Abstract
Obscurin A, a ~720kDa modular protein of striated muscles, binds to small ankyrin 1 (sAnk1, Ank 1.5), an integral protein of the sarcoplasmic reticulum, through two distinct carboxy-terminal sequences, Obsc6316–6436 and Obsc6236–6260. We hypothesized that these sequences differ in affinity, but that they compete for the same binding site on sAnk1. We show that the sequence within Obsc6316–6436 that binds to sAnk1 is limited to residues 6316–6345. Comparison of Obsc6231–6260 to Obsc6316–6345 reveals that Obsc6316–6345 binds sAnk1 with an affinity, (133 ± 43 nM), comparable to that of the Obsc6316–6436 fusion protein, whereas Obsc6231–6260 binds with lower affinity (384±53 nM). Oligopeptides of each sequence compete for binding with both sites at half-maximal inhibitory concentrations consistent with the affinities measured directly. Five of six site-directed mutants of sAnk1 showed similar reductions in binding to each binding site on obscurin, suggesting that they dock to many of the same residues of sAnk1. Circular dichroism (CD) analysis of the synthetic oligopeptides revealed a two-fold greater α-helical content in Obsc6316–6346, ~35%, than Obsc6231–6260, ~17%. Using these data, structural prediction algorithms and homology modeling, we predict that Obsc6316–6345 contains a bent α-helix of 12 amino acids, flanked by short disordered regions, and that Obsc6231–6260 has a short, N-terminal α-helix of 4–5 residues followed by a long disordered region. Our results are consistent with a model in which both sequences of obscurin differ significantly in structure, but bind to the ankyrin-like repeat motifs of sAnk1 in a similar though not identical manner.
Keywords: Obscurin, Ankyrin, Protein Docking, Homology Modeling, Electrostatic Interaction, Surface Plasmon Resonance
A fundamental question in biochemistry and cell biology is how protein complexes organize the internal membrane systems of eukaryotic cells. Skeletal and cardiac muscle are especially opportune tissues in which to address this question because they are so regularly organized. This includes the membranes responsible for excitation-contraction coupling, the transverse tubules and the sarcoplasmic reticulum (SR), that surround each of the basic units of contraction, the sarcomere, in an orderly manner. We and others have presented evidence to suggest that the regular organization of the SR around the contractile apparatus (CA) is mediated by the binding of obscurin, a large modular protein at the periphery of the sarcomere, to small ankyrin 1, a splice variant of the ankyrin-1 gene (sAnk1; also known as Ank1.5)1, 2, 3.
Obscurin A is a ~720 kDa product of the obscurin-MLCK gene that is concentrated around the periphery of myofibrils at the levels of the Z-disk and M-band4,5, 6. Its N-terminal domains are likely to anchor obscurin A to the M-band, through their ability to bind to titin, myomesin and a unique variant of myosin binding protein C slow7, 8, 9. The region of obscurin C-terminal to residue 6211 is adjacent to the network compartment of the SR membrane10, where an integral protein, sAnk1, is concentrated1, 11. Binding of sAnk1 to obscurin is mediated by two domains of sAnk1, resembling ankyrin repeats (ankyrin-like repeats, or ALRs), which associate with two distinct sequences in the C-terminal region of obscurin A, amino acids 6231–6260 and 6316–64366, 12, 13. Both sequences are rich in glutamate and other electronegative amino acid side chains that are likely to bind to the lysine and arginine residues of sAnk1, which we showed are essential for binding to Obsc6316–6436 13. We hypothesize that these two sequences differ in their structures and affinities, but that they compete for a single binding site on sAnk1. Here we compare their binding characteristics to wild type and mutant forms of sAnk1, and use spectroscopic data and homology modeling to predict their structures. Our results suggest that, although both sequences bind to the ALR’s of sAnk1, the more C-terminal sequence contains a 30 residue binding domain with a higher affinity and α-helical content that plays the dominant role in binding when both sequences are present.
EXPERIMENTAL PROCEDURES
Generation of peptides
Peptides were generated and their sequences and purities were verified, by the Biopolymer Core Facility of the University of Maryland School of Medicine.
Generation of GST-Fusion Constructs
PGEX4T-1 constructs of Obsc6316–6436 were generated previously12, and Obsc6316–6345 as well as Obsc6231–6260 were generated using the same methods. PCR was used to amplify the sequence of interest, which was enzymatically digested with BamH1 and EcoR1 and ligated into the pGEX4T-1 vector. Fused constructs were transformed into Top 10 competent cells (Invitrogen, Carlsbad, CA, http://www.invitrogen.com/). After DNA was extracted, the sequence was verified by the Biopolymer Core Facility. Primer sequences for subcloning, as well as for site-directed mutagenesis (see below) can be found in supporting information, Table 1.
Site-directed Mutagenesis
The Quik-Change II mutagenesis kit (Stratagene, Cedar Creek, TX, http://www.stratagene.com/) was used to generate single or serial mutations, following the manufacturer’s instructions. Briefly, primers were made to cover the site of interest and mutations were generated via PCR. Template DNA was removed using Dpn-1 and the mutated plasmid was transformed into XL-1 competent cells. Mutagenesis was verified by sequencing.
Production of Proteins
DNA was transformed into competent BL21* plysS cells, to reduce proteolytic degradation. Cells were grown in sequentially diluted cultures, induced with 1 mM IPTG, and allowed to produce protein for 4 h. Soluble fusion constructs were extracted from sonicated supernatants and purified by affinity chromatography, following procedures recommended by the manufacturer.
Far Western Blots
Blot overlays were performed as previously described12, 13, with one minor modification: to obtain the data shown in Fig. 5, we used a goat anti-mouse 800 IRDye secondary antibody (LI-COR Biosciences, Lincoln, NE, http://www.licor.com/) and we detected bound antibody with a LI-COR Odyssey Infared Imager.
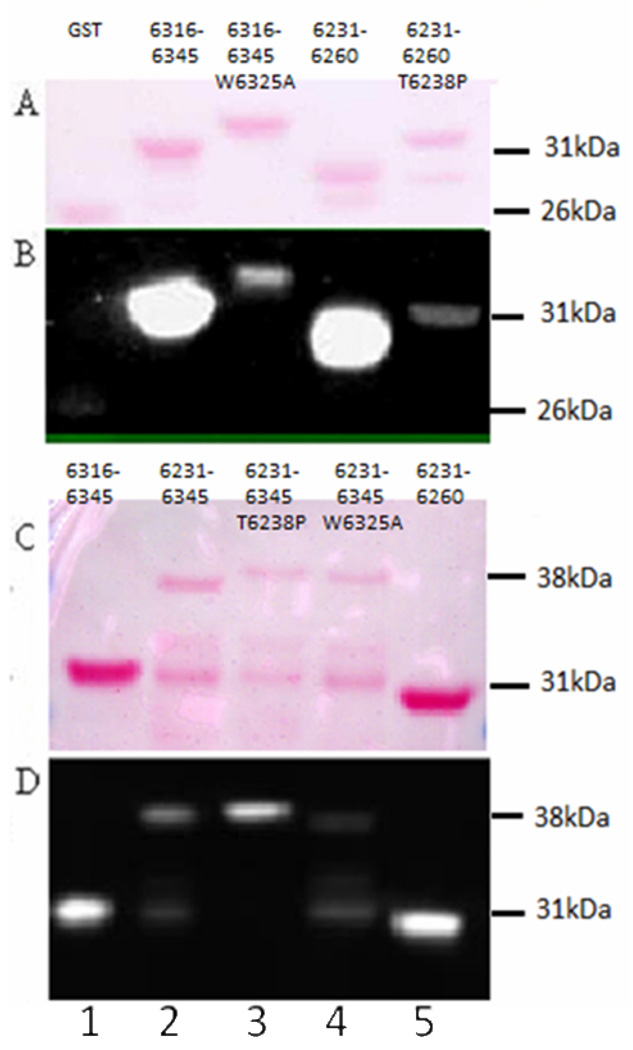
Fig. 5. Residues 6316–6345 of obscurin dominate binding to sAnk1 when both binding sites are present.
All GST constructs following transfer to nitrocellulose are shown in panel A and C with a Ponceau Red stain, which show equal loading. Panels B and D show the results of overlay assays of these nitrocellulose blots. Mutations of W6325 of Obsc6316–6345 to alanine (A,B; Lane 3, GST-Obsc6316–6345 W6325A) and T6328 of Obsc6231–6260 to proline (A,B; Lane 5, GST-Obsc6231–6260T6328P) completely ablate binding of the individual sites to MBP-sAnk129–155 in blot overlays and are barely greater than GST vector alone (A,B; Lane 1). Binding of WT constructs are shown in the adjacent lanes: Lane 2 (GST-Obsc6316–6345), Lane 4 (GST-Obsc6231–6260). These bands run at 31–32 kDa, consistent with the size of the GST fusion constructs. This developed blot in panel B was intentionally overexposed to show the dramatic nature of the inhibition by the mutants which is completely undetectable at low exposures. The GST-Obsc6231–6345 construct, which contains both binding sequences, is difficult to purify and is unstable, so only a small amount runs at the appropriate molecular mass of 38 kDa (C,D; Lane 2); a breakdown product, which retains the binding site within residues 6231–6260, is present at 31 kDa and also binds MBP-sAnk129–155. Mutation of W6325 of GST-Obsc6231–6345 to A (Lane 4: GST-Obsc6231–6345W6325A) reduces binding of the 38kDa band to MBP-sAnk1 almost completely (compare to Lane 2). Mutation of T6328 of GST-Obsc6231–6345 to P (lane 3, GST-Obsc6231–6345T6328P) does not inhibit binding of the 38kDa band to MBP-sAnk129–155 and may in fact enhance it (compared to Lanes 2 and 4). The 31 kDa breakdown product of GST-Obsc6231–6345T6328P does not bind to MBP-sAnk129–155, however, consistent with the effects of this mutation on GST-Obsc6231–6260. Both individual sites, GST-Obsc6316–6345 (Lane 1) and GST-Obsc6231–6260 (Lane 5), like panels A and B bind MBP-sAnk1. This experiment was repeated 5 times, with the same results.
Surface Plasmon Resonance
Quantitative binding studies were performed by surface plasmon resonance (SPR) with a Biacore 3000 (GE Healthcare, http://www.gelifesciences.com), as described12, 13.
After determining that there was no significant difference in binding between Obsc6316–6436 and Obsc6316–6345 by the method outlined below, we compared the ability of Obsc6316–6345 and Obsc6231–6260 to bind sAnk1 (Figure 2A, B), by making GST constructs of both sequences and capturing them with anti-GST (GE Healthcare,) conjugated to a Biacore CM5 chip (GE Healthcare). We then applied MBP-sAnk1 over a wide range of concentrations to determine the dissociation constant, KD. We corrected for non-specific binding by subtracting binding of the MBP fusion proteins to chips bound to GST alone, as well as the signal generated by exposing the chip charged with GST-obscurin to a solution blank. We fitted the data with a 1:1 binding model to determine the kinetic rate constants and KD for the association of each of the GST-obscurin constructs with MBP-sAnk1. The deviation of fits from the binding trace at the beginning of the dissociation phase is a result of a shift in refractive index as well as to non-specific binding to GST alone. Nonetheless all the resulting fits result in Chi2 values less than 5% of peak binding. For competition experiments, solutions containing 1 µM MBP-sAnk1 were pre-mixed with fusion proteins containing each of the binding sites of obscurin with different relative molar amounts of synthetic oligopeptide of either Obsc6316–6345 or Obsc6231–6260.
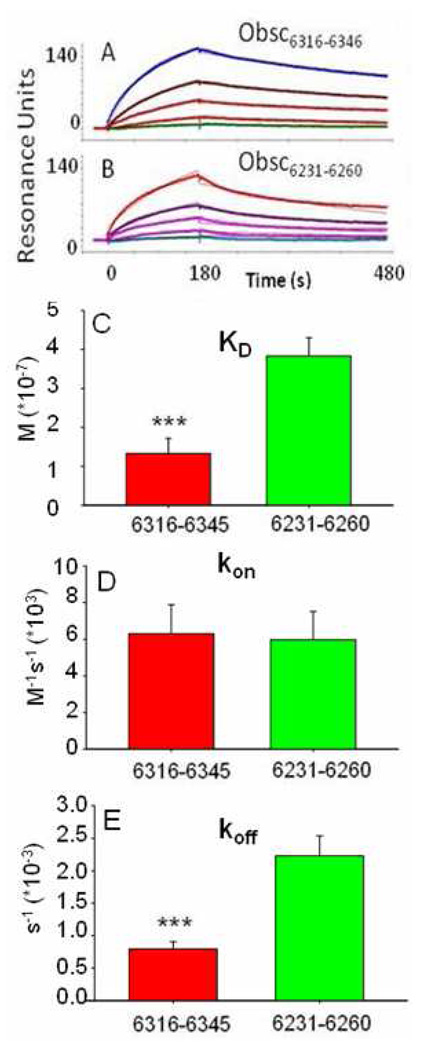
Fig. 2. Kinetics of binding of GST-Obsc6231–6260 and GST-Obsc6316–6345 to MBP-sAnk129–155.
(A, B) Colored curves show binding of serial diluted concentrations of MBP-sAnk1 starting at 3µM to GST-Obsc6316–6345 (A) and GST-Obsc6231–6260 (B). Black curves are fits based on a 1:1 stoichiometry of binding. (C–E) Bar graphs of values of KD, kon, and koff for GST-Obsc6316–6345 A (red) and GST-Obsc6231–6260 (green). *** denotes p < .05. n = 5 for all values.
CD Spectroscopy
CD spectroscopy was performed on a Jasco-810 spectropolarimeter (Jasco, Easton, MD) at 25°C. Spectra were collected from 185 to 260 nm with 0.2 mm resolution and 1.0 cm bandwidth. Spectra for the Obsc6231–6260 and Obsc6316–6346 peptides were obtained at concentrations of 12.5 µM, in a solution containing 20mM sodium tetraborate, 10mM sodium phosphate, pH 7.4, and varying percentages of trifluoroethanol (TFE)23. The background due to buffer alone was subtracted, and the mean residue ellipticity (deg cm2 dmol−1) was calculated using CDPRO software, supplied by the manufacturer. The percent α-helical content was determined from the spectra with TFE at 30% with the CONTINLL methods within the CDPRO analytical software14.
Secondary Structure Prediction Algorithms and Generation of Disorder Plots
We used a number of algorithms to predict secondary structure for both regions of obscurin that are based on the Chou-Fasman approach15. Essentially, these algorithms determine sequence similarity to a prescribed window of amino acids, and then use structural databases to determine a consensus secondary structure. SOPMA, SIMPA, and DSC are all variations of this approach16, 17, 18, differing primarily in the size of the sequence windows within which the sequence-structure relationship is evaluated.
The GOR algorithms take the above approach two steps farther, and integrate the constants for relative abundance of each basic secondary structure state in the phyla of interest, as well as taking hydrophobic triplets into account. GOR IV uses a three-state instead of a four-state model19. A four-state model comprises helix, extended/beta sheet, turn and coil. A three-state model excludes turn.
The PREDATOR algorithm also uses a three state model, but incorporates information about tertiary structure20. In addition to the traditional approach (see above), it incorporates data concerning the hydrogen bonding of beta bridges and alpha helical backbone (i, i+4).
Two other models, PHD and HNN, use hidden neural networks to incorporate evolutionary conservation into their algorithms 21, 22. PHD is widely used, although HNN and DSC (see above) are reported to work better under most circumstances18.
We also used established algorithms to determine the relative structural stability of local regions of both obscurin peptides. These algorithms predict the mobility of α-carbons in the oligopeptide sequences of obscurin at 1000 K, determine regions for which similar short oligopeptide sequences have never been experimentally solved, and predict random coiled domains, a necessary, but not sufficient, condition for disorder (see Results).
Homology Modeling
Obsc6316–6345 was aligned to both the forward and reverse sequence of amino acids 290–319 of RelA. As the alignment to the reverse sequence was closer, with respect to chemical characteristics of amino acids, Obsc6316–6345 was modeled to the sequence in this direction, as well as in the forward direction. As an additional validation of our homology model, we compared its characteristics with those determined by secondary structure prediction algorithms, intrinsic disorder algorithms and circular dichroism experiments.
Reagents
Unless otherwise noted, all reagents were from Sigma Chemical Co. (St. Louis, MO) and were of the highest grade available. All primers for site directed mutagenesis and cloning were made either in the Biopolymer Core Facility or by Integrated DNA Technologies (Coralville, IA, www.IDTDNA.com). The sequences of these primers are tabulated in supplemental Table 1.
RESULTS
Two sets of lysine and arginine residues exposed on the surface of two ALR motifs of sAnk1 have previously been shown to mediate the binding of sAnk1 to Obsc6316–6436 13. The two ALR motifs within sAnk1 are composed of residues 57–122 (see Fig. 1C, underlined in orange). Although these motifs lack several consensus amino acids found in ankyrin repeats, we showed previously that the distribution of charged and hydrophobic residues in this region shares high homology with the AR protein, Notch113. N-terminal to Obsc6316–6436 is another sequence, Obsc6236–6260, that has also been identified as sufficient to bind to sAnk16. Both sequences, Obsc6236–6260 and Obsc6316–6436, have clusters of electronegative residues. Specifically, Obsc6316–6436 has two regions that are rich in electronegative residues, between residues 6316 and 6346 (four glutamates) and residues 6416 and 6436 (nine glutamates and aspartates), whereas Obsc6231–6260 contains two glutamates and three threonines (Fig. 1). Because electrostatic interactions are likely to contribute to binding, we predicted that Obsc6316–6436, with the larger net negative charge, would have a higher affinity for sAnk1 than Obsc6236–6260. We confirmed this in binding experiments, described below, and then investigated the structural differences between these two sequences that may also influence their binding affinities.
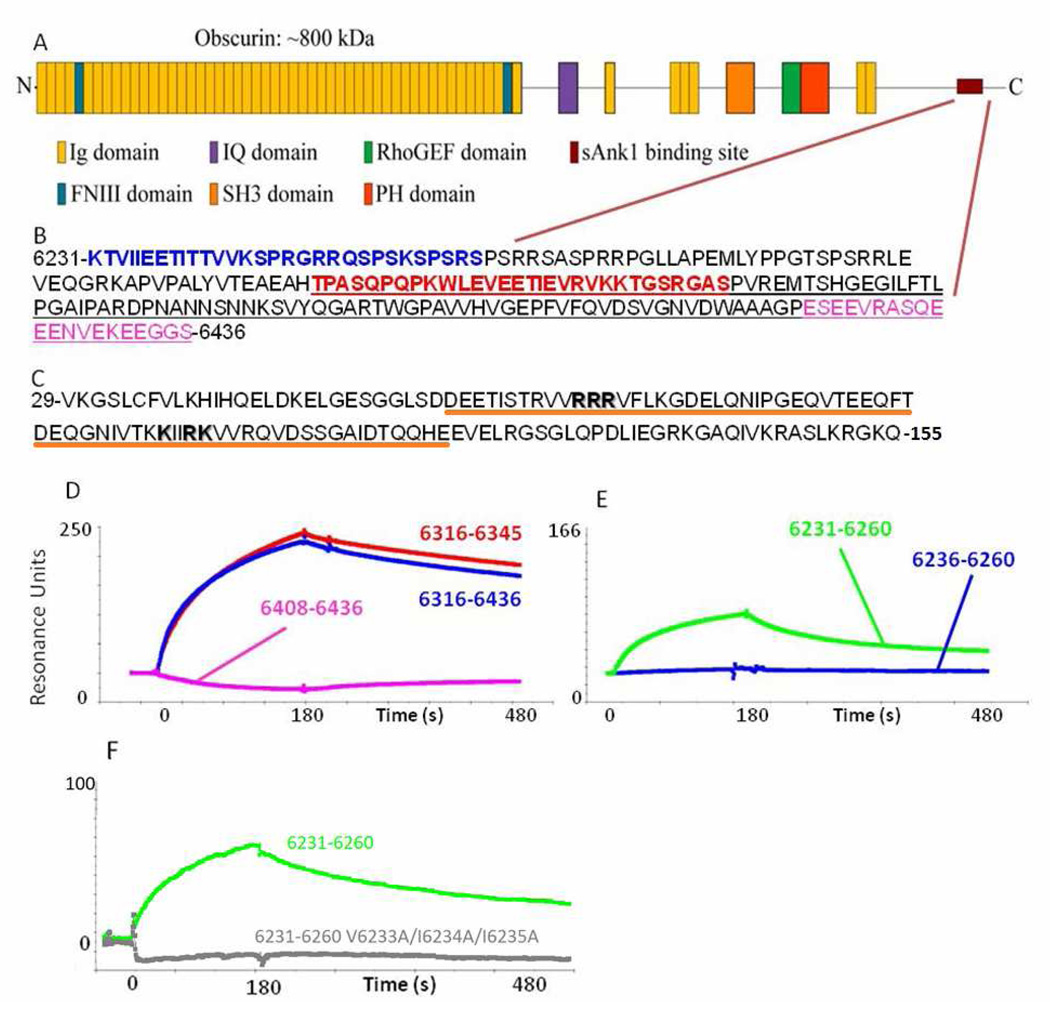
Fig. 1. The sequences of obscurin and sAnk1 that mediate binding.
(A) Cartoon of the domain organization of obscurin A. The burgundy lines indicate the region of the molecule shown in B. (B) The sequence of residues corresponding to amino acids 6231–6436 of obscurin A (Rattus norveigicus). Blue text denotes the sequence identified by Bagnato et al.6. The red sequence denotes the residues identified as the minimal binding domain and used for experiments in this manuscript (see D and E). The underlined sequence is that identified by Kontrogianni-Konstantopoulos et al.12. The sequence in pink shows a sequence rich in electronegative amino acids that is neither necessary nor sufficient to bind sAnk1 (see D, pink). (C) Sequence of residues 29–155 of sAnk1 of the rat. Residues shown by Borzok et al. to mediate binding to obscurin are shown in bold; residues 57–122, underlined in orange have previously been modeled as ankyrin-like repeats13. (D, E) Surface plasmon resonance assays of binding of fusion constructs of sAnk129–155 to different sequences of obscurin. (D) Obsc6316–6345 (red), Obsc6316–6436 (blue), Obsc6408–6436 (pink). (E) Obsc6231–6260 (green), Obsc6236–6260 (blue). (F) Obsc6231–6260 (green), Obsc6231–6260 V6233A/I6234A/I6235A (grey).
Binding Studies
Because we wanted to compare binding under equivalent conditions, we first studied the binding region in Obsc6316–6436 in detail, to learn if binding was mediated by an oligopeptide within this sequence that was closer in size to that of Obsc6236–6260. We used GST fusion constructs to test the ability of different regions of Obsc6316–6436 to bind to an MBP fusion protein of sAnk129–155, which contains the two ALR motifs required for binding obscurin13. GST-Obsc6316–6345 showed tight binding to sAnk129–155, whereas binding of Obsc6408–6436 was insignificant (Figure 1D). Notably, the affinity of the fusion protein containing only residues 6316–6345, 133 ± 43 nM (mean ± S.D.), was similar to the affinity of the fusion protein containing residues 6316–6436, reported by Kontrogianni-Konstantopoulos et al. to be 135nM 12. This suggests that the binding of the latter to sAnk1 is mediated largely by its first 30 amino acids, with the remaining 91 amino acids residues playing no significant role in this interaction. We further tested binding of a construct containing residues 6408–6436, with a similar amount of charged residues to 6316–6345, and found that there was no appreciable binding by far western blot or SPR (data not shown). We therefore focused on the region of obscurin containing residues 6316–6345 for comparison with Obsc6236–6260, first identified and partially characterized by Bagnato et al.6.
We used far western blot and SPR methods to measure the binding of a GST fusion construct of this more N-terminal binding region, GST-Obsc6236–6260, to sAnk1. Binding was minimal on blot overlay (not shown) and insignificant when analyzed by SPR as seen in Figure 1E. As additional residues may be important for binding, we added five flanking amino acids, corresponding to residues 6231–6235 (KTVII), between the GST moiety and residue 6236. Binding of the resultant construct, GST-Obsc6231–6260 was significant (Figure 1E). To confirm that these added residues were necessary for binding, we created a mutant construct with residues 6231–6235 altered to KTAAA. This mutant protein does not bind to sAnk1 (Fig. 1F). Evaluation of the kinetics of binding of wild-type GST-Obsc6231–6260 to MBP-sAnk1 revealed a KD for binding of GST-Obsc6231–6260 to sAnk1 of 384±53 nM, or ~3-fold weaker than the more C-terminal binding site, Obsc6316–6345, (Fig. 2, Table 1). Values of the kinetic constants for association and dissociation indicate that this difference in binding affinity is primarily due to differences in the dissociation rate constant, koff (Table 1).
Table 1.
Kinetic constants of MBP-sAnk1 binding to GST-Obsc6231–6260 and GST-Obsc6316–6345
| Kinetic Constant |
Obsc6231–6260 | Obsc6316–6345 |
|---|---|---|
| kon (1/Ms) | 5.97 × 103 ± 1.54 × 103 | 6.29 × 103 ± 1.60 × 103 |
| koff (1/s) | 2.23 × 10−3 ± 3.07 × 10−4 | 7.92 × 10−4 ± 1.11 × 10−4 |
| KD (M) | 3.83 × 10−7 ± 4.72 × 10−8 | 1.33 × 10−7 ± 3.88 × 10−8 |
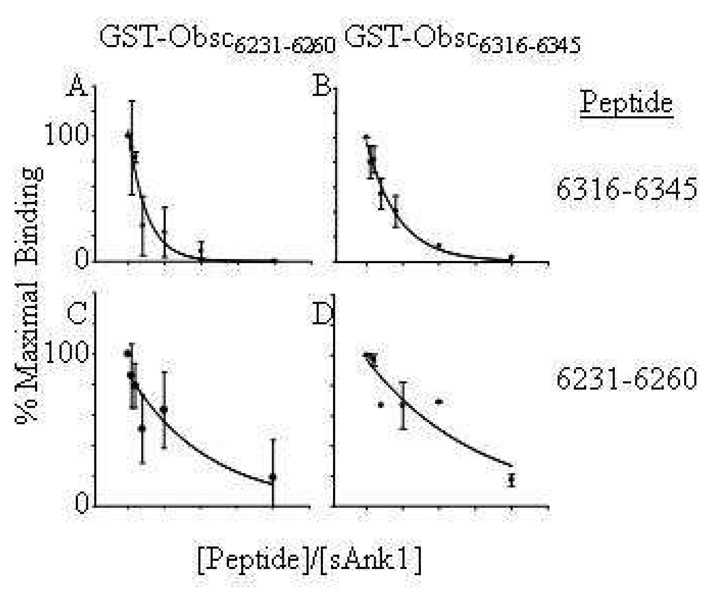
We examined the affinities of these two sequences for sAnk1 further by testing the ability of soluble oligopeptides to inhibit the binding of the fusion proteins. We synthesized oligopeptides of Obsc6231–6260 and Obsc6316–6345 and mixed them at different stoichiometric ratios with MBP-sAnk129–155. We then used SPR to assay the binding of the sAnk1 fusion protein in these mixtures to GST fusion constructs of Obsc6231–6260 or Obsc6316–6346, which were bound to GST antibody covalently attached to the surface of the SPR chip. Consistent with the results of our studies of direct binding, the synthetic oligopeptide containing residues 6316–6345 inhibited binding of MBP-sAnk129–155 to either GST-Obsc6316–6345 or GST-Obsc6231–6260 ~3-fold more effectively than the oligopeptide containing residues 6231–6260 (Fig. 3, Table 2). We derived an “inhibition constant”, defined as the reciprocal of the ratio of the concentrations of the peptide to sAnk1 required to block maximal binding by 50% (Table 2). Although a ~2-fold molar excess of the 6316–6345 peptide was needed to inhibit binding of the GST-Obsc6231–6260 construct to sAnk1 by 50%, a ~6-fold molar excess of the 6231–6260 peptide was required to inhibit binding to the same degree. The relative efficacy of the two peptides to inhibit binding of the fusion proteins differed by 3.3-fold. We obtained a similar ratio for the relative efficiency of the peptides to inhibit binding of sAnk1 to GST-Obsc6316–6345 (Table 2). These results are consistent with our results showing that GST-Obsc6316–6345 binds to MBP-sAnk1 with an affinity ~3-fold higher than that of GST-Obsc6231–6260. An oligopeptide with the sequence Obsc6411–6436, failed to inhibit binding, even at concentrations of 100 µM (not shown), also consistent with our studies of intact fusion proteins.
Fig. 3. Inhibition of binding of sAnk1 to GST-Obsc6316–6345 and GST-Obsc6231–6260 by synthetic oligopeptides.
Solutions containing 1µM MBP-sAnk1 were pre-mixed with fusion proteins containing each of the binding sites of obscurin with different relative molar amounts (shown on ordinate axis) of synthetic oligopeptide of either Obsc6316–6345 or Obsc6231–6260. (A, B) Synthetic oligopeptide 6316–6345 inhibits binding of MBP-sAnk129–155 to GST-Obsc6231–6260 (A) and GST-Obsc6316–6345 (B). (C, D) Synthetic oligopeptide 6231–6260 inhibits binding of MBP-sAnk129–155 to both GST-Obsc6231–6260 (C) and GST-Obsc6316–6345 (D) but at ratios significantly higher than oligopeptide 6316–6345. For all experiments with oligopeptide 6231–6260 experiments, n=5; for experiments with oligopeptide 6316–6345, n=6, except for those at 10:1 and 20:1 ratios, for which n=4.
Table 2.
Relative Inhibitory Activity of Obscurin Oligopeptides
| Construct | sAnk1/Oligopeptide | Ratio | |
|---|---|---|---|
| 6316–6345 | 6231–6260 | ||
| GST-Obsc6231–6260 | 0.5333 | 0.1620 | 3.29 |
| GST-Obsc6316–6345 | 0.2890 | 0.0962 | 3.00 |
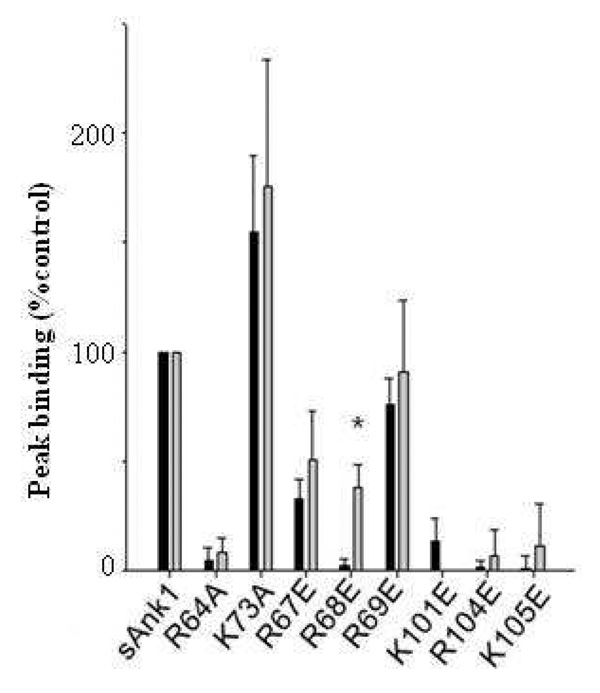
As Obsc6316–6345 and Obsc6231–6260 both contain clusters of electronegative amino acids, we tested their ability to bind to the same sites on sAnk1 by studying a series of site-directed mutants of MBP-sAnk1. These mutants, in which individual lysine or arginine residues, previously modeled as being exposed on the surface of the ALRs, were converted to glutamates (R67E, R68E, R69E, K101E, R104E, K105E), have lower binding affinities for Obsc6316–6436 13. Converting all but one (R68) of these residues to a glutamate inhibited peak binding of both obscurin sequences to a similar degree (Fig. 4). The R68E mutation affected binding of MBP-sAnk1 to Obsc6316–6345 more profoundly than to Obsc6231–6260. These results suggest that, with the exception of R68, binding of Obsc6316–6345 and Obsc6231–6260 require the same set of residues on sAnk1. The binding sites on sAnk1 for both peptides, although not identical, are therefore likely to overlap extensively.
Fig. 4. Site-directed mutants of sAnk1 reduce binding of Obsc6231–6260 and Obsc6316–6345 to a similar extent.
We assayed the effects on binding of mutating the lysine or arginine residues of sAnk1 involved in binding obscurin (see Fig. 1) to glutamates or alanines. Grey bars: Binding to Obsc6231–6260 (normalized to maximal binding, measured with WT sAnk1); black bars: binding to Obsc6316–6345 (normalized similarly). With the exception of sAnk1 R68E, mutations in sAnk1 have similar effects on binding to each of the binding sites on obscurin. n=5 for all experiments. Error bars, S.D.; * indicates a significant difference (p < .05).
We next examined the ability of sAnk1 to bind a polypeptide containing both of obscurin’s binding sites. We generated a GST-Obsc6231–6345 construct and tried to express it at high enough levels in bacteria to purify it for SPR studies. This resulted in a degraded product, with only small levels of the intact polypeptide that could be affinity purified. After purification, the protein was degraded further, with the predominant band containing GST linked to the more N-terminal of the two binding sequences. Because we could not purify enough GST-Obsc6231–6345 for quantitative assays using SPR, we examined binding qualitatively in far western blots. We compared binding of this large construct to sAnk1 in blot overlays to GST fusion constructs of the individual sites, Obsc6231–6260 and Obsc6316–6345, and to site-directed mutants, Obsc6231–6260T6328P and Obsc6316–6345W6325A. Previously, Bagnato et al.6 showed that mutating T6238 to proline inhibits binding to sAnk1. We hypothesized that since tryptophan is a rare, as well the most evolutionary conserved amino acid, mutating the W residue in the middle of obscurin’s high affinity binding site for sAnk1 to alanine would inhibit binding also. These mutations both ablated binding of the GST fusion constructs to MBP-sAnk1 (Fig. 5A,B; Lanes 3 and 5), yielding binding that was barely above the background levels of GST alone (Fig. 5A,B; Lane 1). We introduced these same individual mutations from the 30-mer obscurin fusion proteins into a larger construct containing both sites, to create Obsc6231–6345T6328P and Obsc6231–6345W6325A. The W6325A mutation within the double construct (Fig. 5C, D; Lane 4) significantly decreased binding to sAnk1 compared to control (Fig. 5C, D; Lane 2), due to its ablation of binding of the Obsc6316–6345 binding site to sAnk1 (Fig. 5A, B; Lane 2 and Fig. 5C, D; Lane 1). By contrast, the T6328P mutation (Fig. 5C, D lane 3) slightly increased binding of the Obsc6231–6345 double construct. Binding to the breakdown product of the latter at ~32 kDa disappeared, consistent with complete ablation of binding of sAnk1 to Obsc6231–6260 with the T6328P mutation (Fig 5A, B; Lane 5) compared to Obsc6231–6260 (Fig. 5A, B; Lane 4 and Fig. 5C, D; Lane 5) 6. These results suggest that the more C-terminal site, Obsc6316–6345, is the dominant binding region in the Obsc6231–6345 construct under the conditions used.
Structural Studies
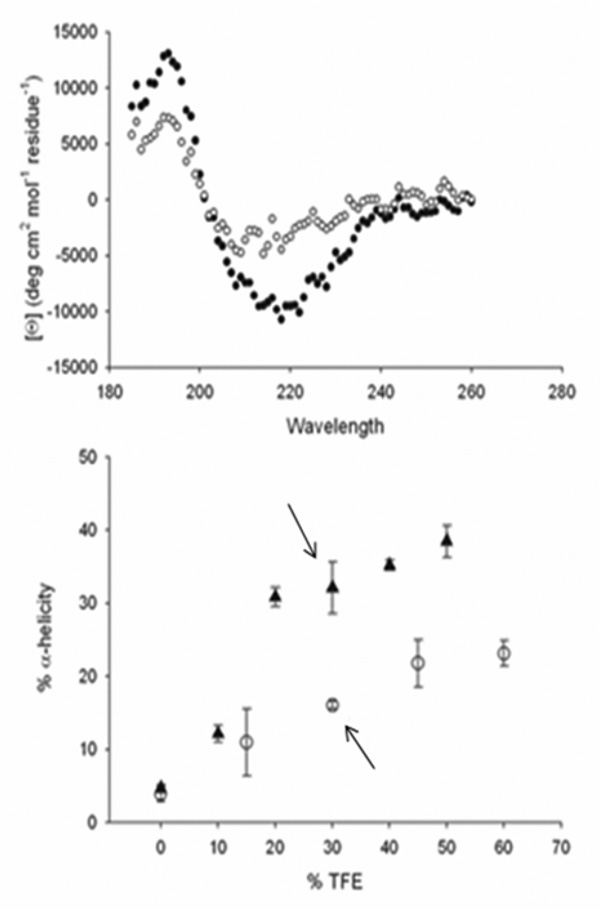
As both Obsc6316–6345 and Obsc6231–6260 associate with many of the same residues on sAnk1 but bind with different affinities, we predicted that they would differ in secondary structure. In initial tests of possible structural differences, we studied each of the synthetic oligopeptides of obscurin containing residues 6231–6260 or 6316–6345 by circular dichroism (CD) spectroscopy. In the presence of 30% TFE, the spectrum of the Obsc6316–6345 oligopeptide consistently showed ~35% α-helicity (Figure 6A, closed triangles). Similar studies of Obsc6231–6260 oligopeptide gave spectra indicating ~17% α-helicity (Figure 6A, open circles). We also examined the helicity of both peptides at other concentrations of TFE (Fig. 6, closed symbols). The α-helical content of residues 6316–6345 was consistently greater than residues 6231–6260 over the entire range of concentrations of TFE assayed, suggesting a significant difference in the secondary structures of these two peptides.
Fig. 6. CD spectra of synthetic oligopeptides of Obsc6231–6260 and Obsc6316–6345.
(A) The CD spectra of both peptides in the presence of 30% TFE are represented by triangles (6316–6345) and open circles (6231–6260) Obsc6316–6345 shows a greater degree of α-helicity, with deeper minima at ~220nm and ~208nm. (B) α-Helicity of the oligopeptides as a function of TFE concentration. Arrows indicate α-helicity at 30% TFE. n = 4.
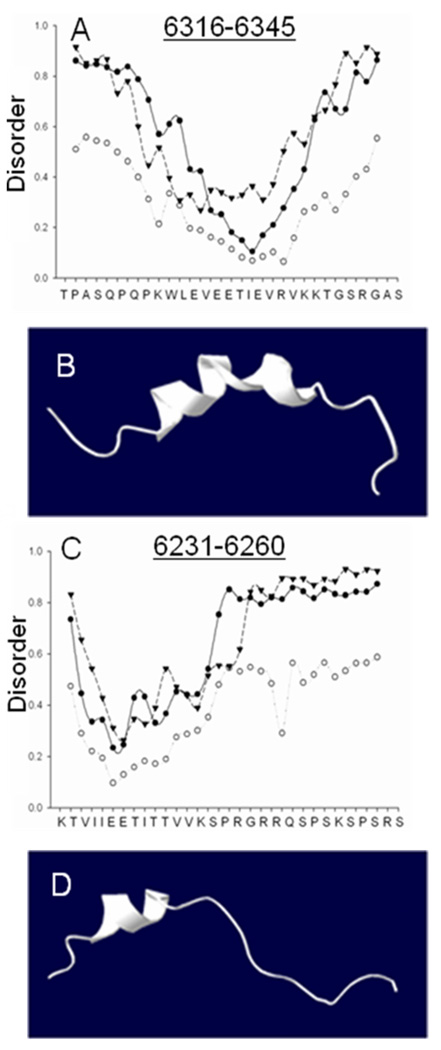
We next analyzed the two oligopeptides with algorithms, designed to predict their local disorder24 and secondary structure (see below), and compared them with the results of our CD studies. The disorder plot for the Obsc6231–6260 (Figure 7B) indicated stability near the N-terminal residues, which in conjunction with the CD data suggested the presence of an α-helix followed by a more disordered C-terminal region. The loops/coils algorithm (closed circles) also indicated that the C-terminal region of Obsc6231–6360 is ‘random coil’. The “Remark 465” algorithm (triangles) predicted that this C-terminal sequence does not assume a fixed conformation. When we used nine different algorithms designed to predict secondary structure (see Methods), only 3 predicted α-helical content (average of 12 % α-helicity) for Obsc6231–6260, with the helix located near the N-terminus of this sequence. The results of these three analyses, though not of the remaining six, suggests the presence of a short, N-terminal α-helix, approximately four residues in length, which is consistent with the CD data.
Fig. 7. Disorder plots and structural models of Obsc6231–6260 and Obsc6316–6345.
(A, C) Disorder plots: circles represent the predicted mobility of α-carbons in the oligopeptide sequences of obscurin at 1000 K; triangles represent regions for which similar short oligopeptide sequences have never been experimentally solved; closed circles represent predicted random coiled domains, a necessary but not sufficient condition for disorder. (B, D) Homology model for Obsc6316–6345 (based on RelA) and a representation of Obsc6231–6260, based on relative alpha helicity and overall similarity to known ankyrin binding motifs such as CDK4, both in good agreement with CD data (Fig. 6) disorder plots (this figure) and secondary structure prediction algorithms (not shown).
The disorder plot for Obsc6316–6345 (Fig. 7A) showed more stability in the central region of this sequence than at the ends. The nine different algorithms designed to predict secondary structure (see Methods) each indicated significant α-helical character in the central region of Obsc6316–6345 (average of 8.4 α-helical residues), with more random structures at the ends. Taken together, these algorithms predict an average of 28% α-helicity for Obsc6316–6345, with the helix located in the center of the peptide, consistent with our CD analyses.
We compared these results with the .pdb file of the crystal structure of RelA (1NFI)25, known to bind to the ankyrin repeats (ARs) of IκB, and found that the residues within it possess similar secondary structural characteristics to those predicted for Obsc6316–6345. Therefore, we used this structure to compose a homology model of Obsc6316–6345 (Fig. 7C). We also developed a model for our predictions of Obsc6231–6260 (Fig. 7D). These models are consistent with the results provided by the algorithms we used to predict structure as well as our CD measurements. If obscurin binds to the ALRs of sAnk1 in the same way that RelA binds to the ARs of IκB, then at least four of the residues identified by Borzok, et al.13 as important for binding would contribute to this interaction.
DISCUSSION
The association of obscurin with the ankyrin-like repeats (ALRs) of sAnk1 is likely to be essential for the SR to organize in a stereotypical fashion around each sarcomere6, 12, 13, 26. Obscurin A has at least two binding sites for sAnk1, located about 60 amino acids apart in its C-terminal region, but their relative roles in binding and the nature of their binding to sAnk1 have not been examined. Using SPR and competition assays, we show that the high affinity binding site on obscurin for sAnk1 is located between residues 6316 and 6345, and that the more N-terminal site, located between residues 6231 and 6260, binds to sAnk1 with an affinity that is about three-fold lower. Qualitative studies of the binding of fusion proteins suggest that residues 6316–6345 dominate binding when both sequences are present. The difference in affinity of these two sequences for sAnk1 is likely to be due in part to differences in conformation, which may determine position-specific electrostatic interactions. This was confirmed by CD spectroscopy and modeling of the secondary structure, which suggested that the higher affinity C-terminal site contains approximately twice as much α-helix as the lower affinity N-terminal site. This stability probably results in a smaller entropic loss when binding occurs, which in turn would yield a higher affinity interaction. Thus, our results indicate that, although they bind to many of the same residues on sAnk1, obscurin’s two binding sites for sAnk1 differ both in affinity and structure,
The A isoform of obscurin is a protein of 6620 amino acids, with ~60 domains that have been identified from their algorithmic homology to canonical domain sequences27, 28. These algorithmically predicted domains include Ig, IQ, FnIII, SH3, RhoGEF, and PH domains, all of which are found in many other eukaryotic proteins29. The size of these domains ranges from ~23 (IQ) to ~112 (SH3) amino acids. Ig domains, by far the most common in this protein, range from 70 to 90 amino acids in size. By definition, these domains fold in characteristic ways. The C-terminal region of obscurin, which was previously thought to be “non-modular9” possesses two short, high affinity sites for sAnk1 between amino acids 6231 and 6345, which we have characterized here. This region also binds to an unusual splice form of AnkB30, but the particular sequences involved have not yet been identified. The binding regions we have studied represent a motif that is considerably smaller in size, which could serve as templates for other proteins that bind to ALRs.
The site we first identified as being present in a ~ 12.7 kDa C-terminal fragment of obscurin, containing residues 6316–6436, binds to sAnk1 with an affinity between 35 nM13 and 135 nM12. These differences in measured affinities are likely due to differences in the stability or folding of the fusion proteins we use in our SPR assays (see Borzok, et al.13 for a discussion). Here we show that only the first ~30 amino acids of this large fragment are needed to achieve binding to sAnk1 with a comparable affinity. Although this sequence contains four glutamate residues, another sequence that follows is also rich in glutamates (residues 6408–6436) but shows no binding activity by itself, suggesting that if electrostatic interactions are involved in the binding of sAnk1 to obscurin, they must be specific. The fact that the affinity for sAnk1 of a fusion protein of Obsc6316–6345 is the same as that of a fusion protein containing the entire 121 amino acid fragment, originally identified as the binding region9, suggests that those 30 residues alone are responsible for all of the binding activity of this region of obscurin to sAnk1. Even shorter sequences may be sufficient for binding sAnk1, but we were unable to produce sufficient quantities of these smaller fusion proteins to test this concept.
As reported first by Bagnato et al.6, the nearby binding site for sAnk1, Obsc6236–6260, located ~60 residues N-terminal to the one our group has identified12, 13, is also small. Their studies were qualitative, however, and did not determine binding affinities by kinetics. For the purposes of this study, we added five additional residues from the previous exon (ENST00000284548, exon 80), just N-terminal to the 25-mer studied by Bagnato et al.6, which significantly increased the binding of the GST fusion construct. Work is under way in our lab to study the role the addition five amino acids that we added, KTVII. Preliminary observations, based on alanine mutants, suggest that the three hydrophobic residues, VII, together provide hydrophobic characteristics necessary for binding to sAnk1.
Direct measurements of binding of sAnk1 to the two sequences of obscurin, determined by SPR of the fusion proteins and oligopeptides in competition assays, consistently demonstrated that Obsc6316–6345 has an affinity for sAnk1 that is ~3-fold higher than that of Obsc6231–6260. The kinetic constants derived from analysis of the SPR curves suggest that this difference in affinity is due to different rates of dissociation. While both sequences have similar “on” rates for binding to sAnk1, the higher affinity site, Obsc6316–6345, has a slower “off” rate. The difference in affinity suggests that the two oligopeptides do not bind identically to sAnk1. This conclusion is supported by our finding that mutation of R68 of sAnk1 to glutamate has a much larger effect on the binding of GST-Obsc6316–6345 than of GST-Obsc6231–6260. The participation of an additional electrostatic interaction, mediated in part by R68, can account in part for the increased affinity of sAnk1 for Obsc6316–6345, as it would provide additional stability to the complex. We acknowledge that these data are derived from studies performed in defined solutions in vitro, which are difficult to extrapolate to the conditions in the small volumes present in the myoplasm between the periphery of the contractile apparatus, where the C-terminal region of obscurin is exposed10, and the surface of the network compartment of the SR, where sAnk1 is concentrated1. Nevertheless, if similar differences in binding exist in situ, and if binding of each of the sequences is independent of the other, then residues 6316–6345 of obscurin are ~3-fold more likely to be bound to sAnk1 at any given time than the more N-terminal residues site.
We cannot rule out the possibility that both sequences may be capable of binding simultaneously, if they can span the distances between the ALRs of neighboring sAnk1 molecules in the disulfide-linked dimers and larger oligomers likely to be present in situ11. This could also be true if sAnk1 molecules are positioned very close to one another in the SR membrane. However, modeling of the ~60 residue sequence that separates residues 6231–6260 from 6316–6345 suggests that it contains significant β-sheet content and is therefore likely to be compact (not shown). This would make it difficult to position two sets of ALRs, present on two independent sAnk1 molecules, close enough to bind both sequences simultaneously.
Since the original sAnk1 structure we proposed was a homology model, we have begun to test the stability of the ALRs using molecular dynamics simulations. Preliminary calculations show that the ALRs stay intact in the presence of computer-applied forcefields. Additional calculations suggest that the center of the high affinity site of obscurin docks on the surface of the refined ALRs via direct electrostatic and hydrophobic contacts (Busby et al., manuscript in preparation). If both sequences do not bind simultaneously, our results suggest that the higher affinity C-terminal site dominates binding, at least in vitro. Our results therefore suggest that the contribution of residues 6231–6260 to binding sAnk1 in vitro is less significant when both sequences are present.
The differences in binding affinity between Obsc6231–6260 and Obsc6316–6345 are likely due both to the differences in their primary sequences and to the secondary structures they assume. Although both are electronegative, the latter has two additional negative charges, while the former has three threonine residues that may play a role in binding6. It is likely that ionic or ion-dipole interactions mediate binding of these sequences to sAnk1, as mutation of each of the threonines in Obsc6231–6260 to proline inhibits binding6, whereas mutation of each of the glutamate residues of Obsc6316–6345 partially inhibits binding, and mutation of several eliminates binding completely (Busby et al., manuscript in preparation). Although the molecular masses of residues 6316–6345 and 6231–6260 are identical (both are 3.3kDa, and both are linked to GST), GST-Obsc6231–6260 consistently runs at a lower apparent molecular weight. As sequence differences can alter mobility, the differences in apparent molecular masses that we measure for the mutants may be expected. In particular, Obsc6316–6345 W6325A migrates more slowly than the WT construct, probably because tryptophan residues are more effective in promoting binding of SDS than alanines, consistent with an earlier report31.
Our evidence further suggests that Obsc6316–6345 and Obsc6231–6260 differ significantly in secondary structure, with Obsc6316–6345 having twice the α-helical content of Obsc6231–6260. Algorithmic predictions and modeling indicate that the location of the helices within these oligopeptides also differs. In Obsc6316–6345, the helix is predicted to be localized centrally and flanked by short stretches of less ordered residues. Our model suggests that at least three of the glutamate residues in this region are exposed on the same face of the helix, where they could interact simultaneously with the lysine and arginine residues of the ALRs of sAnk1 that mediate binding to this region of obscurin13. By contrast, our model of the shorter helix of Obsc6231–6260 shows it to be located near its N-terminus, followed by a disordered region. With so little stable structure predicted, it is perhaps remarkable that Obsc6231–6260 binds to sAnk1 with an affinity as high as ~350 nM. This high affinity may be promoted by the organization of several of its electronegative residues on one face of the predicted α-helix, where they could interact with many of the same lysine and arginine residues of sAnk1 that mediate binding to Obsc6316–6345. This result would be consistent with the results of our assays of binding to site-directed mutants of sAnk1 (Fig. 4; see above).
The distinct structures that we have predicted for obscurin’s two binding sites for sAnk1 are consistent with the results of studies of the structures of the complexes formed by other AR proteins and their ligands, including RelA and IκB, which we have used for comparison. In particular, the Obsc6316–6345 region appears to share some structural homology with RelA290–319 25. Similarly, the Obsc6231–6260 region may share some structural homology with CDK492–111 32. In agreement with our results, the sequences of these proteins that bind to ARs are only 20–30 amino acids in length and contain either short33 or longer25 α-helical regions. This suggests that many proteins that interact with ankyrin or ankyrin-like repeats have short sequences that form at least two distinct kinds of structural domains that mediate binding. Work is in progress in our laboratory to test this idea.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dawn Catino and Mike Murphy for technical assistance with SPR, Suzanne Ventura for administrative support, Dr. Eric Toth for helpful discussions and Dr. Angela Wilks for use of the Jasco CD spectroscope.
This work was supported by NIH grants to RJB (R01-AR056330) and AKK (R01-AR52768), by grants to RJB and AKK from the Muscular Dystrophy Association, and by stipends to BB and CW from two training grants, T32 GM08181 (RJB, P.I.) and T32 AR07592 (Dr. M. Schneider, P.I.).
ABBREVIATIONS
- Obsc
Obscurin
- sAnk1
small ankyrin 1
- PCR
Polymerase Chain Reaction
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- SPR
surface plasmon resonance
- CD
circular dichroism
- GST
glutathione-S-transferase
- MBP
maltose binding protein
- His
deca-histidine epitope tag
- TFE
trifluoroethanol
- ALR
ankyrin-like repeat
Footnotes
SUPPORTING INFORMATION
Primer sequences for subcloning, as well as site-directed mutagenesis can be found in supporting information, Table 1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zhou D, Birkenmeier CS, Williams MW, Sharp JJ, Barker JE, Bloch RJ. Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J. Cell Biol. 1997;136:621–631. doi: 10.1083/jcb.136.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birkenmeier CS, Sharp JJ, Gifford EJ, Deveau SA, Barker JE. An alternative first exon in the distal end of the erythroid ankyrin gene leads to production of a small isoform containing an NH2-terminal membrane anchor. Genomics. 1998;50:79–88. doi: 10.1006/geno.1998.5305. [DOI] [PubMed] [Google Scholar]
- 3.Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81:1353–1392. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 4.Bowman AL, Kontrogianni-Konstantopoulos A, Hirsch SS, Geisler SB, Gonzalez-Serratos H, Russell MW, Bloch RJ. Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett. 2007;581:1549–1554. doi: 10.1016/j.febslet.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlsson L, Yu JG, Thornell LE. New aspects of obscurin in human striated muscles. Histochem. Cell Biol. 2008;130:91–103. doi: 10.1007/s00418-008-0413-z. [DOI] [PubMed] [Google Scholar]
- 6.Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J. Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuzawa A, Lange S, Holt M, Vihola A, Carmignac V, Ferreiro A, Udd B, Gautel M. Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band: implications for hereditary myopathies. J. Cell Sci. 2008;121:1841–1851. doi: 10.1242/jcs.028019. [DOI] [PubMed] [Google Scholar]
- 8.Ackermann MA, Hu LY, Bowman AL, Bloch RJ, Kontrogianni-Konstantopoulos A. Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Mol. Biol. Cell. 2009;20:2963–2978. doi: 10.1091/mbc.E08-12-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J. Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kontrogianni-Konstantopoulos A, Bloch RJ. Obscurin: a multitasking muscle giant. J. Muscle Res. Cell Motil. 2005;26:419–426. doi: 10.1007/s10974-005-9024-7. [DOI] [PubMed] [Google Scholar]
- 11.Porter NC, Resneck WG, O'Neill A, Van Rossum DB, Stone MR, Bloch RJ. Association of small ankyrin 1 with the sarcoplasmic reticulum. Mol. Membr. Biol. 2005;22:421–432. doi: 10.1080/09687860500244262. [DOI] [PubMed] [Google Scholar]
- 12.Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol. Biol. Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borzok MA, Catino DH, Nicholson JD, Kontrogianni-Konstantopoulos A, Bloch RJ. Mapping the binding site on small ankyrin 1 for obscurin. J. Biol. Chem. 2007;282:32384–32396. doi: 10.1074/jbc.M704089200. [DOI] [PubMed] [Google Scholar]
- 14.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 15.Chou PY, Fasman GD. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 16.Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 17.Levin JM. Exploring the limits of nearest neighbour secondary structure prediction. Protein Eng. 1997;10:771–776. doi: 10.1093/protein/10.7.771. [DOI] [PubMed] [Google Scholar]
- 18.King RD, Sternberg MJ. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 1996;5:2298–2310. doi: 10.1002/pro.5560051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 20.Frishman D, Argos P. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 21.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guermeur Y. Ph.D. Thesis. Université Paris; 1997. Combinaison de classifieurs statistiques, application à la prédiction de la structure secondaire des proteins. 6.250. [Google Scholar]
- 23.Polverino de LP, Donadi M, Scaramella E, Frare E, Fontana A. Trifluoroethanol-assisted protein folding: fragment 53--103 of bovine alpha-lactalbumin. Biochim. Biophys. Acta. 2001;1548:29–37. doi: 10.1016/s0167-4838(01)00205-9. [DOI] [PubMed] [Google Scholar]
- 24.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 26.Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Randall WR, Bloch RJ. Obscurin regulates the organization of myosin into A bands. Am. J. Physiol Cell Physiol. 2004;287:C209–C217. doi: 10.1152/ajpcell.00497.2003. [DOI] [PubMed] [Google Scholar]
- 27.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 30.Cunha SR, Mohler PJ. Obscurin targets ankyrin-B and protein phophatase 2A to the cardiac M-line. J. Biol. Chem. 2008;283:31968–31980. doi: 10.1074/jbc.M806050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe T, Ishibashi A, Ariga Y, Hashimoto M, Nikaidou N, Sugiyama J, Matsumoto T, Nonaka T. Trp122 and Trp134 on the surface of the catalytic domain are essential for crystalline chitin hydrolysis by Bacillus circulans chitinase A1. FEBS Lett. 2009;494:74–78. doi: 10.1016/s0014-5793(01)02317-1. [DOI] [PubMed] [Google Scholar]
- 32.Day PJ, Cleasby A, Tickle IJ, O'Reilly M, Coyle JE, Holding FP, McMenamin RL, Yon J, Chopra R, Lengauer C, Jhoti H. Crystal structure of human CDK4 in complex with a D-type cyclin. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4166–4170. doi: 10.1073/pnas.0809645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.