Abstract
RegB is a membrane-spanning sensor kinase responsible for redox regulation of a wide variety of metabolic processes in numerous proteobacterial species. Here we show that full-length RegB purified from Escherichia coli membranes contains bound ubiquinone. Four conserved residues in the membrane-spanning domain of RegB are shown to have important roles in ubiquinone binding in vitro and redox sensing in vivo. Isothermal titration calorimetry measurements, coupled with kinase assays under oxidizing and reducing conditions, indicate that RegB weakly binds both oxidized ubiquinone and reduced ubiquinone (ubiquinol) with nearly equal affinity and that oxidized ubiquinone inhibits kinase activity without promoting a redox reaction. We propose a model in which ubiquinone/ubiquinol bound to RegB readily equilibrates with ubiquinones/ubiquinols in the membrane, allowing the kinase activity to be tuned by the redox state of the ubiquinone pool. This noncatalytic role of ubiquinone in controlling RegB activity is distinct from that of other known ubiquinone-binding proteins, which use ubiquinone as an electron donor or acceptor.
IMPORTANCE
Two-component signaling systems are comprised of a sensor kinase and a cognate response regulator that together control many cellular processes in response to a change in environmental conditions. Many sensor kinases are components of the cell membrane, with a domain that senses a specific environmental stimulus to control the activity of a cytosolic kinase domain. The broadly disseminated RegB/RegA two-component system regulates many energy-related processes in response to changes in the cellular redox state. In this study, the membrane-spanning sensor kinase RegB is shown to regulate its activity by interacting with the ubiquinone pool in a manner that involves novel noncatalytic equilibrium binding of ubiquinone.
INTRODUCTION
As is true for many purple nonsulfur bacteria, Rhodobacter capsulatus exhibits excellent metabolic diversity and is thus capable of growth via photosynthesis, aerobic respiration, and anaerobic respiration (1). R. capsulatus is capable of performing nitrogen fixation, carbon fixation, and hydrogen utilization (2). All of these metabolic/biosynthetic processes are either energy generating or energy utilizing, requiring that R. capsulatus coordinately regulates relevant physiological components to maintain appropriate cellular redox poise. The RegB/RegA two-component system of R. capsulatus coordinates the activation and repression of numerous energy-generating and energy-utilizing systems to control the overall redox poise of the cell (2–4).
RegB is a membrane-spanning sensor kinase that autophosphorylates a conserved histidine under reducing conditions. Phosphorylated RegB (RegB~P) transfers the phosphoryl group to an aspartate in its cognate response regulator, RegA (5, 6), to regulate interactions with promoters and RNA polymerase. The mechanism by which RegB controls kinase activity in response to redox changes has been an active area of investigation since its discovery over a decade ago (3). A previous study demonstrated that RegB has a “redox-active” cysteine, Cys265, located between the site of autophosphorylation and the kinase domain that is partially responsible for redox control of kinase activity. Under oxidizing growth conditions, Cys265 can form an intermolecular disulfide bond to convert active RegB dimers into inactive tetramers (7).
In addition to modulating the redox state of Cys265, the transmembrane domain of RegB is also implicated in redox sensing by interacting with the ubiquinone pool. A highly conserved sequence, GGXXNPF, located in a short periplasmic loop between transmembrane helices 3 and 4, is thought to define a ubiquinone-binding site, as shown by photoaffinity cross-linking with a quinone analog (8). The addition of oxidized ubiquinone to full-length wild-type RegB also inhibits RegB kinase activity in vitro (8). Grammel and Ghosh confirmed a correlation between the in vivo ubiquinone redox state and the expression level of photosynthetic membranes, suggesting that RegB activity is cooperatively regulated by the change of the ubiquinone redox state (9). There is also a model proposed by Kim et al. that suggests that that cbb3 oxidase may be a dominant component regulating RegB kinase activity (10).
The global sensor kinase ArcB from enteric bacteria is also reported to regulate its activity by interacting with ubiquinones (11, 12). Studies of a truncated cytosolic domain of ArcB show that a redox-active cysteine is capable of forming an intermolecular disulfide bond that inhibits kinase activity, not unlike that observed in RegB (12). Disulfide bond formation is stimulated when the cytosolic domain of ArcB is incubated with oxidized ubiquinone in vitro (12). Biochemical analysis of full-length ArcB with its membrane-spanning domain has not been undertaken, so it remains unclear if in vitro formation of a disulfide bond by ubiquinone is simply a nonspecific redox reaction in which ubiquinone is functioning as a simple oxidant. It is also not clear that ArcB has a specific ubiquinone-binding site and if binding of ubiquinone itself affects ArcB kinase activity.
Even though our previous studies established that oxidized ubiquinone inhibits the activity of full-length wild-type RegB in vitro, there remain questions about how ubiquinone interacts with RegB and why only oxidized ubiquinone inhibits kinase activity. For example, it is not known whether RegB preferentially binds oxidized and reduced ubiquinone. It was also not shown whether ubiquinone affects kinase activity irrespective of the redox state of Cys265. To address these issues, we studied the ability of a full-length RegB to bind to ubiquinone under oxidizing and reducing conditions. To dispel issues involving the oxidation and reduction of Cys265, we performed ubiquinone-binding and inhibition studies with a RegB variant that has a Cys265-to-Ser mutation. Here we present evidence that purified full-length RegB C265S contains bound ubiquinone and that alteration of the redox state of the bound ubiquinone indeed regulates kinase activity. Mutations in the conserved ubiquinone-binding site, as well as two highly conserved residues in transmembrane helix 1, disrupt ubiquinone binding in vitro and result in increased aerobic photosystem gene expression in vivo. Interestingly, full-length RegB C265S binds both oxidized and reduced ubiquinone with only oxidized ubiquinone, affecting kinase activity. We propose that the in vivo interaction of RegB with both the reduced and oxidized forms of ubiquinone allows the kinase to monitor the overall redox state of the ubiquinone pool, which allows the kinase activity to be tuned by the cellular energy state.
RESULTS
Purified RegB C265S contains bound ubiquinone.
To test whether RegB is capable of binding ubiquinones, we overexpressed and purified detergent-solubilized full-length RegB from Escherichia coli membrane fractions and then spectrally analyzed the isolated protein for ubiquinone (13). When full-length wild-type RegB is purified under air-oxidizing conditions, the presence of ubiquinone is evidenced by an absorbance maximum at ~275 nm that is reduced after the addition of the reductant sodium borohydride (Fig. 1A, dashed black line). Using the absorption coefficient of coenzyme Q8 (Δ275 = 12.25 mM−1), which is the predominate ubiquinone in E. coli, the amount of oxidized bound ubiquinone present in several independent protein isolates varied from 0.6 to 0.8 mol ubiquinone/mol wild-type RegB. Similar analysis was also undertaken with the full-length RegB C265S mutant that lacks the redox-active Cys265. This protein mutant shows a nearly identical absorption peak at ~275 nm, with the amount of bound ubiquinone in several independent isolates being 0.4 to 0.8 mol ubiquinone/mol RegB C265S. A spectral red shift was also observed from 274 nm to 276.5 nm when comparing the spectrum of RegB C265S that has a small amount of bound ubiquinone (0.44 mol ubiquinone/mol protein) to a preparation that has a larger amount of bound ubiquinone (0.62 mol ubiquinone/mol protein) (Fig. 1B). Similar red shifts have been reported for other ubiquinone-binding proteins upon an increase in ubiquinone content (14, 15). As a negative control, we also assayed for the presence of ubiquinones in a variant of RegB called RegB″ (7), which is a truncated soluble version that lacks the transmembrane domain. Spectral analysis shows no evidence of bound ubiquinone to isolated RegB″ (Fig. 1A, gray lines).
FIG 1 .
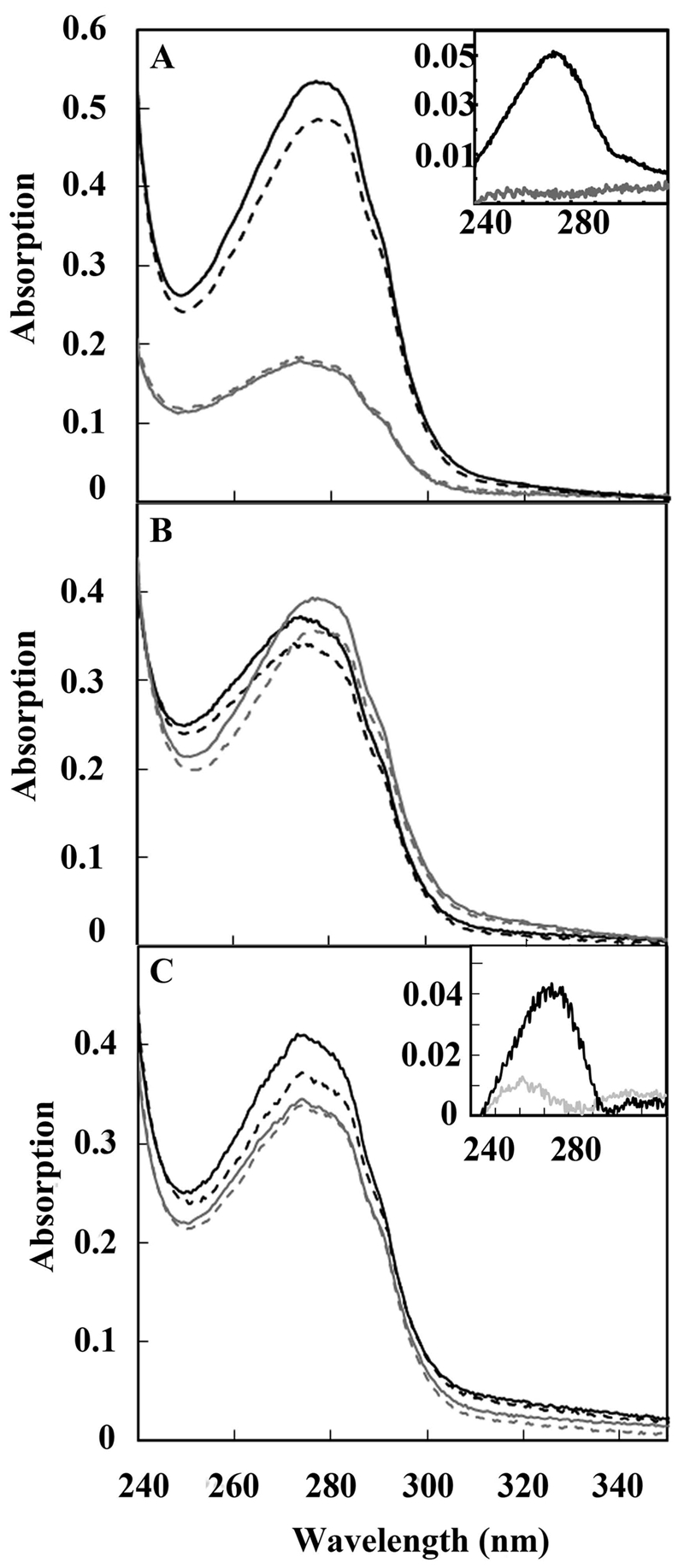
UV spectra of ubiquinone bound to RegB. (A) Spectrum of isolated oxidized full-length wild-type RegB (solid black line) containing 0.82 mol ubiquinone/mol protein that exhibits a decrease in absorbance at 275 nm upon reduction with NaBH4 (dashed black line). A control of truncated RegB″ C265S, which lacks the membrane-spanning domain, shows no reduction in intensity upon addition of NaBH4, thereby indicating the lack of ubiquinone (gray lines). The inset shows a difference spectrum before minus after reduction with NaBH4 of full-length wild-type RegB (black line) and of RegB″ C265S (gray line). (B) A red shift of maximum absorption of ubiquinone caused by an increasing amount of bound ubiquinone. Different batches of purified RegB C265S were scanned before (solid lines) and after (dashed lines) NaBH4 reduction. Batch A (black lines) contains 0.44 mol ubiquinone/mol RegB C265S, while batch B (gray lines) contains 0.62 mol ubiquinone/mol RegB C265S. (C) Isolated RegB C265S was reduced and ultrafiltered in buffer containing NaBH4 as described in the text, and then the sample was divided. The gray scans are of the portion that was scanned before (solid gray line) and after (dashed gray line) reduction with NaBH4. The absence of oxidized ubiquinone in this fraction is evident from the difference scan (inset gray line) of before minus after addition of reductant, which shows only a small peak. The black lines are of the other portion, which was reoxidized by the addition of the oxidant K3Fe(CN)6 at 22°C for 5 min (solid black line), followed by the addition of the reductant NaBH4 (dashed black line). The retention of ubiquinone is evident from the difference spectrum (inset black line) of this preparation before minus after reduction with NaBH4.
Purified RegB binds oxidized and reduced ubiquinone.
To ascertain whether RegB is capable of binding reduced ubiquinone (ubiquinol), we undertook an ultrafiltration dilution assay with full-length RegB C265S in a buffer that contained freshly added crystals of sodium borohydride. Spectral analysis indicated that sodium borohydride effectively reduces bound ubiquinone, so if RegB releases ubiquinone upon reduction, then ubiquinone would be removed by ultrafiltration in the presence of sodium borohydride. However, if RegB is capable of binding both oxidized and reduced ubiquinone, then ubiquinone will be retained even after ultrafiltration in the presence of sodium borohydride. In this assay, 100 µl of 20 µM RegB C265S containing ~0.6 mol ubiquinone/mol RegB C265S was diluted 10-fold by addition of degassed buffer containing freshly added sodium borohydride and then reconcentrated by ultrafiltration to 100 µl. Dilution with buffer containing sodium borohydride followed by concentration with ultrafiltration was repeated three times with the sample and then spectrally assayed for the presence of ubiquinone (Fig. 1C). No decrease at 275 nm was observed after addition of sodium borohydride, indicating either that all ubiquinone was removed by ultrafiltration or that RegB exclusively contained reduced ubiquinone (Fig. 1C, gray lines). To clarify whether the sample still contained bound ubiquinone after ultrafiltration, the oxidant K3Fe(CN)6 was added to oxidize any bound ubiquinone, spectrally analyzed, and then subsequently rereduced with sodium borohydride and rescanned (Fig. 1C, black lines). The subsequent spectral change observed at A275 (ΔA275) indicated that reduced RegB C265S that underwent multiple rounds of reduction and ultrafiltration indeed retained 0.3 mol ubiquinone/mol RegB C265S, thereby indicating that RegB is capable of retaining reduced ubiquinone.
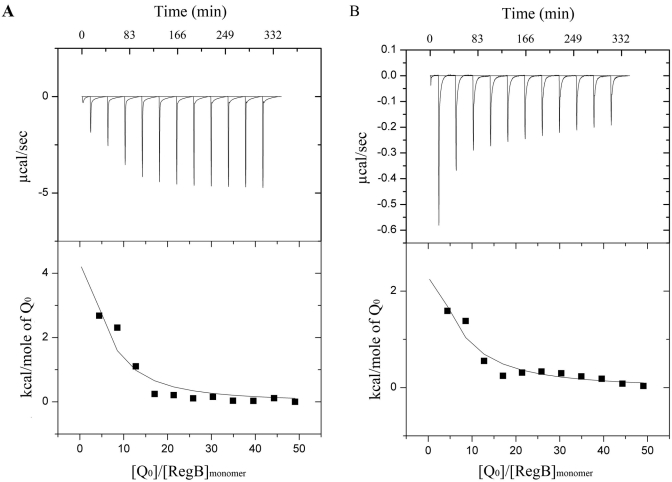
We confirmed that full-length RegB is capable of binding both oxidized and reduced ubiquinone by undertaking analysis of the binding affinity to the soluble ubiquinone analog coenzyme Q0 using isothermal titration calorimetry (ITC). Preliminary data showed weak binding affinities of RegB to both reduced and oxidized Q0 and had c values lower than 1.0. c is the product of protein concentration and binding constant, with a value of 1 often considered the lower limit of ITC experiments. However, it has been demonstrated that a reliable binding constant can be obtained at very low c values using high ligand concentrations and a large volume of injection to drive the binding towards complexation (16, 17). Although accurate enthalpy change (ΔH) and stoichiometry parameter (n) values cannot be defined, the binding constant (Ka) is independent of errors in n at low c values, allowing reliable determination of Ka (17). An optimized titration comprised of eleven injections was used, according to Tellinghuisen (18), where n was fixed at 1 during data fitting. ITC data showed that oxidized ubiquinone has a binding constant of 1 × 104 M−1 (Fig. 2A), while reduced ubiquinone had a similar binding constant of 7.42 × 103 M−1 (Fig. 2B).
FIG 2 .
Ubiquinone binding of RegB studied by isothermal titration calorimetry. (A) Isothermal titration calorimetry of RegB C265S with oxidized ubiquinone. RegB C265S (13 µM) was titrated with 11 consecutive injections of 25 µl 2.5 mM coenzyme Q0 in the presence of 2.5 mM K3Fe(CN)6. (B) Isothermal titration calorimetry of RegB C265S with reduced ubiquinone. RegB C265S (13 µM) was titrated with 11 consecutive injections of 25 µl 2.5 mM coenzyme Q0 in the presence of 5 mM dithionite.
Redox state of bound ubiquinone affects RegB activity in vitro.
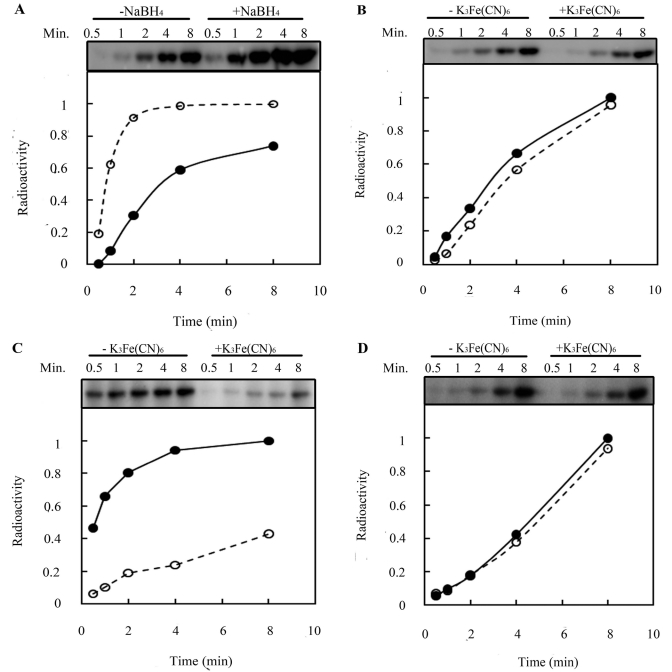
We investigated how ubiquinone regulates RegB kinase activity by assaying the autophosphorylation of RegB C265S under conditions in which the ubiquinone is either reduced or oxidized. For this analysis, bound ubiquinone was either reduced with NaBH4 or oxidized with K3Fe(CN)6 before initiation of the kinase reaction. Analysis of the rate of autophosphorylation demonstrates that the kinase activity of RegB C265S increases 35% by the addition of NaBH4 relative to RegB C265S reactions where no reductant was added (Fig. 3A). A plateau in activity was also reached more quickly in the presence of the reductant. When bound ubiquinone was oxidized by the addition of K3Fe(CN)6, the activity of RegB C265S decreased only slightly (Fig. 3B), likely due to our observation that most of the ubiquinone that is bound to RegB during aerobic purification is already in an oxidized state.
FIG 3 .
Effect of reduction and oxidation of bound ubiquinone on RegB kinase activity. (A) Effect of the reduction of bound ubiquinone on RegB C265S activity. Ten micromolar purified RegB C265S was kept at 22°C for 10 min in the absence (solid line with solid circles) or presence (dashed line with open circles) of NaBH4 before ATP was added to start the kinase reaction. (B) Effect of oxidation of bound ubiquinone on RegB C265S activity. Purified RegB C265S was incubated in the absence (solid line with solid circles) or presence (dashed line with open circles) of 2 mM K3Fe(CN)6 before ATP was added to start the kinase reaction. (C) Effect of the oxidation of reduced bound ubiquinone on RegB C265S activity. Bound ubiquinone in purified RegB C265S was first reduced by NaBH4 at room temperature (RT) for 10 min and then incubated at 22°C for 10 min in the absence (solid line with solid circles) or presence (dashed line with open circles) of 2 mM K3Fe(CN)6 before ATP was added to start the kinase reaction. (D) Effect of oxidation of bound ubiquinone on the activity of RegB″ C265S. RegB″ C265S without the transmembrane domain was treated as described in the legend to panel C as a control.
To clearly demonstrate that oxidization of ubiquinone inhibits activity, we first reduced the bound ubiquinone in RegB C265S with NaBH4 to obtain maximal activity (Fig. 3C, solid line) and then reoxidized the ubiquinone by addition of K3Fe(CN)6 (Fig. 3C, dashed line). In this analysis, the addition of K3Fe(CN)6 to RegB C265S that was first reduced by NaBH4 resulted in a 57% decrease in autophosphorylation activity. As a control, we observed that there is no significant change in kinase activity of the truncated RegB″ C265S cytosolic domain that lacks ubiquinones after similar initial incubation with the reductant NaBH4, followed by treatment with the oxidant K3Fe(CN)6 (Fig. 3D).
Mutations affecting ubiquinone binding.
In a previous study, we identified a quinone binding motif, GGXXNPF, located in a short periplasmic loop between the third and fourth trans-membrane-spanning domains, that is universally conserved among RegB homologs (Fig. 4) (4, 8). Two mutations in this sequence, N110A and F112A, were shown to result in constitutive RegB activity in vivo but were not assayed for their ability to interact with ubiquinones in vitro (8). In this study, we have constructed a more conservative set of substitutions at these positions, N110Q and F112Y, in a RegB C265S background and assayed what effect these new mutations have on in vivo activity and on in vitro ubiquinone binding.
FIG 4 .
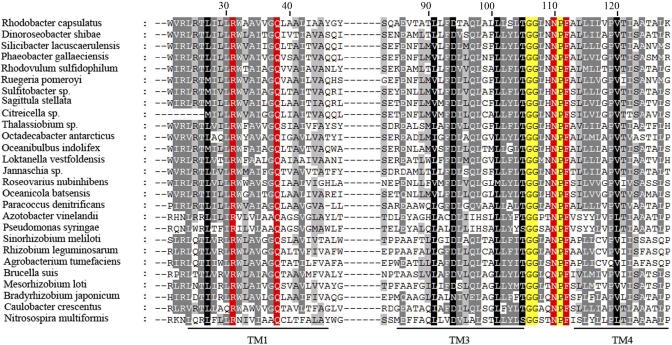
Alignment of the transmembrane domains of representative RegB homologs. Alignment was constructed using Clustal W and edited by GeneDoc. Fully conserved R31, Q38, N110, and F112 residues are highlighted in red. Additional conserved residues constituting part of the GGXXNPF ubiquinone-binding pocket are highlight in yellow. Lines under the alignment denote the transmembrane helixes 1 (TM1), 3, and 4, with helix 2 omitted for brevity.
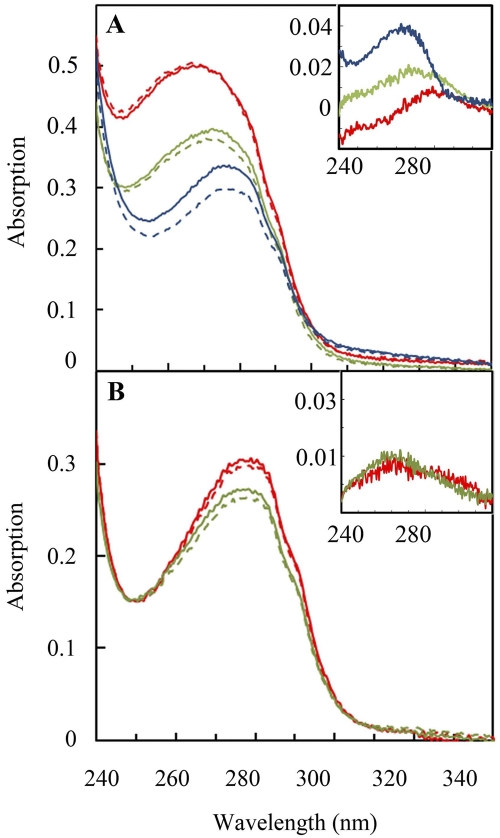
Full-length RegB C265S/N110Q and RegB C265S/F112Y, were expressed, purified, and spectrally assayed for the presence of bound ubiquinone. As shown in Fig. 5A, the ubiquinone content of a representative RegB C265S/N110Q preparation contained only 0.04 mol ubiquinone/mol protein compared to 0.64 mol ubiquinone/mol protein observed for RegB C265S, which does not contain the ubiquinone-binding site mutation. Measurement of protein samples from several independent purifications showed that RegB C265S/N110Q constantly contained considerably less ubiquinone, ranging from 0 to 0.15 mol ubiquinone/mol RegB C265S/N110Q, than RegB C265S, which does not contain the ubiquinone-binding site mutation. Similar analysis of RegB C265S/F112Y shows a more subtle effect on ubiquinone binding, with only a slight reduction of ubiquinone content, ranging from 0.03 to ~0.31 mol ubiquinone/mol RegB C265S/F112Y in several independent isolates (Fig. 5A). Again, there is a red shift of maximum absorption with RegB C265S/N110Q, having a maximal absorption at 269 nm versus 272.5 nm for RegB C265S/F112Y and 277 nm for RegB C265S, due to increasing amounts of bound ubiquinone in these preparations.
FIG 5 .
Involvement of key residues in the membrane-spanning domain that are involved in ubiquinone binding. (A) RegB N110Q/C265S (red), RegB F110Y/C265S (green), and RegB C265S (blue) were scanned before (solid line) and after (dashed line) NaBH4 reduction. The inset shows the difference spectrum before minus after reduction of RegB N110Q/C265S (red line), RegB F110Y/C265S (green line), and RegB C265S (blue line). (B) RegB R31A/C265S (red) and RegB Q38A/C265S (green) were scanned from 240 nm to 340 nm before (solid line) and after (dashed line) NaBH4 reduction. The inset shows the difference spectrum of before minus after reduction of RegB R31A/C265S (red line) and RegB Q38A/C265S (green line). Bound ubiquinone was determined to be 0.16 mol ubiquinone per mol RegB R31A/C265S and 0.20 mol ubiquinone per mol RegB Q38A/C265S.
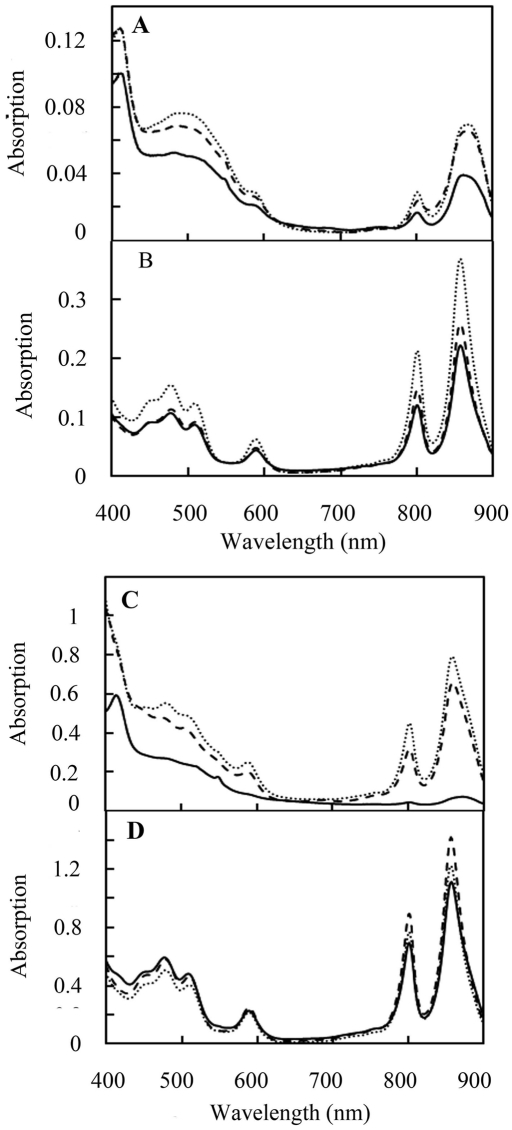
For in vivo analysis, the N110Q and F112Y mutations were recombined to the chromosome of R. capsulatus (Cys265 was not mutated in the in vivo analysis of these ubiquinone-binding site mutants). The amount of photosystem synthesis under different growth conditions was then spectrally assayed, as it has previously been shown that RegB activity affects photosystem synthesis (3, 8). Under semiaerobic conditions, photosystem synthesis increased by 74% in the RegB N110Q mutant strain and by 65% in the RegB F112Y mutant strain (Fig. 6A). Under anaerobic conditions, photosystem expression levels in the RegB N112Q and RegB F112Y mutant strains increased by 67% and 16%, respectively (Fig. 6B).
FIG 6 .
Spectral scans of photopigments in R. capsulatus carrying RegB mutations. (A) Spectral scans of semiaerobically grown wild-type (solid lines), RegB N110Q mutant (dotted lines), and RegB F112Y mutant (dashed lines) cells. (B) Spectrum of anaerobically grown cells as described in the legend to panel A. (C) Spectral scans of semiaerobically grown wild-type (solid lines), RegB R31A mutant (dotted lines), and RegB Q38A mutant (dashed lines) cells. (D) Spectrum of anaerobically grown cells as described in the legend to panel C. For all scans, the cells were grown to the same cell density, collected, sonicated, and scanned from 400 to 900 nm.
An alignment of ~100 RegB homologs revealed that, besides the GGXXNPF motif, there are only two other fully conserved residues, R31 and Q38, in the membrane-spanning region (alignment of representative homologs is shown in Fig. 4) (2, 4). Both of these polar residues are buried in the first transmembrane helix, suggesting a special role in RegB function. To explore the possibility that these two residues may be involved in ubiquinone binding, R31 and Q38 were individually replaced with alanine in the RegB C265S background and spectrally assayed for ubiquinone content upon purification. The bound ubiquinone present in isolated RegB C265S/R31A was 0.12 to 0.21 mol ubiquinone/mol RegB C265S/R31A, while for the C265S/Q38A mutation, there was 0.20 to 0.26 mol ubiquinone/mol RegB C265S/Q38A (Fig. 5B). The decrease in ubiquinone content in these mutants indicates that both R31 and Q38 are likely involved in ubiquinone/ubiquinol binding.
The involvement of R31 and Q38 in controlling RegB activity was confirmed by constructing strains that individually harbor the R31A and Q38A mutations in RegB. Both of these strains exhibit an increase in semiaerobic photosystem expression, with RegB R31A exhibiting an 8.7-fold increase in photopigment biosynthesis and RegB Q38A exhibiting a 7.2-fold increase relative to that of wild-type cells (Fig. 6C). Under anaerobic conditions, photosystem expression levels increased by 9% and 27% in the RegB R31A and RegB Q38A mutant strains, respectively (Fig. 6D).
DISCUSSION
In this study, we provide evidence that RegB binds both oxidized and reduced ubiquinone at nearly equivalent affinities and that only oxidized ubiquinone inhibits kinase activity. The observation that the RegB C265S mutant is still redox responding clearly shows that ubiquinone binding is a signal input capable of functioning independently from the oxidation of cysteine 265. This suggests that ubiquinone/ubiquinol regulates RegB activity through an allosteric effect triggered by binding rather than by disulfide bond formation driven by the oxidizing power of ubiquinone. Studies have shown that ubiquinone binding at one catalytic site in the cytochrome bc1 complex can have an allosteric effect on the other catalytic site; however, this allosteric effect is different in the sense that cytochrome bc1 functions as an enzyme and ubiquinone serves as the substrate (19, 20). To our knowledge, the noncatalytic role of ubiquinone binding, such as that which regulates RegB, in any ubiquinone-binding protein has not been previously reported.
Most ubiquinone-binding sites fall into two groups. Type A sites have a tightly bound ubiquinone that serves as a prosthetic group for carrying one electron. The primary acceptor ubiquinone (QA) in the photosynthetic reaction center is an example of this type (21). Type B sites have an affinity only for a reduced or an oxidized ubiquinone. In these cases, ubiquinones leave after they receive or donate two electrons and protons. Examples of type B sites include the secondary ubiquinones (QB) in photosynthetic reaction centers (22–24) that release ubiquinol after ubiquinone is fully reduced and protonated and DsbB that binds oxidized ubiquinone and uses its oxidative power to generate disulfide bonds (13, 25, 26). Many type A and type B ubiquinone-binding sites have a histidine-containing triad comprised of an aliphatic (X)3-H-(X)2/3-(L/T/S) sequence. The histidine triad is often located at the end of the transmembrane helix or in a short periplasmic loop (27, 28). There are exceptions to this triad, such as DsbB, where residues from two adjacent periplasmic loops form a discontinuous, but spatially similar, histidine-containing triad (25, 29).
A simple sequence alignment suggests that the GGXXNPF ubiquinone-binding site in RegB shares little homology with prototypical histidine-containing triads. However, the absence of primary sequence conservation with the histidine triad may be misleading, as there are several similarities between the genetically and biochemically characterized (8) ubiquinone-binding site in RegB and the atypical ubiquinone-binding site present in ubiquinol oxidase (30). The crystal structure of ubiquinol oxidase indicates that the C-4 carbonyl group of the oxidized ubiquinone ring is hydrogen bonded to a conserved polar His-X-X-Gln sequence that is located in a short periplasmic loop at a lipid bilayer/solvent interphase (30). On the other side of the ubiquinone ring, the C-1 carbonyl is hydrogen bonded to the Arg71 and Asp75 residues, which are surprisingly deeply buried in the lipid bilayer. Like the His-X-X-Gln sequence of ubiquinol oxidase, the conserved Gly-Gly-X-X-Asn-Pro-Phe ubiquinone-binding sequence in RegB is located at a short periplasmic loop. Mutation of Asn110 in RegB inhibits ubiquinone binding, indicating that this residue could be hydrogen bonded to a carbonyl residue of ubiquinone. π-π stacking interactions between the aromatic side group of Phe and Trp to the benzoquinone ring have also been shown to be important in ubiquinone binding in many proteins (27, 31–33), so the fully conserved Phe112 in RegB could allow binding of both oxidized and reduced forms of ubiquinone through ring-stacking interactions. Mutations of the two universally conserved Arg31 and Gln38 polar residues in RegB’s transmembrane helix 1 also are critical residues in ubiquinone binding and may thus serve a similar function of forming a hydrogen bond to the C-1 carbonyl of oxidized ubiquinone, not unlike that of Arg71 and Asp75 in ubiquinol oxidase.
Our working model is that the modest ubiquinone-binding affinity of RegB allows ubiquinone bound to the kinase to readily equilibrate with the ubiquinone pool by diffusion exchange. When the ubiquinone pool becomes more oxidized, then more RegB would contain bound oxidized ubiquinone, presumably with the C-4 carbonyl of the ubiquinone ring hydrogen bonded to Asn110 and the C-1 carbonyl of the ubiquinone ring hydrogen bonded to Arg31 and Gln38. When the ubiquinone pool becomes predominantly reduced to ubiquinol, then equilibration/diffusion would drive more RegB to interact with ubiquinol via stacking interactions with Phe112. Reduction of the C-4 and C-1 carbonyls that occurs in ubiquinol would lead to loss of hydrogen bonding to Asn110, Arg31, and Gln38. This loss of hydrogen bonding would presumably trigger a conformational change in RegB that allows increased kinase activity. Note that the ability of RegB to equilibrate with both oxidized and reduced ubiquinones would allow modest changes in the ubiquinone/ubiquinol ratio to alter the overall average activity of membrane-bound RegB in a cell, not unlike that of a rheostat. The redox state of the ubiquinone pool is not only affected by environmental oxygen tension (9, 34) but also affected by light intensity, the redox state of cytosolic metabolites exchanging electrons, such as NADH and succinate (35), and metabolic processes that use reducing equivalents, such as carbon fixation and nitrogen fixation, all of which being part of the RegB/RegA regulon. Thus, monitoring the redox state of the ubiquinone pool gives RegB the capability to monitor the overall cellular energy and redox state of the cell, and with this capability, it functions as a global regulator to coordinate the synthesis of a complex network of energy-related processes. This model is supported by mathematical modeling by Klamt et al. (35), which indicates that alterations of the redox state of ubiquinone is a suitable signal for the regulation of photosynthetic gene expression. Grammel and Ghosh also found a correlation between the redox state of the ubiquinone pool and the expression level of photosystem in Rhodospirillum rubrum (9). They suggested that RegB activity is highly cooperatively regulated by the redox state of the ubiquinone pool (9), although this study did not take into account the fact that there is also a redox-sensing role of cytosolic Cys265. Given that the RegB Cys265 mutant was used in this study, it is likely that the ubiquinone-binding site as defined in this study, and in our previous study (8), is just one of several redox-dependent signal inputs that are ultimately responsible for controlling the kinase activity of RegB. Just how much RegB activity is controlled by ubiquinone binding and Cys265 remains unclear. Whether there is cooperation between ubiquinone binding and the formation of the disulfide bond at Cys265 needs to be further investigated.
MATERIALS AND METHODS
Strains, media, and growth conditions.
E. coli strains DH5α and SM10 λpir were used for cloning and conjugal transfer of plasmids, respectively. The mobilization of plasmids from E. coli into R. capsulatus strains was performed as described in reference 8. E. coli cells were routinely grown in Luria-Bertani (LB) medium at 37°C.
R. capsulatus strain SB1003 was used as the parent strain to generate all chromosomal mutants with a chromosomal in-frame deletion of regB, as previously reported (8). All R. capsulatus strains were grown in peptone-yeast extract (PY) medium supplemented with 10 µg/ml spectinomycin at 34°C as described previously (8).
Plasmid construction.
Plasmid pET28RegBfull that expresses full-length C-terminal His-tagged RegB was described previously (8). An overexpression plasmid for expressing truncated RegB, pET28RegB″, was constructed by cloning a DNA fragment encoding the RegB cytosolic domain starting with M196 and to the native stop codon, using forward and reverse primers (5′-TACCATGTCGGATGCGCTTTTCGCGACA and 5′-ATCTCGAGGGCGGTGATCGGAACATTC) that incorporated NcoI and XhoI sites, respectively. The PCR-amplified truncated RegB gene segment was then cloned into similar sites of pET28.
All point mutations were generated using the QuikChange mutagenesis kit obtained from Stratagene. For C265S mutations, primer 5′-TGAACAGGCCGAACGCTCCCGCGACAT and a complementary primer were used. For N110Q mutations, primer 5′-CGGGCGGGCTGAACCAACCCTTCGCGCTTTT and a complementary primer were used. For F112Y mutations, primer 5′-GCGGGCTGAACCCCTACGCGCTTTTG and a complementary primer were used. For R31A mutations, primer 5′-GACATTGATCCTGTTGGCATGGGCGGCGGTCGTC and a complementary primer were used. For Q38A mutations, primer 5′-GGCCGCGGTCGTCGGTGCGCTGGCGGC-GCTGAT and a complementary primer were used.
Overexpression and purification of full-length RegB.
E. coli cells that overexpressed full-length wild-type or mutant versions of RegB were grown in Terrific broth (TB) supplemented with 25 µg/ml kanamycin at 37°C until A600 reached 0.4 to 0.6, induced by the addition of 75 µM mM isopropyl β-d-thiogalactopyranoside (IPTG), and then grown overnight at 16°C. The cells were harvested by centrifugation, resuspended in 20 mM sodium phosphate, pH 7.6, and 100 mM NaCl, and then lysed by the M-110L microfluidizer processor (Microfluidics). The supernatant of the cell lysate was centrifuged at 150,000 × g for 1.5 h at 4°C to pellet membranes. The membranes were solubilized in 20 mM sodium phosphate at pH 7.6, 300 mM NaCl, 40 mM imidazole, 20% glycerol, and 1% n-dodecyl-β-d-maltoside (DDM) (Pierce) to a final concentration of 3 to 5 mg protein/milliliter using the RC DC protein assay kit (Bio-Rad) for protein concentration determination. The solubilized membrane fraction was rotated in a 50-ml conical tube at 4°C for 2 h and then centrifuged at 150,000 × g to remove insoluble material. The resultant supernatant was loaded onto a 1-ml HisTrap column (GE), washed with 50-column-volume wash buffer containing 20 mM sodium phosphate at pH 7.6, 300 mM NaCl, 40 mM imidazole, 10% glycerol, and 0.05% DDM. RegB was eluted with 20 mM sodium phosphate at pH 7.6, 300 mM NaCl, 250 mM imidazole, 10% glycerol, and 0.05% DDM.
BL21(λDE3) cells that overexpress RegB″ were grown in TB at 37°C until A600 reached 0.4 to 0.6, induced by the addition of 0.4 mM IPTG, and then grown overnight at 16°C. The cell pellet was collected and resuspended in 20 mM Tris, pH 7.9, and 150 mM NaCl. The cells were lysed by passing them through an M-110L microfluidizer processor (Microfluidics) three times. The supernatant of the cell lysis was loaded into a 1-ml HisTrap column, washed with 30 ml 20 mM Tris at pH 7.9, 150 mM NaCl, and 30 mM imidazole, and then eluted with 20 mM Tris at pH 7.9, 150 mM NaCl, and 250 mM imidazole.
Spectral analysis.
The analysis of oxidized bound ubiquinone involved suspending 5 µM RegB or RegB″ in 50 mM sodium phosphate at pH 7.6, 300 mM NaCl, 10% glycerol, and 0.05% DDM and then scanning from 240 to 340 nm. A few grains of solid sodium borohydride (NaBH4) were then added to the cuvette to reduce ubiquinone, incubated at 22°C for 5 min, and then rescanned from 240 to 340 nm to see the optical changes centered at A275. The amount of oxidized bound ubiquinone was calculated using Δε275 of 12.25 mM−1 (13). For wild-type RegB, RegB C265S, or each of the binding site mutants, at least three independent preparations were measured to confirm that the results are reproducible.
Spectral scans used to analyze photopigments in R. capsulatus cell membranes were performed as previously described (7). The absorption at 850 nm, which is the wavelength of the light-harvesting bacteriochlorophyll (BChl) complex, was measured and compared.
Kinase assays.
Kinase assays were performed as described previously (6). To alter the redox state of bound ubiquinone, a few grains of solid NaBH4 or 2 mM potassium ferricyanide [K3Fe(CN)6] were added to the reaction mixture and incubated at 22°C for 5 min before 1.0 mM ATP and 200 to 400 µCi [γ-32P]ATP (7,000 Ci/mmol; MP Biomedicals) were added.
ITC analysis.
To determine the binding affinity of reduced Q0 to RegB C265S, 5 mM dithionite was included in a degassed buffer containing 50 mM sodium phosphate at pH 7.6, 300 mM NaCl, 10% glycerol, and 0.05% DDM in an anaerobic hood filled with 90% nitrogen, 5% carbon dioxide, and 5% hydrogen. The same buffer was later used to make a 2.5 mM coenzyme Q0 solution and to exchange the buffer of RegB C265S by passing it through a 10DG column (Bio-Rad). The ITC chamber (MicroCal LLC) was purged and filled with argon gas, and the sample cell was then filled with 13 µM RegB C265S and titrated with 2.5 mM Q0. Eleven consecutive 25-µl Q0 injections were then performed at 30-min intervals. The dilution heat of Q0 was obtained by injecting Q0 solution into buffer using the same parameters in a separate experiment. For the binding affinity of oxidized Q0 to RegB, 2.5 mM K3Fe(CN)6 was added to the buffer. Buffer containing 50 mM sodium phosphate at pH 7.6, 300 mM NaCl, 10% glycerol, 0.05% DDM, and 2.5 mM K3Fe(CN)6 was used to exchange the buffer of the RegB C265S sample and to make a 2.5-mM solution of Q0. The Q0 solution was rotated at 4°C for 2 h, and the oxidation of Q0 was confirmed by the solution turning dark brown. The same parameters were used for ITC experimental analysis of binding of reduced Q0 to RegB C265S to obtain the binding and dilution heat of oxidized Q0.
ACKNOWLEDGMENT
This work was supported by National Institutes of General Medical Sciences grant GM40941 awarded to C.E.B.
Footnotes
Citation Wu, J., and C. E. Bauer. 2010. RegB kinase activity is controlled in part by monitoring the ratio of oxidized to reduced ubiquinones in the ubiquinone pool. mBio 1(5):e00272-10. doi:10.1128/mBio.00272-10.
REFERENCES
- 1. Madigan M. T., Gest H. 1978. Growth of a photosynthetic bacterium anaerobically in darkness, supported by “oxidant-dependent” sugar fermentation. Arch. Microbiol. 117:119–122 [DOI] [PubMed] [Google Scholar]
- 2. Elsen S., Swem L. R., Swem D. L., Bauer C. E. 2004. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 68:263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mosley C. S., Suzuki J. Y., Bauer C. E. 1994. Identification and molecular genetic characterization of a sensor kinase responsible for coordinately regulating light harvesting and reaction center gene expression in response to anaerobiosis. J. Bacteriol. 176:7566–7573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu J., Bauer C. E. 2007. RegB/RegA, a global redox-responding two-component system, p. 131–148 In Utsumi R., Bacterial signal transduction: network and drug targets. Landes Bioscience Eurekah, Georgetown, TX [Google Scholar]
- 5. Inoue K., Kouadio J. L., Mosley C. S., Bauer C. E. 1995. Isolation and in vitro phosphorylation of sensory transduction components controlling anaerobic induction of light harvesting and reaction center gene expression in Rhodobacter capsulatus. Biochemistry 34:391–396 [DOI] [PubMed] [Google Scholar]
- 6. Bird T. H., Du S., Bauer C. E. 1999. Autophosphorylation, phosphotransfer, and DNA-binding properties of the RegB/RegA two-component regulatory system in Rhodobacter capsulatus. J. Biol. Chem. 274:16343–16348 [DOI] [PubMed] [Google Scholar]
- 7. Swem L. R., Kraft B. J., Swem D. L., Setterdahl A. T., Masuda S., Knaff D. B., Zaleski J. M., Bauer C. E. 2003. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 22:4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swem L. R., Gong X., Yu C. A., Bauer C. E. 2006. Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J. Biol. Chem. 281:6768–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grammel H., Ghosh R. 2008. Redox-state dynamics of ubiquinone-10 imply cooperative regulation of photosynthetic membrane expression in Rhodospirillum rubrum. J. Bacteriol. 190:4912–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim Y. J., Ko I. J., Lee J. M., Kang H. Y., Kim Y. M., Kaplan S., Oh J. I. 2007. Dominant role of the cbb3 oxidase in regulation of photosynthesis gene expression through the PrrBA system in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 189:5617–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Georgellis D., Kwon O., Lin E. C. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314–2316 [DOI] [PubMed] [Google Scholar]
- 12. Malpica R., Franco B., Rodriguez C., Kwon O., Georgellis D. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. U. S. A. 101:13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bader M. W., Xie T., Yu C. A., Bardwell J. C. 2000. Disulfide bonds are generated by quinone reduction. J. Biol. Chem. 275:26082–26088 [DOI] [PubMed] [Google Scholar]
- 14. Yu L., Deng K., Yu C. A. 1995. Cloning, gene sequencing, and expression of the small molecular mass ubiquinone-binding protein of mitochondrial ubiquinol-cytochrome c reductase. J. Biol. Chem. 270:25634–25638 [DOI] [PubMed] [Google Scholar]
- 15. Shenoy S. K., Yu L., Yu C. 1999. Identification of quinone-binding and heme-ligating residues of the smallest membrane-anchoring subunit (QPs3) of bovine heart mitochondrial succinate:ubiquinone reductase. J. Biol. Chem. 274:8717–8722 [DOI] [PubMed] [Google Scholar]
- 16. Turnbull W. B., Daranas A. H. 2003. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 125:14859–14866 [DOI] [PubMed] [Google Scholar]
- 17. Tellinghuisen J. 2008. Isothermal titration calorimetry at very low c. Anal. Biochem. 373:395–397 [DOI] [PubMed] [Google Scholar]
- 18. Tellinghuisen J. 2005. Optimizing experimental parameters in isothermal titration calorimetry. J. Phys. Chem. B 109:20027–20035 [DOI] [PubMed] [Google Scholar]
- 19. Cooley J. W., Ohnishi T., Daldal F. 2005. Binding dynamics at the quinone reduction (Qi) site influence the equilibrium interactions of the iron sulfur protein and hydroquinone oxidation (Qo) site of the cytochrome bc1 complex. Biochemistry 44:10520–10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cooley J. W., Lee D. W., Daldal F. 2009. Across membrane communication between the Q(o) and Q(i) active sites of cytochrome bc(1). Biochemistry 48:1888–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okamura M. Y., Paddock M. L., Graige M. S., Feher G. 2000. Proton and electron transfer in bacterial reaction centers. Biochim. Biophys. Acta 1458:148–163 [DOI] [PubMed] [Google Scholar]
- 22. Rich P. R. 1984. Electron and proton transfers through quinones and cytochrome bc complexes. Biochim. Biophys. Acta 768:53–79 [DOI] [PubMed] [Google Scholar]
- 23. Skulachev V. P. 1988. Membrane bioenergetics. Springer, Berlin, Germany [Google Scholar]
- 24. Ke B. 2001. Photosynthesis: photobiochemistry and photobiophysics. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 25. Inaba K., Murakami S., Suzuki M., Nakagawa A., Yamashita E., Okada K., Ito K. 2006. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell 127:789–801 [DOI] [PubMed] [Google Scholar]
- 26. Zhou Y., Cierpicki T., Jimenez R. H., Lukasik S. M., Ellena J. F., Cafiso D. S., Kadokura H., Beckwith J., Bushweller J. H. 2008. NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Mol. Cell 31:896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fisher N., Rich P. R. 2000. A motif for quinone binding sites in respiratory and photosynthetic systems. J. Mol. Biol. 296:1153–1162 [DOI] [PubMed] [Google Scholar]
- 28. Gong X., Xie T., Yu L., Hesterberg M., Scheide D., Friedrich T., Yu C. A. 2003. The ubiquinone-binding site in NADH:ubiquinone oxidoreductase from Escherichia coli. J. Biol. Chem. 278:25731–25737 [DOI] [PubMed] [Google Scholar]
- 29. Xie T., Yu L., Bader M. W., Bardwell J. C., Yu C. A. 2002. Identification of the ubiquinone-binding domain in the disulfide catalyst disulfide bond protein B. J. Biol. Chem. 277:1649–1652 [DOI] [PubMed] [Google Scholar]
- 30. Abramson J., Riistama S., Larsson G., Jasaitis A., Svensson-Ek M., Laakkonen L., Puustinen A., Iwata S., Wikstrom M. 2000. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat. Struct. Biol. 7:910–917 [DOI] [PubMed] [Google Scholar]
- 31. Albury M. S., Elliott C., Moore A. L. 2010. Ubiquinol-binding site in the alternative oxidase: mutagenesis reveals features important for substrate binding and inhibition. Biochim. Biophys. Acta. 1797:1933–1939 [DOI] [PubMed] [Google Scholar]
- 32. Jordan P., Fromme P., Witt H. T., Klukas O., Saenger W., Krauss N. 2001. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411:909–917 [DOI] [PubMed] [Google Scholar]
- 33. Andrade S. L., Patridge E. V., Ferry J. G., Einsle O. 2007. Crystal structure of the NADH:quinone oxidoreductase WrbA from Escherichia coli. J. Bacteriol. 189:9101–9107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parson W. 1978. Role of the reaction center in photosynthesis, p. 317–322 In Clayton R. K., Sistrom W. R., The photosynthetic bacteria. Plenum Press, New York, NY. [Google Scholar]
- 35. Klamt S., Grammel H., Straube R., Ghosh R., Gilles E. D. 2008. Modeling the electron transport chain of purple non-sulfur bacteria. Mol. Syst. Biol. 4:156–174 [DOI] [PMC free article] [PubMed] [Google Scholar]