nuclear transcripts. Mol Cell Biol. 1981;1:21–34. doi: 10.1128/mcb.1.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang M, et al. The C-protein tetramer binds 230 to 240 nucleotides of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol Cell Biol. 1994;14:518–533. doi: 10.1128/mcb.14.1.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol. 2000;12:340–345. doi: 10.1016/s0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- 8.Amero SA, et al. Independent deposition of heterogeneous nuclear ribonucleoproteins and small nuclear ribonucleoprotein particles at sites of transcription. Proc Natl Acad Sci U S A. 1992;89:8409–8413. doi: 10.1073/pnas.89.18.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 10.Ule J, Jensen K, Mele A, Darnell RB. CLIP: A method for identifying protein–RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 11.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urlaub H, Hartmuth K, Lührmann R. A two-tracked approach to analyze RNA-protein crosslinking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods. 2002;26:170–181. doi: 10.1016/S1046-2023(02)00020-8. [DOI] [PubMed] [Google Scholar]
- 14.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JH, et al. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol Cell Biol. 2003;23:708–720. doi: 10.1128/MCB.23.2.708-720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaidi SH, Malter JS. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J Biol Chem. 1995;270:17292–17298. doi: 10.1074/jbc.270.29.17292. [DOI] [PubMed] [Google Scholar]
- 17.Gorlach M, Wittekind M, Beckman RA, Mueller L, Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992;11:3289–3295. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorlach M, Burd CG, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 19.Wan L, Kim JK, Pollard VW, Dreyfuss G. Mutational definition of RNA-binding and protein–protein interaction domains of heterogeneous nuclear RNP C1. J Biol Chem. 2001;276:7681–7688. doi: 10.1074/jbc.M010207200. [DOI] [PubMed] [Google Scholar]
- 20.Hockensmith JW, Kubasek WL, Vorachek WR, von Hippel PH. Laser cross-linking of nucleic acids to proteins. Methodology and first applications to the phage T4 DNA replication system. J Biol Chem. 1986;261:3512–3518. [PubMed] [Google Scholar]
- 21.Hockensmith JW, Kubasek WL, Vorachek WR, von Hippel PH. Laser cross-linking of proteins to nucleic acids. I. Examining physical parameters of protein-nucleic acid complexes. J Biol Chem. 1993;268:15712–15720. [PubMed] [Google Scholar]
- 22.Whitson SR, LeStourgeon WM, Krezel AM. Solution structure of the symmetric coiled coil tetramer formed by the oligomerization domain of hnRNP C: implications for biological function. J Mol Biol. 2005;350:319–337. doi: 10.1016/j.jmb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Barnett SF, Friedman DL, LeStourgeon WM. The C proteins of HeLa 40S nuclear ribonucleoprotein particles exist as anisotropic tetramers of (C1)3 C2. Mol Cell Biol. 1989;9:492–498. doi: 10.1128/mcb.9.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAfee JG, Soltaninassab SR, Lindsay ME, LeStourgeon WM. Proteins C1 and C2 of heterogeneous nuclear ribonucleoprotein complexes bind RNA in a highly cooperative fashion: support for their contiguous deposition on pre-mRNA during transcription. Biochemistry. 1996;35:1212–1222. doi: 10.1021/bi951974k. [DOI] [PubMed] [Google Scholar]
- 25.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 26.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 27.Gooding C, Roberts GC, Moreau G, Nadal-Ginard B, Smith CW. Smooth muscle-specific switching of alpha-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 1994;13:3861–3872. doi: 10.1002/j.1460-2075.1994.tb06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oberstrass FC, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 29.McAfee JG, Shahied-Milam L, Soltaninassab SR, LeStourgeon WM. A major determinant of hnRNP C protein binding to RNA is a novel bZIP-like RNA binding domain. RNA. 1996;2:1139–1152. [PMC free article] [PubMed] [Google Scholar]
- 30.Choi YD, Grabowski PJ, Sharp PA, Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986;231:1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- 31.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbard TJ, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ule J, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.