Abstract
Coilin is a nuclear protein that plays a role in Cajal body formation. The function of nucleoplasmic coilin is unknown. Here we report that coilin interacts with Ku70 and Ku80, which are major players in the DNA repair process. Ku proteins compete with SMN and SmB′ proteins for coilin interaction sites. The binding domain on coilin for Ku proteins cannot be localized to one discrete region, and only full-length coilin is capable of inhibiting in vitro non-homologous DNA end joining (NHEJ). Since Ku proteins do not accumulate in CBs, these findings suggest that nucleoplasmic coilin participates in the regulation of DNA repair.
Keywords: Coilin, Ku70, Ku80, NHEJ
1. Introduction
The highly organized eukaryotic nucleus allows for regulated gene expression and appropriate processing of RNA transcripts. Small nuclear ribonucleoproteins (snRNPs) play a pivotal role in the splicing of pre-mRNA. The biogenesis of snRNPs is a multistep process, and in cells with high transcription demands, some of these steps take place in subnuclear domains known as Cajal bodies (CBs) [1]. CBs are present in yeast, plants, insects and mammals and, in addition to the snRNP maturation, also participate in telomerase formation [2,3]. Interestingly, mammalian CBs share components with histone locus bodies and thus may impact histone gene transcription [2,4,5].
Coilin is the CB signature protein and plays a major role in bringing all components of the CB together to facilitate its various functions [6–8]. For example, coilin directly interacts with the survival of motor neuron (SMN) protein and several Sm proteins [9–11]. The phosphorylation status of coilin influences CB formation, self-interaction and association with SMN and Sm proteins [12–15]. Interestingly, 70% of coilin is not found in the CB, but is nucleoplasmic [16]. In comparison to our knowledge about coilin activity in the CB, essentially nothing is known about the function of nucleoplasmic coilin, where the vast majority of the protein resides.
To explore the functional importance of nucleoplasmic coilin, we have conducted coilin pulldown assays coupled with MS/MS analysis and identified the Ku proteins as interaction partners. Both the Ku proteins, Ku80 and Ku70, associate with coilin in vivo and directly in vitro. The association of Ku80 or Ku70 with coilin modulates its interaction with SMN and SmB′. The addition of recombinant coilin inhibits in vitro non-homologous DNA end joining (NHEJ), and thus demonstrates a possible role for nucleoplasmic coilin in regulating DNA repair.
2. Materials and methods
2.1. Cell culture and DNA constructs
HeLa cells were obtained from the American Type Culture Collection (ATCC) and cultured as previously described [17]. GST-coilin constructs and purification have been previously described [15]. Ku80 and Ku70 cDNAs were purchased (Open Biosystems) and cloned in frame into pET28a (Novagen) using standard molecular biology techniques. GST-N-terminal coilin (N-362) and His-N-terminal Ku80 (N-565) were prepared using the Quick Change Mutagenesis kit (Stratagene) and verified by sequencing.
2.2. In vitro binding assays and co-immunoprecipitation
GST-pulldown assays and immunoprecipitations were conducted as described [11,15]. An antibody to GFP was used as a negative control for the immunoprecipitation reactions. Proteins were detected using antibodies to coilin (H300, Santa Cruz), SMN (BD BioSciences), Ku80 (Abcam), SmB (Sigma-Aldrich) or the T7-tag (Novagen).
2.3. Identification of coilin interacting proteins
HeLa cells were flash frozen and lysed in 1 mL modified RIPA [13]. Lysate was then pre-cleared with 50 uL 50% glutathione sepharose beads (GE Healthcare). The supernatant was next incubated with GST or Coilin-GST fusion proteins (on beads) at 4°C for 4 hours, followed by extensive washing and SDS-PAGE. The gel was silver stained and bands were excised. Proteins in the bands were identified by the LCMS facility at Yale University (New Haven, CT).
2.4. Non-homologous end joining assay
NHEJ assays were carried out as described [18,19] with a few modifications. Briefly, HeLa cells were harvested, lysed by 3 cycles of freeze-thawing in liquid nitrogen and resuspended in hypotonic lysis buffer (10 mM Tris-HCl pH 8, 60 mM KCl, 1 mM EDTA pH 8, 1 mM DTT, protease inhibitor cocktail (Roche). The substrate for NHEJ (250 ng/reaction) was pBluescript digested using EcoRI/SalI (to generate non-homologous ends). GST- fusion proteins used in the NHEJ assays were eluted from glutathione beads with reduced glutathione per standard protocols and added to the reaction along with the HeLa lysate at the start of the reaction.
3. Results and discussion
3.1. Coilin interacts with Ku proteins
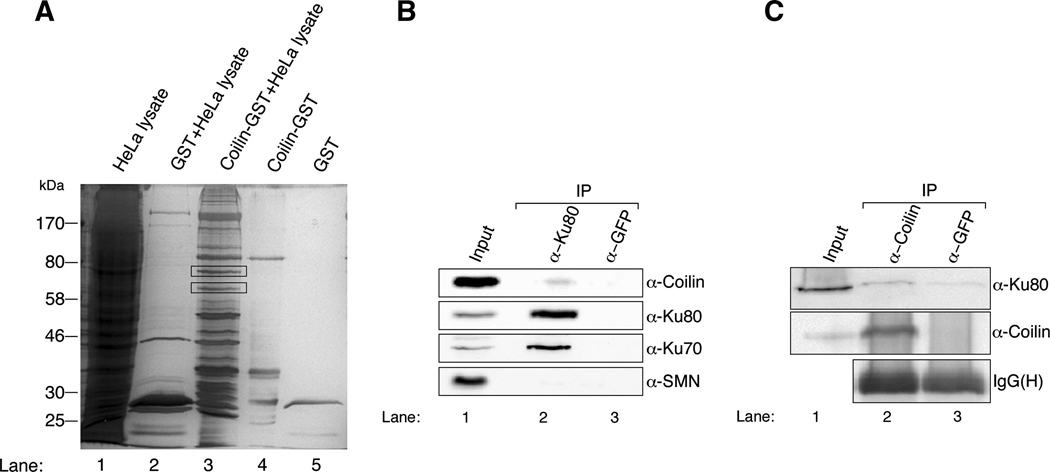
To identify coilin interaction proteins, we conducted coilin-GST pulldown assays with HeLa lysate (Fig 1A). The two bands marked in lane 3 were analyzed by mass spectroscopy and found to contain the ATP-dependent DNA helicase 2 subunit 2 (Ku80) and ATP-dependent DNA helicase 2 subunit 1 (Ku70) (Table 1). Ku proteins are nucleoplasmic and do not accumulate in CBs, suggesting that the interaction between coilin and Ku proteins takes place in the nucleoplasm. These interactions were verified with endogenous proteins by co-immunoprecipitation using Ku80 (Fig 1B) or coilin (Fig 1C) antibodies. Since SMN interacts with coilin and is found in CBs, its absence in the Ku80 complex (Fig 1B) may again indicate that nucleoplasmic coilin, rather than coilin in the CB, forms an association with Ku proteins.
Fig. 1.
Ku70 and Ku80 interact with coilin. (A) HeLa lysate incubated with GST or coilin-GST beads was subjected to 10% SDS-PAGE and silver stained. The gel shows input (lane 1), GST incubated with lysate (lane 2), coilin-GST incubated with lysate (lane 3), coilin-GST beads (lane 4) and GST beads (lane 5). The protein bands marked with rectangles in lane 3 were subjected to mass spectroscopic analysis. (B and C) Ku70 and Ku80 form a complex with coilin that does not include SMN. HeLa lysate was subjected to immunoprecipitation with anti-Ku80, anti-coilin or anti-GFP antibodies (as a negative control), followed by SDS-PAGE and western transfer. The blot was separately probed with antibodies to coilin, Ku80, SMN and Ku70. The immunoglobulin heavy chain is shown to indicate that approximately equal amounts of anti-coilin and anti-GFP antibodies were used. The input lane for both (A) and (B) represents 3% of the lysate used.
Table 1.
Mass spectroscopic analysis data of coilin interacting molecules. Ku80 and Ku70 were recovered from the higher and lower molecular weight protein bands, respectively.
| Protein ID | Gene Symbol | Protein Name | Molecular Weight | % Coverage |
|---|---|---|---|---|
| IPI00220834 | XRCC5 | ATP-dependent DNA helicase 2 subunit 2 (Ku80/Ku86) | 82652 | 67.1 |
| IPI00644712 | XRCC6 | ATP-dependent DNA helicase 2 subunit 1 (Ku70) | 69799 | 61.9 |
3.2. Ku proteins directly interact with coilin
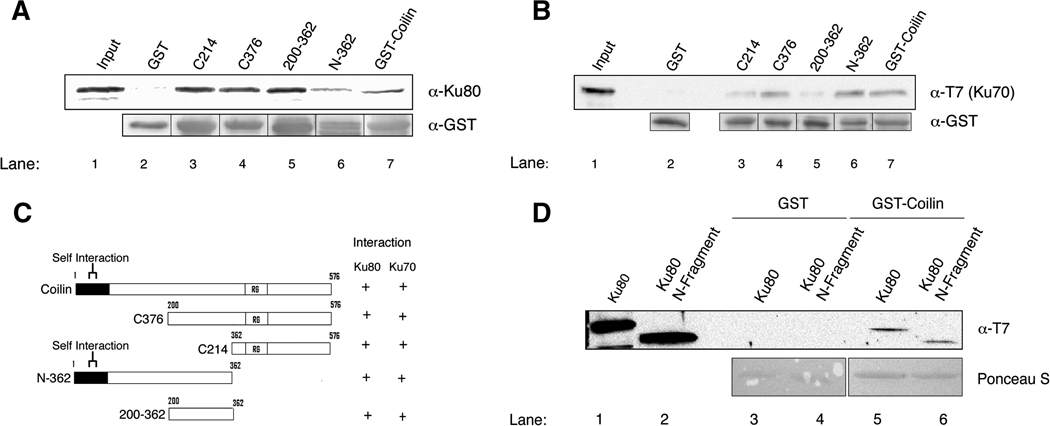
To investigate if the interaction between coilin and Ku proteins is direct, we conducted GST-pulldown assays using bacterially purified proteins (Fig 2). Full-length (576 aa) human coilin fused to GST and soluble, T7-tagged Ku proteins were used in the assays. Various GST-coilin fragments were used to delimit the binding region for Ku proteins on coilin (Fig 2C). As shown in Fig 2A (lane 7) and Fig 2B (lane 7), full-length GST-coilin interacted directly with Ku80 and Ku70, respectively. Ku proteins were also recovered with different coilin fragments (Fig 2A, B), indicating that there are several distinct Ku protein binding sites along the coilin protein (Fig 2C). Deletion of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) interaction domain of Ku80 (aa 565–732) [20] did not impact coilin association (Fig 2D), indicating that coilin binds on Ku80 at a different location than DNA-PKcs.
Fig. 2.
Coilin directly interacts with Ku70 and Ku80. GST-pulldown of Ku80 and Ku70 by GST-fused full-length coilin or mutants thereof. (A) Bacterially purified GST or GST fused to various coilin fragments was incubated with bacterially purified soluble Ku80, followed by extensive washing of the beads, SDS-PAGE, and western transfer. The blots were probed separately with anti-Ku80 and anti-GST antibodies. The input lane accounts for 10% of Ku80 used in the pulldown reactions. (B) Soluble, bacterially expressed T7-tagged Ku70 was incubated with GST and GST-coilin fusion proteins and treated as described above. The blot was probed with antibodies to the T7 tag to detect Ku70, and GST antibodies to monitor the level of GST fusion protein used in each reaction. (C) Schematic representation of the different coilin fragments that were fused to GST and used in the pulldown reactions. The coilin self-interaction domain and RG box, which mediates coilin interaction with SMN, are indicated. (D) The C-terminus of Ku80 is not necessary for coilin interaction. Soluble full length T7-tagged Ku80 (N-732aa) and an N-terminal T7-tagged Ku80 (N-565aa) fragment were incubated with GST (lanes 3 and 4) or GST-coilin (lanes 5 and 6) beads. The washed beads were subjected to SDS-PAGE, western blotted and probed with T7 antibodies to detect Ku80 or the N-565 fragment. Ponceau S (bottom panels) was used to detect the amount of GST proteins in the reactions, and the input lanes account for 10% of the soluble protein used in the pulldown reactions.
3.3. Ku proteins compete with SMN and SmB′ for binding sites on coilin
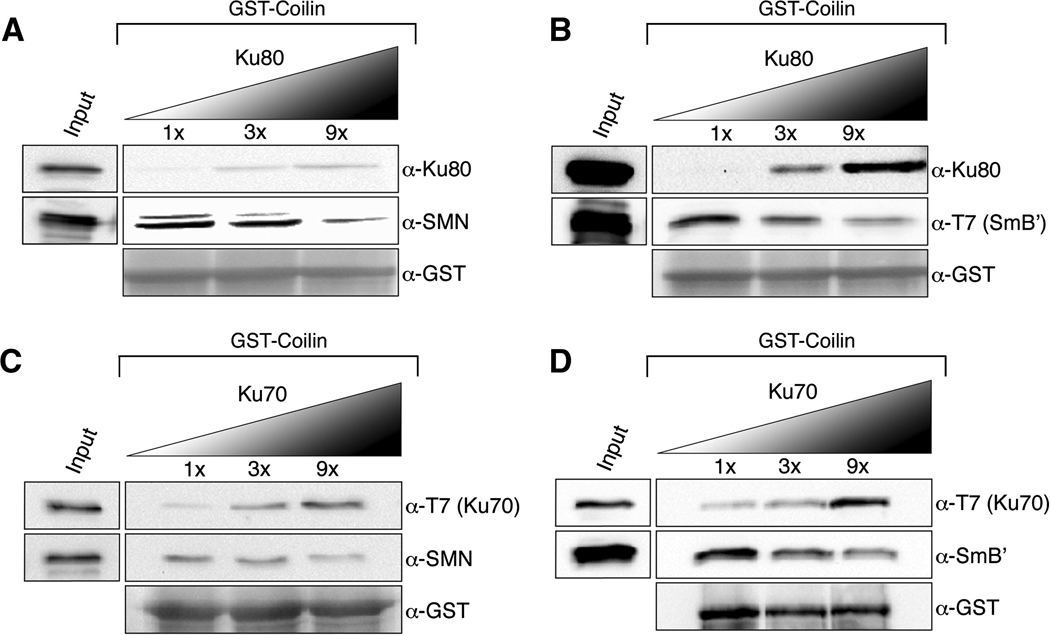
Previous studies showed that SMN and SmB′ compete for binding sites on coilin [9]. The interaction of Ku proteins with coilin drove us to investigate whether the Ku proteins also compete with SMN and SmB′ for binding sites on coilin. Competition GST pulldown experiments were conducted by incubating a constant amount of GST-coilin with a fixed amount of SMN or SmB′ and increasing concentrations of Ku proteins in separate reactions (Fig 3). The binding of both SMN and SmB′ to coilin was reduced upon increasing amounts of Ku80 (Fig 3AB) or Ku70 (Fig 3CD). The competition pulldown findings provide a mechanism whereby coilin in the CB is more likely to be complexed with SMN and Sm proteins, whereas these associations are reduced in the nucleoplasm due to coilin interaction with Ku proteins.
Fig. 3.
SMN and SmB′ compete with Ku80 and Ku70 for binding sites on coilin. GST-pulldown assays were conducted with a fixed amount of GST-coilin and SMN or SmB′ and increasing amounts (1X to 9X) of Ku80 or Ku70. Input lanes accounts for 20% of the protein used in the 1X reaction. (A) SMN and Ku80 are competing for binding sites on coilin. The western blot was probed with Ku80, SMN and GST antibodies. (B) Ku80 and SmB′ are competing for binding sites on coilin. The western blot was probed with Ku80, T7 (to detect SmB′) and GST antibodies. (C) Ku70 and SMN compete for binding sites on coilin. The western blot was probed with T7 (to detect Ku70), SMN and GST antibodies. (D) Ku70 and SmB′ compete for binding sites on coilin. The western blot was probed with anti-T7 to detect Ku70, anti-SmB to detect SmB′ and anti-GST to detect GST-coilin.
3.4. Coilin inhibits in vitro NHEJ
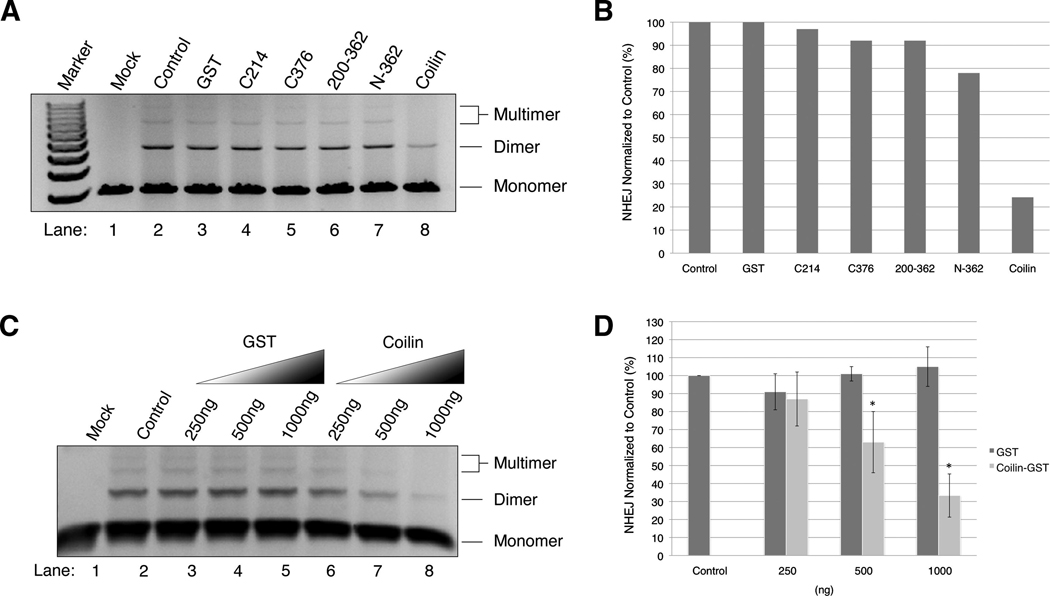
The Ku proteins Ku70 and Ku80 (also known as Ku86), were discovered as an autoantigen in patients with scleroderma-polymyositis overlap syndrome [21]. Ku proteins play a major role in NHEJ along with DNA-PKcs, Artemis, DNA ligase IV and XRCC4 [22]. The interaction of coilin with Ku proteins suggests that coilin might have a role in some aspect of the NHEJ pathway. To test this, we conducted in vitro NHEJ using HeLa lysate and EcoRI/SalI digested pBluescript with or without the addition of bacterially expressed coilin or coilin fragments (Fig 4). Compared to the formation of dimers and multimers in the control reaction (Fig 4A, lane 2 and quantified in Fig 4B), full-length coilin inhibited NHEJ but the other coilin constructs had no effect. Increasing concentrations of coilin correlated with decreasing amounts of NHEJ (Fig 4C and quantified in Fig 4D). The NHEJ efficiency was significantly (p<0.05) inhibited with increasing concentrations (500 and 1000ng) of soluble coilin. These observations suggest that coilin might be playing a regulatory role in the DNA repair mechanism. Since Ku proteins form a heterodimer to circle the broken DNA ends and recruit DNA-PKcs and other proteins needed to carry out end joining [23], we speculate that increasing coilin in the NHEJ reactions may prevent efficient recruitment of these other components. Interestingly, coilin localizes to centromeres damaged by herpes simplex virus infection [24] and UV-C treatment fragments CBs [25]. Therefore, the connection between DNA repair and coilin is not without precedence in the literature. Intriguingly, Ku80’s essential role in human cells is not DNA repair but telomere loss [26]. Since CBs participate in certain aspects of telomerase biogenesis and delivery, it is possible that the interaction between coilin and Ku80 influences telomere length.
Fig. 4.
Coilin inhibits NHEJ efficiency. In vitro NHEJ reactions were carried out with pBluescript which had non-homologous ends and HeLa lysate plus bacterially purified GST or GST-fused full length or coilin fragments. Mock represents the NHEJ reaction without lysate. Control represents the NHEJ reaction with DNA and lysate. The rest of the NHEJ reactions represent DNA, lysate and addition of respective proteins. (A) Full length coilin inhibits NHEJ. GST, full length coilin-GST and different fragments of GST-coilin (1000ng each) were added to the in vitro NHEJ reaction. The reaction mixture was subjected to agarose gel (0.7%) electrophoresis. (B) Graphical representation of NHEJ efficiencies of Fig A. (C) Increasing amounts of coilin correlate with NHEJ inhibition. GST and coilin-GST were added in increasing levels (250, 500 and 1000 ng) in separate NHEJ reactions along with mock and control. The reaction mixtures were subjected to agarose gel (0.7%) electrophoresis. (D) Graphical representation of NHEJ efficiencies of Fig C. The inhibition of NHEJ was significantly (p<0.05) different from the control at 500 and 1000ng of soluble coilin addeded reactions.
Acknowledgements
We thank Andrew Gilder and Hanna J. Broome for their critical reading of this manuscript. This work was supported by NIH grant R01GM081448 (to MDH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cioce M, Lamond AI. Cajal Bodies: A Long History of Discovery. Annu Rev Cell Dev Biol. 2005 doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 2.Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 4.Hebert MD. Phosphorylation and the Cajal body: modification in search of function. Arch Biochem Biophys. 2010;496:69–76. doi: 10.1016/j.abb.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barcaroli D, et al. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci U S A. 2006;103:14808–14812. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker KE, et al. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS One. 2009;4:e6171. doi: 10.1371/journal.pone.0006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol. 2010;17:403–409. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- 9.Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001;15:2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell. 2002;3:329–337. doi: 10.1016/s1534-5807(02)00222-8. [DOI] [PubMed] [Google Scholar]
- 11.Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schumperli D, Matera AG. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma. 2005;114:155–166. doi: 10.1007/s00412-005-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing Coiled Bodies Is Regulated in Interphase and Mitosis - Evidence that the Coiled Body Is a Kinetic Nuclear Structure. J. Cell Biol. 1993;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hearst SM, et al. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci. 2009;122:1872–1881. doi: 10.1242/jcs.044040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyota CG, Davis MD, Cosman AM, Hebert MD. Coilin phosphorylation mediates interaction with SMN and SmB'. Chromosoma. 2010;119:205–215. doi: 10.1007/s00412-009-0249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam YW, Lyon CE, Lamond AI. Large-scale isolation of Cajal bodies from HeLa cells. Mol Biol Cell. 2002;13:2461–2473. doi: 10.1091/mbc.02-03-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, Xu H, Subramony SH, Hebert MD. Interactions between Coilin and PIASy partially link Cajal bodies to PML bodies. J Cell Sci. 2005;118:4995–5003. doi: 10.1242/jcs.02613. [DOI] [PubMed] [Google Scholar]
- 18.Pastwa E, Somiari RI, Malinowski M, Somiari SB, Winters TA. In vitro non-homologous DNA end joining assays--the 20th anniversary. Int J Biochem Cell Biol. 2009;41:1254–1260. doi: 10.1016/j.biocel.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Perrault AR, Takeda Y, Qin W, Iliakis G. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31:5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 21.Mimori T, Akizuki M, Yamagata H, Inada S, Yoshida S, Homma M. Characterization of a high molecular weight acidic nuclear protein recognized by autoantibodies in sera from patients with polymyositis-scleroderma overlap. J Clin Invest. 1981;68:611–620. doi: 10.1172/JCI110295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morency E, Sabra M, Catez F, Texier P, Lomonte P. A novel cell response triggered by interphase centromere structural instability. J Cell Biol. 2007;177:757–768. doi: 10.1083/jcb.200612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cioce M, Boulon S, Matera AG, Lamond AI. UV-induced fragmentation of Cajal bodies. J Cell Biol. 2006;175:401–413. doi: 10.1083/jcb.200604099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci U S A. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]