Abstract
Muscle specific miRNAs, myomiRs, have been shown to control muscle development in vitro and are differentially expressed at rest in diabetic skeletal muscle. Therefore, we investigated the expression of these myomiRs, including miR-1, miR-133a, miR-133b and miR-206 in muscle biopsies from vastus lateralis of healthy young males (n = 10) in relation to a hyperinsulinaemic–euglycaemic clamp as well as acute endurance exercise before and after 12 weeks of endurance training. The subjects increased their endurance capacity, 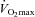 (l min−1) by 17.4% (P < 0.001), and improved insulin sensitivity by 19% (P < 0.01). While myomiR expression remained stable during a hyperinsulinaemic–euglycaemic clamp, an acute bout of exercise increased mir-1 (P < 0.05) and mir-133a (P < 0.05) expression before, but not after, training. In resting biopsies, endurance training for 12 weeks decreased basal expression of all four myomiRs (P < 0.05). Interestingly, all myomiRs reverted to their pre-training expression levels 14 days after ceasing the training programme. Components of major pathways involved in endurance adaptation such as MAPK and TGF-β were predicted to be targeted by the myomiRs examined. Tested predicted target proteins included Cdc42 and ERK 1/2. Although these proteins were downregulated between post-training period and 2 weeks of cessation, an inverse correlation between myomiR and target proteins was not found. In conclusion, our data suggest myomiRs respond to physiological stimuli, but their role in regulating human skeletal muscle adaptation remains unknown.
(l min−1) by 17.4% (P < 0.001), and improved insulin sensitivity by 19% (P < 0.01). While myomiR expression remained stable during a hyperinsulinaemic–euglycaemic clamp, an acute bout of exercise increased mir-1 (P < 0.05) and mir-133a (P < 0.05) expression before, but not after, training. In resting biopsies, endurance training for 12 weeks decreased basal expression of all four myomiRs (P < 0.05). Interestingly, all myomiRs reverted to their pre-training expression levels 14 days after ceasing the training programme. Components of major pathways involved in endurance adaptation such as MAPK and TGF-β were predicted to be targeted by the myomiRs examined. Tested predicted target proteins included Cdc42 and ERK 1/2. Although these proteins were downregulated between post-training period and 2 weeks of cessation, an inverse correlation between myomiR and target proteins was not found. In conclusion, our data suggest myomiRs respond to physiological stimuli, but their role in regulating human skeletal muscle adaptation remains unknown.
Introduction
Skeletal muscle is a highly plastic organ, capable of altering phenotype in response to changes in neuromuscular activity, mechanical loading, and metabolic perturbations (Hoppeler & Fluck, 2002). It is well established that both endurance exercise and endurance training activate many signalling pathways to improve skeletal muscle function, while physical inactivity, a risk factor for many chronic diseases, is characterized by skeletal muscle atrophy and insulin resistance (Ferrando et al. 1996; Krogh-Madsen et al. 2010).
While the molecular mechanisms regulating muscle adaptation are not yet fully clear, one candidate feature is the coordinated expression of muscle-specific microRNAs (myomiRs). MicroRNAs (miRNA) are short non-coding RNAs that regulate protein abundance (Lee & Ambros, 2001). Primary miRNA transcripts (pri-miRNAs) are cleaved into 70 bp stemloop structures (pre-miRNAs), transported to the cytoplasm and cleaved again by the enzyme Dicer into mature miRNAs (∼19–22 bp). Mature miRNAs are incorporated into a protein complex, called the RNA-induced silencing complex (RISC) (Wienholds & Plasterk, 2005). The RISC acts by hybridizing either perfectly or partially to complementary binding sites located in the 3′ untranslated region (UTR) of target mRNAs, inhibiting translation by mRNA cleavage or steric hindrance (Bartel, 2004; Xie et al. 2005).
A single miRNA can regulate the expression of hundreds of mRNAs and proteins (Lee et al. 1993; Baek et al. 2008a; Check, 2008) and this has been shown to play a crucial role in cell cycle regulation and developmental processes, leading to a tissue specific miRNA profile during normal development (Chen et al. 2006; Kim et al. 2006).
In skeletal muscle, mir-1, mir-133a, mir-133b and mir-206 together account for nearly 25% of all miRNA expression and are as a group often referred to as myomiRs (McCarthy et al. 2009). The expression of myomiRs is dramatically increased during myogenesis (Chen et al. 2006). Furthermore, differential expression of myomiRs following resistance exercise and in diabetic skeletal muscle suggest that myomiRs play a role in human health and disease (McCarthy & Esser, 2007; Drummond et al. 2008; Gallagher et al. 2010). The differences in miRNA regulation in diabetic and healthy skeletal muscle following a hyperinsulinaemic–euglycaemic clamp may result from differences in insulin sensitivity or signalling, which can be improved by endurance training. These observations raised the hypothesis that the coordinated increase in myomiR expression contributes to skeletal muscle adaptation to acute and chronic endurance exercise.
To test this hypothesis, we measured myomiR expression in response to acute endurance exercise, before and after 12 weeks of endurance training and in response to hyperinsulinaemic–euglycaemic clamp before and after the 12 weeks training programme as we hypothesized that insulin, as a growth factor, would regulate myomiR expression.
Methods
Subjects
Ten healthy, trained men participated in the study. Subject characteristics are listed in Table 1. Before inclusion in the study, a medical examination with blood test screening, a test for maximal power output (Pmax), and an oral glucose tolerance test were performed. Exclusion criteria included regular physical exercise less than 2 and more than 4 times per week, BMI > 30 kg m−2, smoking and impaired glucose tolerance. The purpose of the study, possible risks, and discomforts were explained to the subjects before written consent was obtained. The study was approved by the local Ethical Committee of Copenhagen and Frederiksberg and was in accordance with the Declaration of Helsinki.
Table 1.
Subject characteristics
| Subject characteristics (n = 10) | |
|---|---|
| Age (years) | 30.5 ± 5.5 |
| Weight (kg) | 79.2 ± 9.0 |
| Height (cm) | 180.2 ± 4.8 |
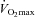 (l min−1) (l min−1) |
4.2 ± 0.5 |
| Pmax (W) | 328 ± 33 |
Data are means ± s.d.
Body composition
Before and after the training period whole body fat and fat-free tissue mass measurements were performed using a dual-energy X-ray absorptiometry (DXA) scanner (Lunar Prodigy, GE Healthcare, WI, Madison USA, software v. 8.8).
Maximal oxygen consumption test (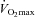 )
)
An incremental exercise test to volitional fatigue was performed on an electrically braked cadence-independent cycle ergometer (Monark 839E, Monark Ltd, Varberg, Sweden). When the subjects were unable to maintain a cadence of 60 r.p.m. for more than 15 s, the test was stopped. Inhaled oxygen and expired carbon dioxide were recorded on line (Quark b2, CosMed; Rome, Italy).
Exercise trial
Before and after the training period the subjects performed a 60 min cycle ergometer exercise bout at 65% of Pmax (Monark 839E, Monark Ltd, Varberg, Sweden). On the experimental day, subjects arrived, in a fasted state, at the lab between 07.30 and 08.00 h. After exercise the subjects rested on a hospital bed for 180 min.
Hyperinsulinaemic–euglycaemic clamp
Hyperinsulinaemic–euglycaemic clamps were done as previously described (Yfanti et al. 2010). Briefly, the participants reported at the laboratory at 08.00 h after an overnight fast 9 days prior to beginning training and 3–5 days after their last training bout. An intravenous catheter was placed in an antecubital vein of one arm for infusion of insulin and glucose. A second intravenous catheter was placed in a dorsal hand vein of the contralateral arm for blood sampling. After baseline blood samples were obtained, infusion of insulin (Actrapid; Novo Nordisk Insulin, Copenhagen, Denmark; 100 IU ml−1) started at a constant rate of 80.0 mU min−1 m−2 body surface area. Euglycaemia was achieved by co-infusion of glucose (200 g (1000 ml)−1) at a variable rate. Arterialized blood was analysed for glucose and potassium concentrations every 10 min.
Muscle biopsies
Muscle biopsies from vastus lateralis were taken at time points 0 and 180 min during the insulin clamp, and at time points 0 (before exercise), 60 (immediately after exercise) and 240 min (3 h after the end of exercise) during the exercise trial, before and after the training period (Fig. 1). Tissue samples were obtained using the percutaneous needle method with suction under local anaesthesia, using 3–5 ml of 20 mg ml−1 lidocaine (SAD, Denmark Copenhagen). Muscle tissue was immediately frozen in liquid nitrogen and stored at −80°C until further analysis.
Figure 1. Schematic overview of the study design.
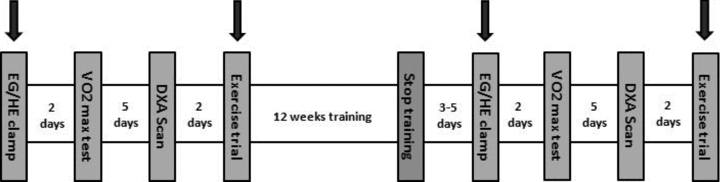
Downward arrows indicate the days during which biopsies were taken. Resting biopsies on trial exercise before training were compared to before the euglycaemic–hyperinsulinaemic clamp to obtain training effect and compared to the acute exercise trial post-training to obtain a comparison following cessation.
Training protocol
The subjects trained, under supervision, on a cycle ergometer (Monark 839E) with a 5 times per week frequency for 12 weeks. The participants were instructed to maintain their habitual diets, which were registered 3 days (including a weekend day) at the beginning, after 6 weeks of training, and at the end of the training period. Furthermore the subjects were instructed not to eat 2 h previous to the daily exercise bouts. To determine the intensity of the training for the following days of the week, a Pmax test was performed every Monday. Tuesday and Thursday the training consisted of intervals, Tuesday's at intensities of 85–91% of Pmax for 70–80 min and Thursday's at intensities of 75–81% of Pmax for 75–81 min. Wednesday and Friday consisted of continuous biking, Wednesday's at 60–66% of Pmax for 60–80 min, and Friday's at intensities of 55–61% of Pmax for 120–150 min. The participants were allowed to miss only 5% of the total amount of training.
MyomiR quantification
Approximately 30 mg skeletal muscle biopsy samples were homogenized in TRIzol (Invitrogen, Carlsbad, CA, USA) using a Tissuelyser (Qiagen, Valencia, CA, USA) and total RNA was isolated according to the manufacturer's protocol. Total RNA was dissolved in RNase-free water and quantified using a Nanodrop ND 1000 (Thermo Scientific, Wiklmington, DE, USA). The abundance of mature miRNAs was measured using TaqMan miRNA assays for miR-1, -133a/b and -206 according to the manufacturer's directions (Applied Biosystems Inc., Foster City, CA, USA). Briefly, reverse transcription was performed with miRNA-specific RT primers and 10 ng of total RNA for 30 min at 37°C followed by 10 min incubation at 95°C. Quantitative real-time PCR (qPCR) was performed using an ABI-PRISM 7900 Sequence Detection System (Applied Biosystems) according to the manufacturer's protocol in triplicates, using the RT product and microRNA-specific PCR primers for 40 cycles (two steps: 95°C for 15 s followed by 60°C for 30 s). RNU48, a small nuclear RNA, was measured in parallel, using the same type of detection assay (Applied Biosystems), and was used as an endogenous control for miRNA expression analyses. RNU48 expression was not regulated by acute endurance exercise (P > 0.05), 12 weeks of endurance training (P > 0.05) or a euglycaemic–hyperinsulinimic clamp (P > 0.05). Fold changes were calculated using the ΔΔCt method.
MyomiR target prediction
The web-based computational tool DIANA-mirPath was used to identify molecular pathways potentially altered by the coordinated change in expression of all four myomiRs (Papadopoulos et al. 2009). DIANA mirpath in itself is not a prediction tool, but rather a web-based platform that combines the prediction tool Targetscan 5.1 and the pathway tool KEGG (Kyoto Encyclopedia of Genes and Genomes). DIANA-mirPath was set to use TargetScan Human 5.1 to identify possible mRNA targets. The level of significance was set at P < 0.05.
Western blot analysis
Approximately 25 mg of skeletal muscle samples were homogenized using a Tissue-lyser (Qiagen, Crawley, West Sussex, UK) in ice cold 50 mm Tris, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 50 mm sodium fluoride, 5 mm sodium pyrophosphate, 2 mm sodium orthovanadate, 1 mm PMSF, 1 mm dithiotreitol, 0.5% (v/v) protease inhibitor cocktail and 1% (v/v) Nonidet P-40. Protein lysates were then centrifuged at 13,000 g for 15 min at 4°C and the pellet was discarded. Protein concentration was measured using a Bio-Rad protein assay. Samples were diluted in Laemmli buffer and boiled for 2 min before loading 20 μg onto a 4–12% gradient bis-Tris NuPage gel (Invitrogen). The gel was run for 70 min at 125 V and protein was transferred onto a PVDF membrane using the Invitrogen Iblot system for 7 min. The membrane was blocked for 1 h at room temperature in 5% milk and incubated with primary antibody overnight at 4°C. Antibody dilutions were: p44/42 MAPK (ERK1/ERK2) Rabbit mAb (Cell Signaling Technology) at 1:1000 in 5% bovine serum albumin (BSA)/Tris-buffered saline Tween-20 (TBST); CDC42 Rabbit mAb (Cell Signaling Technology) at 1:1000 in 5% BSA/TBST; SMAD2 rabbit mAb (Cell Signaling Technology) at 1:1000 in 5% skimmed milk/TBST at 1:1000. Blots were washed and incubated with anti-rabbit IgG horse radish peroxidase-conjugated antibody diluted 1:5000 (Cell Signaling Technology) for 1 h at room temperature. The signal was detected using Supersignal West Femto Luminal/Enhancer Solution (Thermo Scientific, Waltham, MA, USA) by a charge-coupled device camera (Bio-Rad, Hemel Hempstead, UK). Following exposure, blots were briefly rinsed in TBST and then incubated in 0.5% Reactive Brown (Sigma-Aldrich) for 15 min. Blots were analysed and quantified using ImageQuant software (GE Healthcare/Amersham, Little Chalfont, UK), with Reactive Brown utilized to quantify total protein, as a control for equal loading and transfer.
Statistical analysis
The statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA). Results are presented as means ± s.e.m. All data were tested for normality of distribution before further analysis using the Kolmogorov–Smirnov test. Student's t test for paired data or one-way ANOVA was applied as stated in the figure legends. When a significant P value was achieved, a Bonferroni post hoc analysis was performed to make comparisons between time points.
Results
Physiological improvements in response to 12 weeks of endurance training
Ten healthy young men (Table 1) participated in the study. In response to a 12 week endurance training programme the subjects increased their 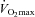 (l min−1) by 17.4% (P < 0.01). During the training period the subjects gained 2.0 ± 0.3 kg (s.e.m.) fat free mass and lost 3.6 ± 0.6 kg fat mass (s.e.m.) (P < 0.05). The glucose infusion rate (GIR) during a hyperinsulinaemic–euglycaemic clamp increased by 19% (P < 0.01) in response to 12 weeks of training. A summary of the physiological improvements is given in Table 2.
(l min−1) by 17.4% (P < 0.01). During the training period the subjects gained 2.0 ± 0.3 kg (s.e.m.) fat free mass and lost 3.6 ± 0.6 kg fat mass (s.e.m.) (P < 0.05). The glucose infusion rate (GIR) during a hyperinsulinaemic–euglycaemic clamp increased by 19% (P < 0.01) in response to 12 weeks of training. A summary of the physiological improvements is given in Table 2.
Table 2.
Physiological response to 12 weeks of endurance training
| Pre-training | Post-training | |
|---|---|---|
| Fitness | ||
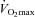 (l min−1) (l min−1) |
4.2 ± 0.5 | 4.9 ± 0.5*** |
| Body Composition | ||
| Fat mass (kg) | 16.3 ± 6.9 | 12.8 ± 6.8*** |
| Fat free mass (kg) | 61.3 ± 5.2 | 63.1 ± 5.3** |
| Fat free mass leg (kg) | 21.1 ± 2.3 | 21.8 ± 2.3* |
| Glucose metabolism | ||
| Glucose (mmol l−1) | 4.9 ± 0.5 | 4.9 ± 0.3 |
| Insulin (pmol l−1) | 23.2 ± 7.9 | 19.8 ± 6.3 |
| Glucose infusion rate (mU min−1 m−2) | 22.3 ± 4.1 | 26.5 ± 5.4** |
Data are means ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001.
MyomiRs are not regulated by insulin
We used a 3 h hyperinsulinaemic–euglycaemic clamp to test whether insulin regulated myomiRs or whether training-induced improvements in insulin sensitivity affected the myomiR expression pattern. None of the myomiRs was significantly altered by the clamp, either before or after the training period (Fig. 4) (P > 0.05). Thus, while 3 h of a hyperinsulinaemia failed to alter myomiR levels in the current study, alternative insulin infusion protocols and additional time points should be tested to determine whether insulin could contribute to the regulation of myomiRs in young healthy subjects.
Figure 4. Myomir expression in response to euglycaemic–hyperinsulinaemic before and after endurance training.
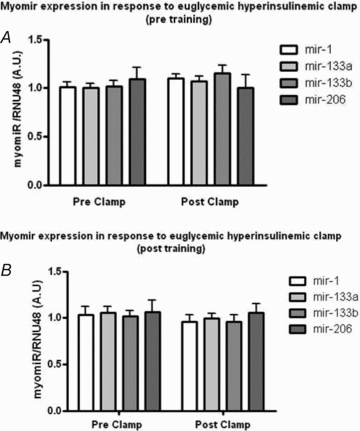
MyomiR expression in response to euglycaemic–hyperinsulinaemic clamp before and after a 12 week training period. A paired t test did not show a significant effect of insulin infusion in the expression of all four myomiRs (P > 0.05) before (A) or after the training period (B).
MyomiRs are induced by endurance exercise in untrained individuals
To test whether myomiR expression was regulated by endurance exercise, subjects underwent 60 min of cycling at 65% of Peak Watt. MyomiR expression was measured in vastus lateralis biopsies immediately before, immediately after, and 3 h after exercise, before and after 12 weeks of training. Before the training period a one-way (RM) ANOVA revealed a significant increase in the myomiRs mir-1 (P < 0.05) and mir-133a (P < 0.05) but not in mir-133b and mir-206 expression (Fig. 2A). After the training period, there was no acute exercise induced effect on the measured myomiR expression (Fig. 2B). Hence, as the myomiRs are induced in response to acute exercise in untrained individuals but not in trained, this suggests that myomiRs indirectly affect protein abundance by targeting the mRNA transcripts following acute exercise only in untrained individuals.
Figure 2. Myomir expression in response to endurance exercise.
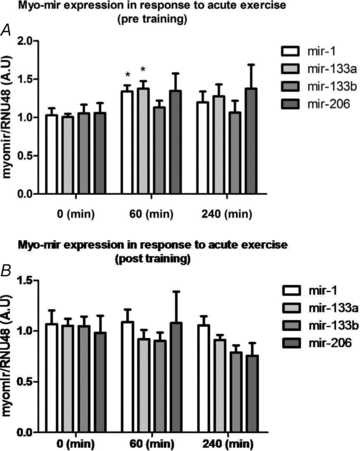
MyomiR expression in response to a single 60 min bout at 65% Pmax of endurance exercise before and after 12 weeks of high intensity endurance training. A one-way (RM) ANOVA demonstrated a significant effect of exercise for two myomiRs (mir-1: P < 0.05; mir-133a: P < 0.05) before the training period. None of the myomiRs were differentially altered in response to acute endurance exercise after 12 weeks of training. *Main effect of exercise, P < 0.05.
MyomiRs are decreased after 12 weeks of endurance training
While acute exercise response predominately resembles a stress response (in particular in untrained individuals) (Keller et al. 2007), endurance training reflects a new homeostasis within skeletal muscle. We measured the myomiR expression in resting vastus lateralis muscle biopsies obtained before and after 12 weeks of endurance exercise. Interestingly, in contrast to the response observed to acute exercise in untrained subjects, endurance training for 12 weeks, measured in biopsies obtained at rest, resulted in decreased expression of all myomiRs: mir-1 (32%, P < 0.05), mir-133a (23%, P < 0.01), mir-133b (19%, P < 0.05) and mir-206 (49%, P < 0.01) (Fig. 3). These data emphasize the differences in gene expression in acute response to exercise and expression at rest following a long period of repeated exercise.
Figure 3. Myomir expression at rest in response to endurance training.
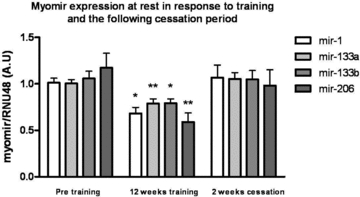
MyomiR expression in resting samples following 12 weeks of training and 2 weeks of cessation of training. Healthy, young men (n = 10) performed a 12 week endurance-training programme. Muscle biopsies were obtained at rest before and after the training period. A one way (RM) ANOVA demonstrated a significant effect of training in the expression of all four myomiRs. A Bonferroni multiple comparison post hoc test revealed a training induced downregulation of mir-1 (P < 0.05), mir-133a (P < 0.01), mir-133b (P < 0.05) and mir-206(P < 0.01) and an upregulation of mir-1 (P < 0.05), mir-133a (P < 0.01), mir-133a (P < 0.05), and mir-206 (P < 0.05) in the following cessation period. *Main effect of exercise, P < 0.05; **main effect of exercise, P < 0.01.
Two weeks following training cessation, myomiR levels are reversed to untrained levels
To investigate whether the downregulated myomiR expression was adjusted to cessation of exercise, we measured the resting myomiR expression levels again after 2 weeks of normal daily activity (no exercise training). The expression of myomiRs was reversed to a non-significant level when compared to before the training period (P > 0.05). MyomiRs significantly increased from the post-training time point compared to expression levels after 14 days of cessation (mir-1 (36%, P < 0.05), mir-133a (25%, P < 0.05), mir-133b (23%P < 0.05) and mir-206 (41%, P < 0.05)) (Fig. 3). Thus, myomiRs seems to rapidly adjust to the current physical activity level.
Targeted pathways by myomiRs
Using the bioinformatic prediction tool DIANA-mirPath (Papadopoulos et al. 2009) we identified 949 potential mRNA targets of the myomiRs. The KEGG pathway database recognized 226 of those mRNAs and placed them in 121 different biological pathways. Among the most significant affected pathways, important for muscle metabolism, was the TGF-β pathway (P < 0.0001) and the MAPK pathway (P < 0.05).
Specific targets of myomiRs
Based on our bioinformatics analysis we selected Cdc42 and ERK1/2 as proteins in the MAPK pathway and Smad2 as a protein in the TGF-β pathway. As predicted both Cdc42 and ERK1/2 were regulated in response to training and the following cessation period (Fig. 5A and B) (P < 0.05), while post hoc analysis demonstrated a significant difference between the post-training and cessation periods where a 25% and 30% decrease in expression occurred respectively. Contrary to our hypothesis Smad2 did not differ between groups (data not shown). While, no significant correlation between the levels of myomiRs and target proteins was present, the changes in myomiR and target protein levels from 3–5 days post-training to 14 days post-training were positively correlated (data not shown) contrary to the hypothesis that miRNAs are negatively regulating target protein levels.
Figure 5. MyomiR targeted protein levels at rest in response to endurance training.
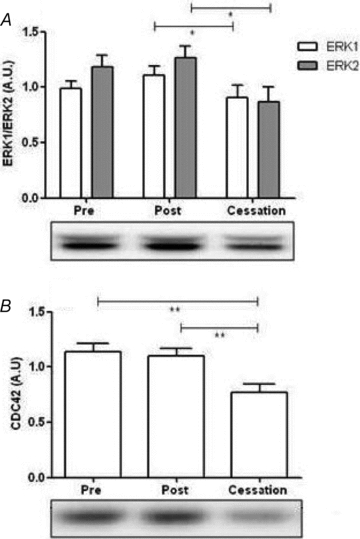
Protein expression at rest of Cdc42, representing the MAPK pathway and ERK1/2, representing the TGF-β pathway, based on the prediction analysis of the myomiRs. A one way (RM) ANOVA demonstrated a significant change in the Cdc42 and ERK1/2 levels (A and B) (P < 0.05) in the experimental period. A post hoc analysis demonstrated a significant difference between the post training and cessation period. Main effect of endurance training period is indicated: *P < 0.05, **P < 0.01.
Discussion
In the current study, we profiled myomiR expression in human in vivo models of alterations in skeletal muscle physiology. We demonstrated that while myomirs were not regulated by insulin administration, two out of four myomiRs were induced in response to an acute bout of endurance exercise, before, but not after, the initiation of an endurance training program. In contrast, the abundance of all four myomiRs decreased at rest following 12 weeks of endurance training, while just 14 days after cessation of regular training myomiRs returned to pre-training levels. Bioinformatics prediction analyses and subsequent western blot analyses only partially supported our findings at the protein level, and did not support a role for myomiRs to directly interact with the mRNA of a few of the predicted proteins.
As we observed miR-1 and miR-133a increasing following a single bout of endurance exercise we hypothesized that repeated bouts of endurance training would lead to an increase in these myomiRs in human skeletal muscle. However, following 12 weeks of high intensity endurance training all four myomiRs were downregulated. The pattern of increased expression during acute exercise, but downregulation at rest following chronic training is not unique to myomiRs. For example, acute endurance exercise increases interleukin-6 mRNA expression in skeletal muscle (Pedersen et al. 2004), but endurance training reduces resting mRNA levels of interleukin-6 (Lambert et al. 2008). Importantly, we cannot rule out the possibility that myomiRs are decreased at later time points during recovery, as one limitation of our design is that we have only measured myomiR expression immediately and 3 h following acute endurance exercise. However, our human data is in partial agreement with a study performed in mice in which the expression of miR-1, was increased following forced treadmill running in mice (Safdar et al. 2009). The increase in miR-1 and miR-133a immediately after acute endurance exercise is in line with the increase in the transcription factors MyoD, myogenin, and MRF4 that also occur following acute endurance exercise in skeletal muscle, and have been shown to be likely regulators of myomiR expression (Kadi et al. 2004; Coffey et al. 2006; Sweetman et al. 2008).
In addition to showing that high intensity endurance training decreases myomiR expression, we also established a time window for how long this adaptation remains in the absence of further training. When measured 3–5 days following the endurance training myomiR expression was significantly reduced. The time window of 3–5 days allowed us to avoid any effects of acute exercise, which can last up to 48 h post exercise, while still maintaining many of the training adaptations such as insulin sensitivity (Table 2) and markers of mitochondrial activity (Yfanti et al. 2010). However, within 14 days of ceasing the endurance training program myomiRs expression no longer significantly differed from pre-training levels. This time course is particularly interesting in the light of our recent study indicating that 14 days of reducing daily steps from 8000 to 1500 reduces insulin sensitivity and leg lean mass (Olsen et al. 2008). Conversely, leg lean mass in our subjects increased (Table 2), suggesting a role for myomiRs in regulating muscle mass during endurance exercise in addition to their already established role in muscle hypertrophy and resistance exercise training (McCarthy & Esser, 2007; Drummond et al. 2008). Additional endurance training adaptations such as increased oxidative enzymes, 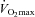 , and stroke volume are also partially reduced within 12 days of ceasing training in fit subjects (Coyle et al. 1985). Combined, these data suggest that myomiRs might be regulated in a similar fashion as established exercise adaptations.
, and stroke volume are also partially reduced within 12 days of ceasing training in fit subjects (Coyle et al. 1985). Combined, these data suggest that myomiRs might be regulated in a similar fashion as established exercise adaptations.
One of the most well characterized adaptations to endurance exercise is an improvement in insulin sensitivity (Wojtaszewski & Richter, 2006), which was observed with our training protocol. In addition to regulating glucose transport insulin is a potent growth factor in skeletal muscle capable of regulating muscle mass (Sinha et al. 1996). However, myomiR expression was not regulated by an hyperinsulinaemic–euglycaemic clamp in our subjects. Our results are in contrast to a recent finding by Granjon and colleagues (Granjon et al. 2009) who performed miRNA microarray in human skeletal muscle before and immediately following an hyperinsulinaemic–euglycaemic clamp. The authors found that 39 miRNAs, including the myomiRs, miR-1 and miR-133b were downregulated in response to the hyperinsulinaemic conditions. While both studies used similar, but differentially normalized, insulin infusion rate (2 mU min−1 kg−1 in Granjon's study and 80.0 mU min−1 m−2 in the current study) and the same time points for muscle biopsies, there are several discrepancies that could explain the differences in myomiR expression. Granjon et al. used older men of unknown fitness level, whereas our subjects were young and healthy. In addition, the insulin concentrations achieved during the clamps differed in the two studies (1299.9 pmol l−1 by Granjon et al. and approximately 300 pmol l−1 lower in the current study). However, both of these insulin concentrations are supraphysiological and should provide a maximal stimulus for insulin induced transcription. While both studies measured the expression levels at the same time points following the clamps, it is plausible that the kinetics of myomiR expression differs in young and old people.
Thus, further studies on the kinetics of myomiR expression in response to a hyperinsulinaemic–euglycaemic clamp in both young and old subjects should help resolve the observed discrepancies. Interestingly, it was recently shown that 62 miRNAs, including miR-206 and miR-133a, were differentially expressed at rest in human diabetic muscle (Gallagher et al. 2010). Together with our observations, this further emphasizes that the coordinated miRNA expression, rather than the expression of one single miRNA, is critical to the skeletal muscle in the development of a complex phenotype such as diabetes or endurance training.
One approach to predict the most targeted pathways by a specific set of miRNAs is to use bioinformatics algorithms. We utilized the DIANA mirPath software (Papadopoulos et al. 2009) and found that myomiRs are predicted to target several key proteins within the MAPK and TGF-β pathways. The MAPK pathway has already been established as an important exercise stress induced pathway (Kramer & Goodyear, 2007) while the TGF-β pathway is known to alter satellite cell proliferation and recruitment (Schabort et al. 2009).
We identified three predicted target proteins within the MAPK pathway, Cdc42 and ERK1/2, which changed in protein expression partly in accordance with the myomiR expression profile. In contrast, Smad2, one of the predicted targets in the TGF-β pathway, remained unchanged. However, no significant inverse correlation between myomiR level and target protein was found when all time points where pooled as was hypothesized. Interestingly, contrary to our hypothesis the change in myomiR and target protein levels from post training to 14 days after training was positively, not negatively correlated (data not shown). Thus, at this time the data do not support a direct role for the measured myomiRs to interact with the mRNA of the selected target proteins at this time. One alternative possibility is that the myomiRs or other miRNAs may target negative regulators of the targets proteins we measured, which would lead to the observed positive correlation observed. Furthermore, the complexity of miRNA targeting of mRNAs is exemplified by the hundreds of mRNA transcripts an individual miRNA targets, and that multiple miRNAs can bind a given mRNA (Selbach et al. 2008). The individual effects of miRNA in cell culture models are relatively modest and may act to fine tune protein concentrations (Baek et al. 2008b; Selbach et al. 2008). Thus, predicting target proteins based on the effect of only a few miRNA may not accurately reflect the net effect of all miRNAs, especially in vivo in response to a physiological stimulus. Lastly, while myomiRs are only expressed in myocytes (Chen et al. 2006; McCarthy, 2008) the targets selected for western blot measurements are also expressed in non-muscle cells present in the skeletal muscle biopsy. Thus non-muscle cells which do not express myomiRs may dilute any direct effects of myomiRs on target mRNA in muscle, or may have altered expression of the target proteins measured.
In this study we have used both acute and chronic endurance exercise to provide the first evidence that in human skeletal muscle myomiRs rapidly adjust to physical activity level. However, the details by which myomiRs maybe directly altering human physiology in response to endurance exercise remain unknown.
Acknowledgments
Ruth Rousing, Hanne Villumsen and Remie Mounier are acknowledged for their technical assistance. We thank James A. Timmons for scientific discussions and advice. The Centre of Inflammation and Metabolism is supported by a grant from the Danish National Research Foundation (no. 02-512-55). This study was further supported by the Danish Medical Research Council, the Commission of the European Communities (Grant Agreement no. 223576), Rigshospitalet and as a part of the research program of the UNIK: Food, Fitness & Pharma for Health and Disease (see http://www.foodfitnesspharma.ku.dk). The UNIK project is supported by the Danish Ministry of Science, Technology and Innovation. The Copenhagen Muscle Research Centre (CMRC) is supported by a grant from the Capital Region of Denmark.
Author contributions
Study conception and design: T.Å., S.N., Sample collection and data analysis: S.N., C.Y., A.R., T.Å. Manuscript Preparation: S.N., M.L., C.S., B.K.P. Manuscript Editing: S.N., M.L., B.K.P., A.R., T.Å., C.S., and C.Y. All authors approved the final version for publication. This work was completed at Rigshospitalet, University of Copenhagen, Denmark.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008a;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008b;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Check HE. Thousands of proteins affected by miRNAs. Nature. 2008;454:562. doi: 10.1038/454562b. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey VG, Shield A, Canny BJ, Carey KA, Cameron-Smith D, Hawley JA. Interaction of contractile activity and training history on mRNA abundance in skeletal muscle from trained athletes. Am J Physiol Endocrinol Metab. 2006;290:E849–E855. doi: 10.1152/ajpendo.00299.2005. [DOI] [PubMed] [Google Scholar]
- Coyle EF, Martin WH, III, Bloomfield SA, Lowry OH, Holloszy JO. Effects of detraining on responses to submaximal exercise. J Appl Physiol. 1985;59:853–859. doi: 10.1152/jappl.1985.59.3.853. [DOI] [PubMed] [Google Scholar]
- Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab. 2008;295:E1333–E1340. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando AA, Lane HW, Stuart CA, Davis-Street J, Wolfe RR. Prolonged bed rest decreases skeletal muscle and whole body protein synthesis. Am J Physiol Endocrinol Metab. 1996;270:E627–E633. doi: 10.1152/ajpendo.1996.270.4.E627. [DOI] [PubMed] [Google Scholar]
- Gallagher I, Scheele C, Keller P, Nielsen A, Remenyi J, Fischer C, Roder K, Babraj J, Wahlestedt C, Hutvagner G, Pedersen B, Timmons J. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med. 2010;2:9. doi: 10.1186/gm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granjon A, Gustin MP, Rieusset J, Lefai E, Meugnier E, Guller I, Cerutti C, Paultre C, Disse E, Rabasa-Lhoret R, Laville M, Vidal H, Rome S. The microRNA signature in response to insulin reveals its implication in the transcriptional action of insulin in human skeletal muscle and the role of a sterol regulatory element-binding protein-1c/myocyte enhancer factor 2C pathway. Diabetes. 2009;58:2555–2564. doi: 10.2337/db09-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppeler H, Fluck M. Normal mammalian skeletal muscle and its phenotypic plasticity. J Exp Biol. 2002;205:2143–2152. doi: 10.1242/jeb.205.15.2143. [DOI] [PubMed] [Google Scholar]
- Kadi F, Johansson F, Johansson R, Sjostrom M, Henriksson J. Effects of one bout of endurance exercise on the expression of myogenin in human quadriceps muscle. Histochem Cell Biol. 2004;121:329–334. doi: 10.1007/s00418-004-0630-z. [DOI] [PubMed] [Google Scholar]
- Keller P, Vollaard N, Babraj J, Ball D, Sewell DA, Timmons JA. Using systems biology to define the essential biological networks responsible for adaptation to endurance exercise training. Biochem Soc Trans. 2007;35:1306–1309. doi: 10.1042/BST0351306. [DOI] [PubMed] [Google Scholar]
- Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer HF, Goodyear LJ. Exercise, MAPK, and NF-κB signaling in skeletal muscle. J Appl Physiol. 2007;103:388–395. doi: 10.1152/japplphysiol.00085.2007. [DOI] [PubMed] [Google Scholar]
- Krogh-Madsen R, Thyfault JP, Broholm C, Mortensen OH, Olsen RH, Mounier R, Plomgaard P, Hall GV, Booth FW, Pedersen BK. A 2-wk reduction of ambulatory activity attenuates peripheral insulin sensitivity. J Appl Physiol. 2010;108:1034–1040. doi: 10.1152/japplphysiol.00977.2009. [DOI] [PubMed] [Google Scholar]
- Lambert CP, Wright NR, Finck BN, Villareal DT. Exercise but not diet-induced weight loss decreases skeletal muscle inflammatory gene expression in frail obese elderly persons. J Appl Physiol. 2008;105:473–478. doi: 10.1152/japplphysiol.00006.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102:306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA, Peterson CA, Dupont-Versteegden EE. Evidence of MyomiR network regulation of β-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol Genomics. 2009;39:219–226. doi: 10.1152/physiolgenomics.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen RH, Krogh-Madsen R, Thomsen C, Booth FW, Pedersen BK. Metabolic responses to reduced daily steps in healthy nonexercising men. JAMA. 2008;299:1261–1263. doi: 10.1001/jama.299.11.1259. [DOI] [PubMed] [Google Scholar]
- Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- Pedersen M, Steensberg A, Keller C, Osada T, Zacho M, Saltin B, Febbraio MA, Pedersen BK. Does the aging skeletal muscle maintain its endocrine function? Exerc Immunol Rev. 2004;10:42–55. [PubMed] [Google Scholar]
- Safdar A, Abadi A, Akhtar M, Hettinga BP, Tarnopolsky MA. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS One. 2009;4:e5610. doi: 10.1371/journal.pone.0005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schabort EJ, Van Der Merwe M, Loos B, Moore FP, Niesler CU. TGF-β's delay skeletal muscle progenitor cell differentiation in an isoform-independent manner. Exp Cell Res. 2009;315:373–384. doi: 10.1016/j.yexcr.2008.10.037. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sinha A, Formica C, Tsalamandris C, Panagiotopoulos S, Hendrich E, DeLuise M, Seeman E, Jerums G. Effects of insulin on body composition in patients with insulin-dependent and non-insulin-dependent diabetes. Diabet Med. 1996;13:40–46. doi: 10.1002/(SICI)1096-9136(199601)13:1<40::AID-DIA991>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, Munsterberg A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol. 2008;321:491–499. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Wojtaszewski JF, Richter EA. Effects of acute exercise and training on insulin action and sensitivity: focus on molecular mechanisms in muscle. Essays Biochem. 2006;42:31–46. doi: 10.1042/bse0420031. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yfanti C, Akerstrom T, Nielsen S, Nielsen AR, Mounier R, Mortensen OH, Lykkesfeldt J, Rose AJ, Fischer CP, Pedersen BK. Antioxidant supplementation does not alter endurance training adaptation. Med Sci Sports Exerc. 2010;42:1388–1395. doi: 10.1249/MSS.0b013e3181cd76be. [DOI] [PubMed] [Google Scholar]


