Abstract
Despite their extensive clinical and pathological heterogeneity, all malignant germ cell tumors (GCTs) are thought to originate from primordial germ cells. However, no common biological abnormalities have been identified to date. We profiled 615 microRNAs (miRNAs) in pediatric malignant GCTs, controls and GCT cell lines (48 samples in total) and re-analyzed available miRNA expression data in adult gonadal malignant GCTs. We applied the bioinformatic algorithm Sylamer to identify miRNAs that are of biological importance by inducing global shifts in mRNA levels. The most significant differentially expressed miRNAs in malignant GCTs were all from the miR-371~373 and miR-302 clusters (adjusted p<0.00005), which were over-expressed regardless of histological subtype [yolk sac tumor (YST)/seminoma/embryonal carcinoma (EC)], site (gonadal/extragonadal) or patient age (pediatric/adult). Sylamer revealed that the hexamer GCACTT, complementary to the 2-7 nucleotide miRNA seed AAGUGC shared by six members of the miR-371~373 and miR-302 clusters, was the only sequence significantly enriched in the 3′untranslated region (3′UTR) of mRNAs down-regulated in pediatric malignant GCTs (as a group), YSTs and ECs; and in adult YSTs (all versus non-malignant tissue controls; p<0.05). For the pediatric samples, down-regulated genes containing 3′UTR GCACTT showed significant over-representation of Gene Ontology (GO) terms related to cancer–associated processes, whereas for down-regulated genes lacking GCACTT, GO terms generally represented metabolic processes only, with few genes per term (adjusted p<0.05). We conclude that the miR-371~373 and miR-302 clusters are universally over-expressed in malignant GCTs and coordinately down-regulate mRNAs involved in biologically significant pathways.
Keywords: AAGUGC, embryonic stem cell, germ cell tumor, miRNA, mRNA
Introduction
Germ cell tumors (GCTs) are clinically and pathologically complex neoplasms that occur from the neonatal period through to late adulthood (1). Benign forms show extensive somatic differentiation and are referred to as mature (MT) and immature (IT) teratoma, while malignant GCTs are classified into seminomas and non-seminomatous tumors [yolk sac tumors (YSTs) and embryonal carcinomas (ECs)] (2).
Despite their heterogeneity, the germ cell theory of tumorigenesis states that all GCTs arise from totipotent primordial germ cells (3). However, biological abnormalities that are conserved across the age and histological range of malignant GCTs have not yet been identified. Here, we investigate the patterns and consequences of microRNA (miRNA) expression across the spectrum of GCTs. miRNAs regulate gene expression via translational repression and mRNA destabilization (4-6), the latter being detectable from mRNA expression changes (7, 8). This regulation is principally determined by the miRNA seed region, which binds to the seed complementary region (SCR) in the 3′ untranslated region (3′UTR) of mRNA targets (9). The seed region comprises nucleotides (nt) 2-8 of the miRNA, with 2-7nt being most critical for binding specificity (10). Importantly, miRNAs play a key role in cancer development, both as oncogenes and as tumor suppressor genes (TSGs) (4, 8, 11-13).
Little information is available concerning miRNA profiles in GCTs. One study used quantitative reverse transcription PCR (qRT-PCR) to determine levels of a restricted range of 156 miRNAs in adult gonadal GCTs, compared to three cases of normal testis (patient age uncertain) (14). This work suggested that the miR-371~373 gene cluster is highly expressed in adult malignant GCTs, a view supported by a genetic screen of primary human cells that implicated miR-372~373 as oncogenes in testicular GCTs (TGCTs), acting through inhibition of large tumor suppressor homolog 2 (LATS2) (15).
To date, there is no published miRNA profiling data for GCTs of pediatric patients, nor of those arising at extragonadal sites in any age-group. It is therefore unknown whether particular changes in miRNA expression represent a fundamental feature of malignant GCTs. The present study had two principal aims. First, we sought to determine global miRNA profiles in pediatric GCTs arising at both gonadal and extragonadal sites, and to compare the changes observed with those reported for adult gonadal malignant GCTs. Second, we applied the bioinformatic algorithm Sylamer (16) to identify miRNA changes that are of biological significance by inducing global shifts of mRNA expression. Our data indicate that the miR-371~373 and miR-302 clusters, of which six members (miR-372~373 and miR-302a~302d) share the identical 2-7nt seed region AAGUGC, have a fundamental role in the pathogenesis of malignant GCTs by down-regulating functionally significant target genes.
Materials and Methods
Tumor samples
The study received Multicenter Research Ethics Committee (ref:02/4/071) and Local Research Ethics Committee (ref:01/128) approval. We performed miRNA expression profiling on 48 samples, representing 32 pediatric GCTs from 22 female and 10 male patients (12 YSTs, 11 seminomas, three ECs, three MTs, three ITs), two testicular seminomas from young adults, eight control samples and six GCT cell lines [CLs; authenticated using short tandem repeat profiling (17); Supplementary Figure S1] (Supplementary Table S1). To avoid confusion with data from our re-analysis of miRNA expression in adult GCTs (14), all of these samples are henceforth referred to as ‘pediatric’. We use ‘seminoma’ to refer to all tumors with seminomatous histology, regardless of site (i.e., testicular seminoma, ovarian dysgerminoma, and extragonadal germinoma) (18, 19). The eight control tissues represented four normal gonadal specimens (one case each of pre- and post-pubertal male and female gonad) and four developmental samples (two fetal yolk sacs and two fetal female gonads).
miRNA microarray expression profiling
Total RNA was isolated as described previously (2). Sample and human reference (20) RNA were hybridized to the miRCURY LNA array platform v9.2 (Exiqon, Vedbaek, Denmark). The miRNA GAL file was updated to miRBase v13.01 which annotated 615 probes on the array. The 48 raw miRNA (.txt) data files [Gene Expression Omnibus (GEO) accession number: GSE18155] were processed using the Bioconductor packages limma and ArrayQualityMetrics in R (21, 22). The median expression value of the quadruplicate spots for each miRNA was calculated after subtraction of background intensities. Within-array (global-loess) and between-array (Aquantile method) normalization was performed (23) before a contrast matrix defining all pairwise comparisons was fitted (24). The data were filtered to exclude low variability probes (median expression value inter-quartile-range <0.6) and subsequently used for unsupervised hierarchical clustering, using a distance measure of 1 minus the Pearson correlation coefficient between samples. For heatmaps, values for each probe were centered by subtracting the mean expression value across samples.
Differential expression was assessed using a moderated t-statistic and p-values adjusted for multiple testing using Benjamini and Hochberg’s method (25). miRNAs with adjusted p-values <0.01 were considered statistically significant and differentially expressed. Lists of differentially expressed miRNAs generated for four different comparisons (pediatric malignant GCTs, YSTs, seminomas and EC versus non-malignant control tissue) were subsequently used for Sylamer analysis. Only the most significantly differentially expressed miRNAs (adjusted p<1×10−5) were represented on heatmaps, to enhance visualization of key miRNAs. Taqman qRT-PCR validation of miRNA levels, normalized to RNU24, was performed as previously described (20).
We compared our findings with published miRNA expression data for adult gonadal GCTs, as obtained by qRT-PCR (14). The raw cycle threshold (CT) data file was downloaded from the journal website2. After removal of the four spermatocytic seminoma (SS) samples, which do not occur in the pediatric population, miRNA CT values for the remaining 60 adult tissue samples and five GCT CLs were normalized to let-7a (which displayed the least variable expression across all samples), to obtain Δ CT values. The mean of all ΔCT values for all samples was then subtracted to obtain ΔΔCT values, which were used to perform unsupervised hierarchical clustering analysis and to generate lists of differentially expressed genes, employing the criteria used for the pediatric samples.
mRNA expression analysis
Matching global mRNA expression profiles were available for 21 of the 42 pediatric tissue samples examined by miRNA microarray. These represented 17 malignant GCTs (10 YSTs, six seminomas, one EC) and four non-malignant controls, comprising one MT and three normal gonads (one pre- and one post-pubertal testis and one post-pubertal ovary) (Supplementary Table S1). Profiling had previously been performed using the HG-U133A GeneChip (Affymetrix, Santa Clara, CA), comprising 22,283 probe sets corresponding to 13,042 genes. Data for 16 samples had previously been published (2); the EC, MT and three normal controls were previously unreported (GEO accession: GSE18155). In addition, we re-analyzed published data from a study of adult TGCTs that also used the HG-U133A GeneChip [(26); GEO accession: GSE3218], excluding two suboptimal YST samples (K14 and K18) (2). We used data from 25 such specimens, representing eight pure YSTs, 12 pure seminomas and five normal adult testis controls (26).
Raw mRNA (.CEL) files were processed and quantile normalized using Robust Multi-array Average (RMA) in R (6, 21, 27), using the Affymetrix annotation of March 2009. RMA-transformed expression values were analyzed for differential expression (24) with significance studied by t-test and adjusted for multiple testing (25). Pathway enrichment analysis was performed using the Gene Ontology (GO) algorithm3, as it permitted comparison of differentially expressed genes (log2 fold-change <−1.5 and adjusted p<0.01) grouped by the presence or absence of the SCR corresponding to the common 2-7nt seed of the miR-372~373 and miR-302a~d clusters. NCBI Entrez Gene identifiers were evaluated for biological process category over-representation within a total gene universe defined by the HG-U133A annotation library, using the hyperGTest function within the Bioconductor GOstats package (28). GOterms with adjusted p<0.01 (25) were considered statistically significant.
Sylamer algorithm
Full details of Sylamer are provided elsewhere4 (16). In brief, the algorithm assesses enrichment and/or depletion of nucleotide words of specific length, complementary to elements of the seed region (nucleotide positions 1-8) of miRNAs (i.e. SCRs), in the 3′UTRs of genes within ranked lists, with significance calculated using hypergeometric statistics. The primary aim of Sylamer is to identify whether changes in miRNA expression are of biological significance, with the secondary aim of producing lists of target genes for further validation.
For each ranked genelist derived from the mRNA expression data, we undertook Sylamer analysis for the six SCR elements of increasing size: three hexamers (corresponding to miRNA seed positions 1-6, 2-7 and 3-8); two heptamers (positions 1-7 and 2-8); and one octamer (position 1-8). Due to over-representation of conserved adenosines flanking SCRs in mRNAs (10), the complementarity criterion was discarded for SCR position 8 (seed position 1), where the nucleotide was always set to be adenosine, irrespective of the actual nucleotide at that position. For each comparison analyzed, the mRNA genelist was first ranked from down-regulated (to the left) to up-regulated (to the right). For each SCR under consideration, an enrichment/depletion p-value was computed at different cut-offs in the ranked gene list. At each cut-off, an SCR was either enriched in the 3′UTRs of the genes to the left and accordingly depleted in the 3′UTRs of the genes to the right, or conversely depleted on the left and enriched on the right. An event of enrichment on one side and corresponding depletion on the other side of the cut-off is associated with a single p-value. Varying the cut-off resulted in a set of p-values for each SCR (y-axis) visualized on a landscape plot (16), in which the log10-transformed p-values were sign-adjusted and plotted against the ranked genelist (x-axis). Sign-adjustment depended on the specific enrichment/depletion status of the pertinent SCR. A point plotted along the positive y-axis signifies that the SCR is enriched in the genes to the left and depleted in the genes to the right, whereas a point plotted along the negative y-axis conversely signifies depletion to the left and enrichment to the right. The displacement along the y-axis identifies the significance of the joint enrichment/depletion p-value for the SCR at that cut-off, according to the sign-adjusted log10-transformation.
For the present study, we combined Sylamer significance scores for different elements of each SCR to obtain a single summed significance score for each group of miRNAs sharing the same seed region. To do this, miRNAs were assigned to groups, defined by a common seed, and a single score was produced for the combined Sylamer results for the set of SCR hexamers, heptamers and octamer particular to each group. This approach integrated signals from different word lengths and increased method sensitivity compared to standard Sylamer analysis. The same analysis was applied to all possible words of 8nt length (all with an adenosine at position 8), including those that do not represent SCRs. The resulting scores followed an extreme value distribution by the nature of the scoring criteria employed. By fitting this distribution, p-values were assigned to the scores, with values <0.01 considered to be significant. As a filtering step, we only considered miRNA groups that contained at least one significantly differentially expressed miRNA.
Results
miRNA expression profiles in malignant GCTs
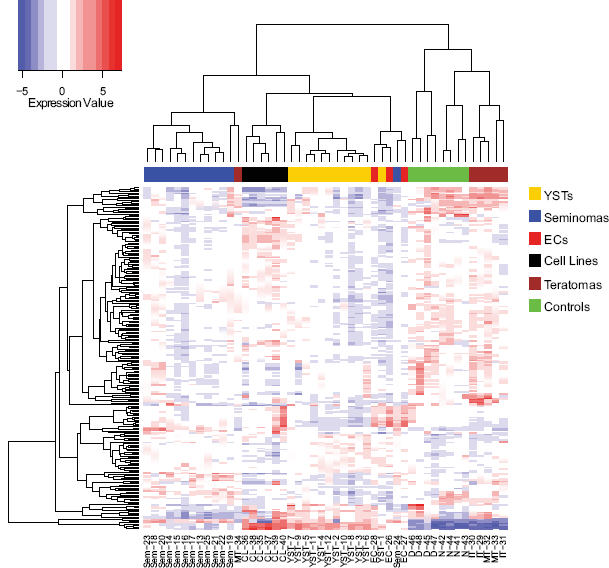
In initial unsupervised hierarchical clustering analysis, normalized microarray expression data for the 246 miRNAs that showed variable expression in the 48 pediatric samples and CLs were used to generate a heatmap (Figure 1). The dendrogram divided into two main branches, one containing the pediatric malignant GCT tissues and CLs, the other containing the non-malignant tissue, i.e. the teratomas (MT and IT) and the normal and developmental control samples. Only one non-malignant sample clustered with the malignant GCTs - MT tissue (MT-34) from a mixed GCT that also contained a malignant element. The pediatric malignant GCTs subdivided principally by histological subtype, with dendrogram subdivisions comprising seminomas, CLs, YSTs and ECs. The non-malignant samples subdivided into two branches, with developmental controls (fetal yolk sac and gonads) in one branch and normal gonadal tissue (pre- and post-pubertal ovary and testis) with the teratomas in the other.
Figure 1. Unsupervised hierarchical clustering analysis of miRNA expression in 42 pediatric tissue samples and 6 GCT cell lines.
Sample numbers refer to those in Supplementary Table S1.
In further analyses of our dataset we focused on the 42 tissue specimens only, removing the six CLs. No miRNA showed significant differential expression (adjusted p<0.01) between the MT and IT samples. When comparing the six teratoma samples and eight normal control specimens, only two of the 615 miRNAs were differentially expressed, with p-values that only just reached significance (miR-9 and miR-9*; adjusted p=0.0098 and 0.0091 respectively). Consequently, all 14 non-malignant samples were combined for subsequent comparisons with the pediatric malignant GCT tissues. Comparing all malignant GCTs versus the non-malignant samples produced a list of 170 significantly differentially expressed miRNAs, of which 44 (25.9%) were over-expressed in malignant GCTs and 126 (74.1%) under-expressed (Supplementary Table S2A). A heatmap based on the most significantly differentially expressed miRNAs (adjusted p<1×10−5; n=65) showed complete segregation between the pediatric malignant GCT and non-malignant samples (Supplementary Figure S2). The top 10 differentially expressed miRNAs in this comparison are shown in Table 1A. The top nine were from just two miRNA clusters - miR-371~373 and miR-302 (the latter including miR-367) - and all were over-expressed in the malignant GCTs.
Table 1. Over-expression of miR-371~373 and miR-302 clusters in malignant GCTs.
A) The top 10 differentially expressed miRNAs segregating pediatric and adult malignant GCTs from non-malignant tissue, ranked by adjusted p-value. miRNAs in bold are members of the miR-371~373 and miR-302 clusters. Note that miR-371, reported in the adult study, is now annotated as miR-371-3p. B) Chromosomal location and seed sequence of the miR-371~373 and miR-302 clusters. The common 2-7nt seed region is underlined. C) Transcription factors (TFs) over-expressed in malignant GCTs overall, and at least one histological subtype (all versus non-malignant tissues), in both the pediatric and adult datasets. The columns show the rankings, fold-change and p-values of genes differentially expressed in malignant GCTs overall. Also shown is the ranking in the main histological subtypes of GCT, seminoma (Sem) and YST. In all cases, where no ranking is given, the TF was not on the relevant list of differentially expressed genes. While all six TFs were over-expressed in seminomas, SOX17 and TEAD4 were also over-expressed in YSTs
| A | |||||||
|---|---|---|---|---|---|---|---|
| Pediatric Dataset | Adult Dataset | ||||||
| Rank | miRNA | Log2 Fold Change |
Adjusted p-value |
Rank | miRNA | ΔΔ Ct | Adjusted p-value |
| 1 | miR-302a | +4.41 | 6.28E-15 | 1 | miR-371 (-371-3p) | −11.18 | 1.18E-24 |
| 2 | miR-373 | +5.40 | 2.55E-14 | 2 | miR-373 | −10.26 | 1.66E-22 |
| 3 | miR-367 | +5.10 | 2.55E-14 | 3 | miR-372 | −10.79 | 4.87E-22 |
| 4 | miR-302c | +5.07 | 2.55E-14 | 4 | miR-302b | −12.45 | 1.65E-20 |
| 5 | miR-371-3p | +4.31 | 2.55E-14 | 5 | miR-302d | −12.41 | 1.65E-20 |
| 6 | miR-372 | +4.91 | 4.09E-14 | 6 | miR-373* | −8.72 | 2.85E-20 |
| 7 | miR-302d | +5.08 | 6.85E-14 | 7 | miR-367 | −12.38 | 8.08E-20 |
| 8 | miR-302b | +4.80 | 4.65E-11 | 8 | miR-302a | −12.14 | 1.30E-19 |
| 9 | miR-373* | +1.65 | 5.26E-11 | 9 | miR-302c | −11.41 | 5.62E-17 |
| 10 | miRPlus_17892 | +1.07 | 1.38E-10 | 10 | miR-302b* | −9.61 | 8.97E-16 |
| B | |||
|---|---|---|---|
| miRNA | Chromosome Location | miRBase Accession |
5′ to 3′ Sequence |
| hsa-miR-371-3p | 19q13.41 | MIMAT0000723 | AAGUGCCGCCAUCUUUUGAGUGU |
| hsa-miR-372 | 19q13.41 | MIMAT0000724 | AAAGUGCUGCGACAUUUGAGCGU |
| hsa-miR-373 | 19q13.41 | MIMAT0000726 | GAAGUGCUUCGAUUUUGGGGUGU |
| hsa-miR-302a | 4q25 | MIMAT0000684 | UAAGUGCUUCCAUGUUUUGGUGA |
| hsa-miR-302b | 4q25 | MIMAT0000715 | UAAGUGCUUCCAUGUUUUAGUAG |
| hsa-miR-302c | 4q25 | MIMAT0000717 | UAAGUGCUUCCAUGUUUCAGUGG |
| hsa-miR-302d | 4q25 | MIMAT0000718 | UAAGUGCUUCCAUGUUUGAGUGU |
| hsa-miR-367 | 4q25 | MIMAT0000719 | AAUUGCACUUUAGCAAUGGUGA |
| C | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pediatric Dataset | Adult Dataset | ||||||||
| Transcription Factor |
Chromosome Location |
Overall Rank (n=347) |
Overall Log2 Fold Change |
Overall Adjusted p-value |
GCT Subtype Rank n=523 (Sem) n=647 (YST) |
Overall Rank (n=1019) |
Overall Log2 Fold Change |
Overall Adjusted p-value |
GCT Subtype Rank n=1104 (Sem) n=1196 (YST) |
| NANOG | 12p13.31 | 5 | +3.23 | 7.00E-08 | 1 (Sem) | 250 | +1.85 | 6.54E-09 | 1 (Sem) |
| TEAD4 (TEF-3) | 12p13.33 | 8 | +3.06 | 1.00E-04 | 34 (Sem) 276 (YST) |
42 | +2.77 | 2.71E-06 | 19 (Sem) 352 (YST) |
| POU5F1(OCT3/4) | 6p21.33 | 26 | +2.35 | 1.52E-07 | 13 (Sem) | 254 | +1.85 | 3.26E-07 | 23 (Sem) |
| TFAP2C | 20q13.2 | 45 | +2.13 | 5.96E-07 | 11 (Sem) | 195 | +1.96 | 3.38E-08 | 9 (Sem) |
| SOX17 | 8q11.22 | 114 | +1.67 | 3.01E-04 | 48 (Sem) 39 (YST) |
10 | +3.60 | 1.44E-11 | 41 (Sem) 42 (YST) |
| SOX15 | 17p12.3 | 260 | +1.21 | 2.11E-03 | 198 (Sem) | 570 | +1.35 | 1.10E-04 | 86 (Sem) |
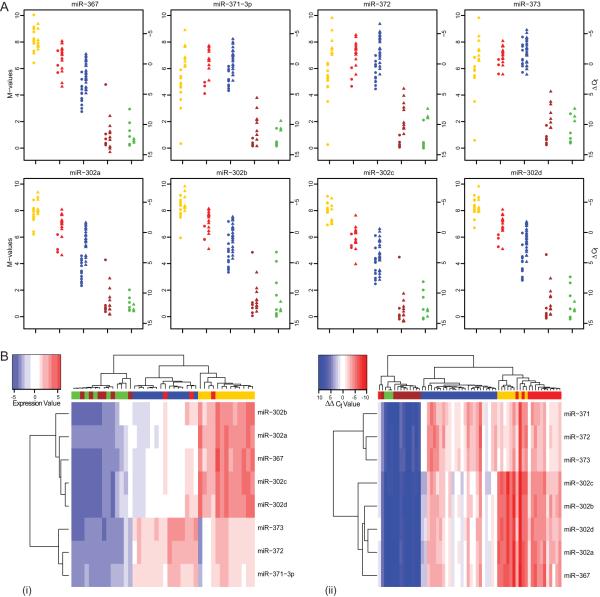
Our re-analysis of published qRT-PCR profiling of miRNA expression in adult gonadal GCTs is described in Supplementary Results (Supplementary Figure S3, Supplementary Table S2B). The top 10 differentially expressed miRNAs (Table 1A) were exclusively over-expressed miRNAs from the miR-371~373 and miR-302 clusters. Due to the different platforms used, data for the pediatric and adult samples could not be compared directly. However, parallel plots of expression values for each paediatric and adult specimen of the eight main members of the miR-371~373 and miR-302 clusters [i.e. non-miR* sequences (29)] confirmed differential expression between the malignant GCTs and the non-malignant (teratoma and control) samples (Figure 2A). Expression patterns were similar within histological subtypes, regardless of patient age.
Figure 2. Differential expression of the miR-371~373 and miR-302 clusters in malignant GCTs.
Samples are color-coded as in Figure 1. A) Expression of the eight main members of the miR-371~373 and miR-302 clusters in all tissue samples in the pediatric and adult datasets. Circles represent microarray expression M-values from individual pediatric samples (normalized within and across arrays), while triangles represent normalized Δ CT values for individual adult samples. B) Hierarchical clustering analysis based on the miR-371~373 and miR-302 clusters in (i) pediatric and (ii) adult samples.
Hierarchical clustering analysis using just the eight main members of the miR-371~373 and miR-302 clusters showed complete segregation of malignant GCTs from non-malignant samples for both the pediatric and adult data [a single outlier (an EC sample) for the latter notwithstanding (Figure 2B)]. Interestingly, six of these eight miRNAs (miR-372~373 and miR-302a~302d) share a common key 2-7nt seed region AAGUGC (Table 1B), which corresponds to the SCR hexamer GCACTT in mRNA targets. There was no evidence that expression of these miRNA clusters was DNA copy number driven in pediatric malignant GCTs, as assessed by 1Mb interval array-based comparative genomic hybridization (30, 31) (data not shown). For details of miRNA expression profiles in malignant GCT subtypes and CLs, and associations between miRNA expression and tumor site, see Supplementary Results, Supplementary Tables S2-S4 and Supplementary Figures S4/S5.
Transcriptional regulation of miRNA clusters
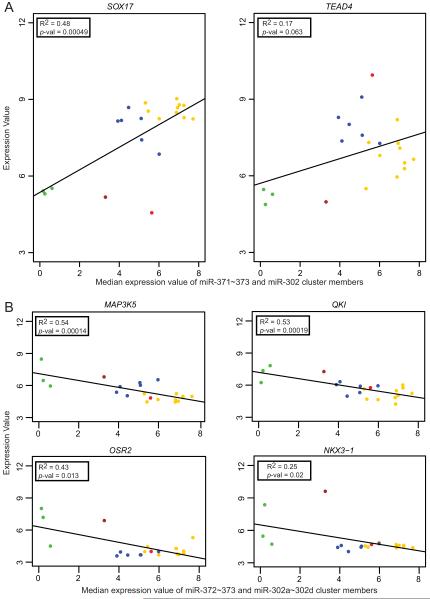
To identify transcription factors (TFs) that may be responsible for miR-302 and miR-371~373 cluster over-expression, we examined gene expression profiles in our pediatric and the published adult (26) GCT datasets. For this screening exercise, we applied less stringent criteria of log2 fold-change >1.0 and adjusted p<0.01. We identified six TFs that were over-expressed in malignant GCTs overall, and in at least one malignant subtype analysis, in both the pediatric and adult datasets (Table 1C). While NANOG, POU5F1, TFAP2C and SOX15 were specifically over-expressed in seminomas, SOX17 and TEAD4 were over-expressed in both seminomas and YSTs. The pediatric EC over-expressed all TFs except SOX17. For the 21 pediatric samples for which matched miRNA and mRNA expression data were available, linear regression analysis showed a positive correlation between the median expression value for the eight main miRNAs from the miR-371~373 and miR-302 clusters and expression levels of SOX17 and TEAD4 (p<0.0005 and p=0.06, respectively) (Figure 3A).
Figure 3. Relationships between expression of miR-302 and miR-371~373 clusters and protein-coding genes.
The graphs show linear regression analysis for 21 pediatric samples with matching mRNA and miRNA expression data, plotting levels of protein-coding gene expression (y-axis) against the matched median expression values for members of the miR-371~373 and miR-302 clusters (x-axis). Panel A shows data for the transcription factors SOX17 and TEAD4, plotted against all eight main members of the miRNA clusters. Panel B shows data for four representative common SCR-containing genes, from 22 under-expressed in both pediatric and adult malignant GCTs, plotted against the six miRNAs that contain the common 2-7nt seed. Analogous plots for the remaining 18 genes are shown in Supplementary Figure S9. Samples are color-coded as in Figure 1.
Sylamer analysis of mRNA profiles in malignant GCTs
Having validated expression levels of selected miRNAs using qRT-PCR (Supplementary Results, Supplementary Figure S5), we next performed Sylamer analysis on complete mRNA genelists ranked according to differential expression between pediatric malignant GCTs and non-malignant tissues. We focused on SCRs corresponding to seeds in miRNAs that we had identified as being differentially expressed, as the majority of pediatric malignant GCT samples in which miRNA profiling had been performed had matched mRNA profiling data. For further details of the analysis model, and the demonstration of the advantages of miRNA target identification using Sylamer rather than standard prediction methods, see Supplementary Results and Supplementary Figures S6/S7.
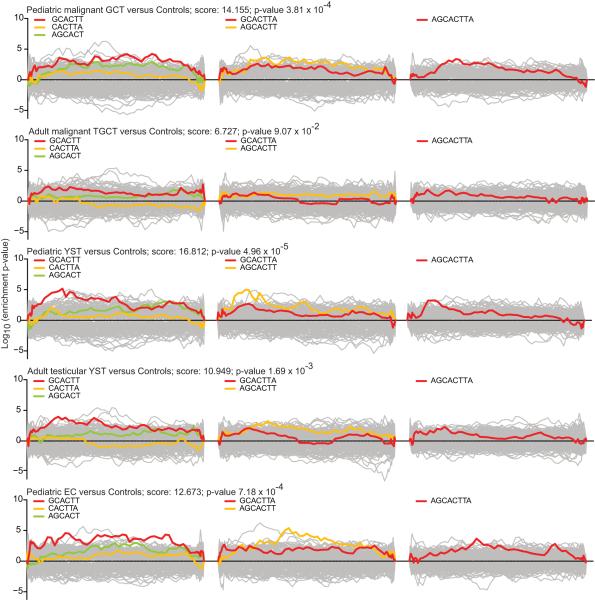
We observed that the SCR hexamer GCACTT, complementary to the common 2-7nt seed region AAGUGC of miR-372~373 and miR-302a~302d, was the most enriched in the mRNAs down-regulated in pediatric malignant GCTs (single p-value for all SCR elements of different length=3.81×10−4), YSTs (p=4.96×10−5) and ECs (p=7.18×10−4), compared to non-malignant tissues (Figure 4). No enrichment for this common SCR was seen when pediatric seminomas were compared with controls. Similar analysis of published data for adult TGCTs (26) also revealed that the common SCR hexamer GCACTT was the most enriched in genes under-expressed in adult testicular YSTs versus normal adult testis (single p-value for all SCR elements=1.69×10−3) (Figure 4). This hexamer showed non-significant enrichment in down-regulated genes when considering all adult TGCTs (p=0.09) (Figure 4) and adult testicular seminomas compared to normal adult testis. No other SCRs corresponding to the seeds of significantly over-expressed miRNAs were over-represented (p<0.01) in the 3′UTRs of genes under-expressed in malignant GCTs in either the pediatric or adult datasets. For Sylamer analysis of global shifts in mRNA profiles corresponding to miRNAs under-expressed in malignant GCTs, see Supplementary Results and Supplementary Figure S8.
Figure 4. Sylamer landscape plot analysis for the SCR words corresponding to the common seed of miR-372~373 and miR-302a~302d.
Log10-transformed and sign-adjusted enrichment p-values for each SCR word, relative to p-values of all other words, are plotted on the y-axis, against the ranked genelist on the x-axis (down-regulated genes to left; up-regulated genes to right). For each comparison, the left hand plot shows the data for the three hexamers complementary to 1-8nt in the common seed region, the central plot the two heptamers and the right hand plot the octamer. The single summed significance score and p-value for all six SCR words in each comparison is given.
Pathway enrichment analysis of down-regulated mRNAs in malignant GCTs
Of 120 mRNAs that we identified as significantly down-regulated in pediatric malignant GCTs versus non-malignant tissue samples, transcript and 3′UTR information were available for 102. Sylamer identified that while the common 2-7nt SCR GCACTT was present in the 3′UTR of 17.4% (n=30) of the 172 up-regulated mRNAs in this comparison (log2 fold-change >1.5 and adjusted p<0.01) and a similar percentage of all 13,042 genes covered by the Affymetrix U133A GeneChip (16.3%; n=2,125), it was enriched in the 102 down-regulated mRNAs, being present in 41 (40.2%) (Table 2A). The 41 mRNAs showed significant over-representation of the GO terms ‘regulation of cellular and biological processes’, ‘intracellular signaling cascade’ and the three related terms ‘regulation of GTPase activity’, ‘regulation of Ras protein signal transduction’ and ‘regulation of small GTPase signal transduction’ (Table 2B). In contrast, for the remaining 61 down-regulated mRNAs in which the common SCR was absent, the over-represented GO terms were generally related to a small number of metabolic processes only, with small numbers of genes per term (Table 2C).
Table 2. Biological significance of the miR-371~373 and miR-302 clusters in malignant GCTs.
A) The common 2-7nt SCR GCACTT is enriched in down-regulated mRNAs in pediatric malignant GCTs. The Table shows the numbers of down-regulated genes in which the sequence complementary to the common 2-7nt seed of miR-372~373 and miR-302a~302d is either present or absent. B) and C) Gene Ontology analysis for mRNAs down-regulated in pediatric malignant GCTs versus non-malignant samples, according to C) the presence or D) the absence of the SCR corresponding to the common 2-7nt seed of miR-372~373 and miR-302a~302d. D) The 22 down-regulated gene targets, common to both pediatric and adult malignant GCTs, for which the 3′UTR contains the common 2-7nt SCR. The 19 genes containing the common 2-7nt SCR that were significantly down-regulated in the pediatric samples only are listed in Supplementary Table S5
| A | ||||
|---|---|---|---|---|
| Pediatric GCT Dataset |
Number of Down- regulated Genes |
Down-regulated Genes with Transcript and 3′UTR Information |
Common SCR Present in 3′UTR |
Common SCR Absent in 3′UTR |
|
Malignant GCT versus Controls |
120 | 102 (85.0%) | 41 (40.2%) | 61 (59.8%) |
|
YST versus Controls |
146 | 126 (86.3%) | 47 (37.3%) | 79 (62.7%) |
|
Seminoma versus Controls |
189 | 159 (84.1%) | 51 (32.1%) | 108 (67.9%) |
| B | |||||
|---|---|---|---|---|---|
| Pediatric malignant GCT versus non-malignant controls; common SCR present | |||||
| GO Term | GOBPID | Total Genes in GO Term |
Gene Count | Expected Gene Count |
Adjusted p- value |
|
Regulation of small GTPase signal transduction |
GO:0051056 | 115 | 5 | 0.499 | 9.90E-04 |
| Regulation of cellular process | GO:0050794 | 2943 | 22 | 12.78 | 3.20E-03 |
| Regulation of Ras protein signal transduction | GO:0046578 | 92 | 4 | 0.4 | 5.20E-03 |
| Regulation of biological process | GO:0050789 | 3033 | 22 | 13.171 | 5.50E-03 |
| Regulation of Ras GTPase activity | GO:0032318 | 44 | 3 | 0.191 | 7.20E-03 |
| Intracellular signaling cascade | GO:0007242 | 769 | 10 | 3.339 | 8.00E-03 |
| C | |||||
|---|---|---|---|---|---|
| Pediatric malignant GCT versus non-malignant controls; common SCR absent | |||||
| GO Term | GOBPID | Total Genes in GO Term |
Gene Count | Expected Gene Count |
Adjusted p- value |
| Malate metabolic process | GO:0006108 | 5 | 2 | 0.031 | 2.60E-03 |
|
Fat-soluble vitamin metabolic process |
GO:0006775 | 6 | 2 | 0.037 | 3.90E-03 |
| Vitamin A metabolic process | GO:0006776 | 6 | 2 | 0.037 | 3.90E-03 |
| Golgi organization and biogenesis | GO:0007030 | 8 | 2 | 0.05 | 7.20E-03 |
| D | ||||||
|---|---|---|---|---|---|---|
| Gene Information | Pediatric Dataset | Adult Dataset | Function OMIM: www.ncbi.nlm.nih.gov/omim/ and GENATLAS: genatlas.medecine.univ-paris5.fr/ |
|||
| Accession | Name | Rank (n=102) |
Log2 Fold Change |
Rank (n=212) |
Log2 Fold Change |
|
| NM_000849 | GSTM3 | 1 | −3.38 | 3 | −5.03 | Metabolic, mutated in cancer |
| NM_053001 | OSR2 | 3 | −2.69 | 95 | −2.19 | Zinc finger protein, transcription factor, development |
| NM_006379 | SEMA3C | 4 | −2.51 | 78 | −2.41 | Immunoglobulin domain, short basic domain |
| NM_181847 | AMIGO2 | 5 | −2.50 | 161 | −1.76 | Adhesion molecule |
| NM_006167 | NKX3-1 | 7 | −2.47 | 19 | −3.68 | Transcription factor; down-regulated in TGCT / prostate ca |
| NM_207304 | MBNL2 | 8 | −2.35 | 114 | −2.07 | Zinc finger protein, regulates alternative splicing |
| NM_018013 | SOBP | 9 | −2.16 | 42 | −3.02 | Nuclear zinc finger protein; cell fate and patterning |
| NM_001023567 | GOLGA8B | 10 | −2.00 | 75 | −2.45 | Golgi autoantigen, golgin subfamily a, 8B |
| NM_178140 | PDZD2 | 11 | −2.02 | 31 | −3.33 | Transmembrane receptor binding protein |
| NM_002736 | PRKAR2B | 12 | −2.01 | 127 | −2.00 | cAMP-dependent protein kinase |
| NM_001015045 | FAM13A1 | 17 | −1.94 | 113 | −2.08 | Family with sequence similarity 13, A1. Function unknown |
| NM_015230 | ARAP2 | 18 | −1.92 | 145 | −1.85 | ArfGAP protein that regulates focal adhesion |
| NM_005491 | MAMLD1 | 22 | −1.78 | 136 | −1.94 | Transactivates Hes3 promoter |
| NM_005923 | MAP3K5 | 24 | −1.76 | 210 | −1.51 | Activates MAPK; tumor suppressor gene; pro-apoptotic |
| NM_001116 | ADCY9 | 25 | −1.76 | 174 | −1.70 | Adenylate cyclase |
| NM_001101800 | FAM13B | 26 | −1.71 | 154 | −1.79 | Family with sequence similarity 13, B. Function unknown |
| NM_006022 | TSC22D1 | 27 | −1.67 | 181 | −1.67 | Transcription factor; early-response gene |
| NM_022817 | PER2 | 30 | −1.63 | 104 | −2.13 | Hyper-methylated in cancer; circadian rhythm |
| NM_206853 | QKI | 31 | −1.62 | 76 | −2.43 | RNA binding protein; RNA export and stability |
| NM_006380 | APPBP2 | 36 | −1.55 | 63 | −2.63 | Interacts with microtubules |
| NM_020194 | MFF | 40 | −1.50 | 165 | −1.75 | Membrane protein; apoptosis |
| NM_001017977 | DCAF6 | 41 | −1.50 | 151 | −1.83 | Enhances transcription by nuclear receptors |
Of the 41 mRNAs containing the common 2-7nt SCR in the pediatric comparison, we identified 22 that were also present in the corresponding adult genelist (and therefore most likely to be direct targets of the miR-372~373 and miR-302a~302d families), including numerous cancer-associated genes (Table 2D, Supplementary Results and Supplementary Table S5). Linear regression analysis showed significant negative correlations for 21 of the 22 genes between expression levels and the median expression value for the six miRNAs from the miR-371~373 and miR-302 clusters that contain the common 2-7nt seed AAGUGC, using data from the 21 pediatric samples with matched miRNA and mRNA expression data (Figure 3B and Supplementary Figure S9).
Similar observations were made for mRNAs down-regulated in pediatric YSTs versus non-malignant tissue and for mRNAs down-regulated in pediatric seminomas versus non-malignant tissue. For both comparisons, the common 2-7nt SCR was enriched in down-regulated genes, being present in 37.3% and 32.1% respectively (Table 2A). Likewise, GO terms for SCR-containing mRNAs included a range of cancer-associated processes, while GO terms for mRNAs without the SCR generally represented metabolic processes only (Supplementary Tables S6/S7). For details of enrichment for the common 2-7nt SCR GCACTT in down-regulated mRNAs in adult TGCTs and analogous GO analysis, see Supplementary Results and Supplementary Table S8. For genes down-regulated in YSTs versus non-malignant samples and seminomas versus non-malignant samples for the pediatric and adult datasets, see Supplementary Results and Supplementary Tables S9/S10, respectively.
Discussion
In this study we have demonstrated that the majority of miRNAs differentially expressed in pediatric malignant GCTs are down-regulated, as has been observed for other types of malignancy (32). Nevertheless, the most significant differential expression was up-regulation of the miR-371~373 and miR-302 clusters, regardless of histological subtype, tumor site (ovary, testis or extragonadal) or patient age. Over-expression of these miRNAs appears to be specific to malignant GCTs, with no similar findings for other malignancies or diseases to date, save for miR-372~373 over-expression in an isolated case of an exceptionally rare embryonal brain tumor, at much lower levels than in malignant GCTs (33).
The miR-371~373 cluster was previously reported to be over-expressed in adult gonadal malignant GCTs, based on qRT-PCR (14) and RNase protection assay (15). However, these reports are inconsistent regarding expression of the miR-302 cluster in such tumors. One stated that miR-302a~302d expression was undetectable in many miR-371~373-expressing malignant GCTs, consistent with miR-371~373 over-expression being a selected event in malignant GCT development (15). In contrast, the other appears to illustrate over-expression of miR-302a~302d in all adult gonadal malignant GCTs, although this was not explicitly commented on (14). Our re-analysis of the published qRT-PCR data shows that the miR-302 cluster is as significantly over-expressed as miR-371~373 in adult gonadal malignant GCTs compared to non-malignant tissues (teratomas and controls) (Table 1A), mirroring our observation for pediatric gonadal and extragonadal malignant GCTs. As both miRNA clusters are believed to be ESC-specific pluripotency markers (34-38), our findings suggest that expression of the miR-371~373 and miR-302 clusters in malignant GCTs either represents persistence of an embryonic pattern of miRNA expression that is not present in normal tissues and teratomas (the latter having undergone somatic differentiation), or acquired re-expression, regulated by an as yet undetermined mechanism. We observed associations between miR-371~373/miR-302 levels and TF over-expression, warranting future investigations of their functional relationships, which may be complex. For example, NANOG and POU5F1 have binding sites in the miR-302 (35, 39, 40) and miR-371~373 (40) cluster promoter regions, while POU5F1 is negatively regulated by miR-145 (41) which is significantly down-regulated in both pediatric and adult malignant GCTs (Supplementary Table 2A/B). Our observation of SOX17 over-expression in seminoma and YST, but not in an EC sample, is consistent with previous reports (42-44).
Our Sylamer analysis strongly suggests that over-expression of the miR-371~373 and miR-302 clusters is functionally important in malignant GCTs by globally affecting levels of target mRNAs. We observed significant enrichment of the SCR hexamer GCACTT (complementary to the common miR-372~373 and miR-302a~302d 2-7nt seed AAGUGC) in genes under-expressed in pediatric malignant GCTs, YSTs and EC versus non-malignant controls and in adult YSTs versus testicular controls. Sylamer did not identify over-representation of SCRs corresponding to other over-expressed miRNAs. Nevertheless, such miRNAs may contribute to the clinicopathological heterogeneity of malignant GCTs, by targeting a smaller, more discrete number of mRNAs. For example, each subtype of pediatric malignant GCT showed specific abnormalities of miRNA expression (such as over-expression of miR-182~183 cluster in seminomas, miR-375 in YSTs and miR-515~526 cluster in ECs) and we observed significant differential miRNA expression in intracranial versus extracranial seminomas. Interestingly, however, the striking differences in mRNA expression that we previously observed between pediatric and adult malignant GCTs (2) were not reflected by similar differences in miRNA expression profiles.
Using GO analysis, we demonstrated that for pediatric malignant GCTs, and their main subtypes YST and seminoma, the down-regulated mRNAs containing the SCR corresponding to the common miR-372~373/miR-302a~302d seed mediate cellular processes important in oncogenesis and malignant progression (signal transduction, cell cycle, development and morphogenesis, etc.), in contrast to the small number of metabolic processes identified for down-regulated mRNAs without the common SCR. Together, these findings indicate the generalized functional significance of miR-372~373 and miR-302a~302d in the biology of malignant GCTs. Interestingly, these miRNA clusters, via their common 2-7nt seed AAGUGC, are known to be essential for regulating G1-S transition and promoting rapid proliferation in embryonic stem cells (45, 46). Our data further support the use of GO enrichment analysis to identify groups of genes targeted by the same miRNA seed that share a biological function (47). Of note, considerably weaker signals were obtained from the Sylamer and GO enrichment analysis of the adult mRNA dataset (26), in which controls were normal adult testis samples only, leading to large numbers of differentially expressed genes being related to male reproduction and spermatogenesis. We obtained a more tractable list of differentially expressed genes by selecting a range of control tissues containing normal germ cells at different developmental stages.
In conclusion, our data indicate that the miR-371~373 and miR-302 clusters are universally over-expressed in malignant GCTs and are of functional significance by down-regulating mRNAs involved in biologically significant pathways. It will now be important to translate our findings clinically. The miRNA expression changes we describe may improve tumor diagnosis and post-treatment monitoring, and enable novel therapeutic approaches that target fundamental abnormalities of malignant GCT cells.
Supplementary Material
Acknowledgments
Financial support: Medical Research Council Clinical Research Training Fellowships [RDP and MJM], Glaxo-SmithKline Postdoctoral Fellowship [HKS] and Wellcome Trust Sanger Institute Postdoctoral Fellowship [CA-G], with further support from the Medical Research Council, Cancer Research UK, CLIC Sargent, Parthenon Trust, and Addenbrooke’s Charitable Trust.
Footnotes
References
- 1.Palmer RD, Nicholson JC, Hale JP. Management of germ cell tumours in childhood. Current Paediatrics. 2003;13:213–20. [Google Scholar]
- 2.Palmer RD, Barbosa-Morais NL, Gooding EL, et al. Pediatric malignant germ cell tumors show characteristic transcriptome profiles. Cancer Res. 2008;68(11):4239–47. doi: 10.1158/0008-5472.CAN-07-5560. [DOI] [PubMed] [Google Scholar]
- 3.Teilum G. Classification of endodermal sinus tumour (mesoblatoma vitellinum) and so-called “embryonal carcinoma” of the ovary. Acta Pathol Microbiol Scand. 1965;64(4):407–29. doi: 10.1111/apm.1965.64.4.407. [DOI] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Giraldez AJ, Mishima Y, Rihel J, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770):75–9. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 6.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 7.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25(46):6188–96. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 12.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Molecular cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 14.Gillis AJ, Stoop HJ, Hersmus R, et al. High-throughput microRNAome analysis in human germ cell tumours. J Pathol. 2007;213(3):319–28. doi: 10.1002/path.2230. [DOI] [PubMed] [Google Scholar]
- 15.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124(6):1169–81. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 16.van Dongen S, Abreu-Goodger C, Enright AJ. Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods. 2008;5(12):1023–5. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci U S A. 2001;98(14):8012–7. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calaminus G, Patte K. Germ Cell Tumors in Children and Adolescents. In: Agarwal BR, Perilongo G, Rogers P, Strahlendorf C, Eden OB, editors. Education Book, 37th meeting of SIOP Eindhoven. International Society of Paediatric Oncolgy; The Netherlands: 2005. [Google Scholar]
- 19.Eble JN, Sauter G, Epstein JI, Sesterhenn IA, editors. World Health Organization Classification of Tumours. Pathology and genetics of the urinary system and male genital organs. IARC Press; Lyon: 2004. [Google Scholar]
- 20.Muralidhar B, Goldstein LD, Ng G, et al. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212(4):368–77. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- 21.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics--a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25(3):415–6. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–75. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31(4):265–73. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 24.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc Ser B-Methodol. 1995;57:289–300. [Google Scholar]
- 26.Korkola JE, Houldsworth J, Chadalavada RS, et al. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006;66(2):820–7. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–8. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8. doi: 10.1093/nar/gkm952. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng G, Winder D, Muralidhar B, et al. Gain and overexpression of the oncostatin M receptor occur frequently in cervical squamous cell carcinoma and are associated with adverse clinical outcome. J Pathol. 2007;212(3):325–34. doi: 10.1002/path.2184. [DOI] [PubMed] [Google Scholar]
- 31.Greshock J, Naylor TL, Margolin A, et al. 1-Mb resolution array-based comparative genomic hybridization using a BAC clone set optimized for cancer gene analysis. Genome Res. 2004;14(1):179–87. doi: 10.1101/gr.1847304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 33.Pfister S, Remke M, Castoldi M, et al. Novel genomic amplification targeting the microRNA cluster at 19q13.42 in a pediatric embryonal tumor with abundant neuropil and true rosettes. Acta Neuropathol. 2009;117(4):457–64. doi: 10.1007/s00401-008-0467-y. [DOI] [PubMed] [Google Scholar]
- 34.Lakshmipathy U, Love B, Goff LA, et al. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2007;16(6):1003–16. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barroso-del Jesus A, Romero-Lopez C, Lucena-Aguilar G, et al. Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28(21):6609–19. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laurent LC. MicroRNAs in embryonic stem cells and early embryonic development. J Cell Mol Med. 2008;12(6A):2181–8. doi: 10.1111/j.1582-4934.2008.00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20(16):2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh MR, Lee Y, Kim JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270(2):488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 39.Card DA, Hebbar PB, Li L, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28(20):6426–38. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–33. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 42.de Jong J, Stoop H, Gillis AJ, et al. Differential expression of SOX17 and SOX2 in germ cells and stem cells has biological and clinical implications. J Pathol. 2008;215(1):21–30. doi: 10.1002/path.2332. [DOI] [PubMed] [Google Scholar]
- 43.Looijenga LH. Human testicular (non)seminomatous germ cell tumours: the clinical implications of recent pathobiological insights. J Pathol. 2009;218(2):146–62. doi: 10.1002/path.2522. [DOI] [PubMed] [Google Scholar]
- 44.Nonaka D. Differential expression of SOX2 and SOX17 in testicular germ cell tumors. Am J Clin Pathol. 2009;131(5):731–6. doi: 10.1309/AJCP7MNCNBCRN8NO. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40(12):1478–83. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Blelloch R. Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 2009;69(10):4093–6. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie W, Flamant S, Rasko JE. Predicting microRNA targets and functions: traps for the unwary. Nat Methods. 2009;6(6):397–8. doi: 10.1038/nmeth0609-397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.