Abstract
Growth hormone (GH) is important in the development and maintenance of bone; however, the IGF-dependent and -independent molecular pathways involved remain to be established. We used microarray analysis to evaluate GH signaling pathways in 4-wk-old GH-deficient mice following a single injection of GH (4 mg/kg body wt) or PBS (n = 6/group) at 6 or 24 h after treatment. Six thousand one hundred sixty genes were differentially expressed at P ≤ 0.05, and 17% of these genes were identified at both time points. Several of the genes differentially expressed were expressed sequence tags, and the remaining genes fell into 49 Gene Ontology categories. For subsequent studies, we focused on T-box (Tbx)3, a novel transcription factor, which increased more than twofold at both time points. Real-time RT-PCR analysis determined that pretreatment with IGF-binding protein-4 did not block GH-induced Tbx3 expression in vitro. Pretreatment with TNF-α blocked GH-induced Tbx3 expression. Tbx3 expression increased during osteoblast differentiation and following BMP-7 and Wnt3a treatment (P ≤ 0.05). Blocking Tbx3 expression by small interfering RNA decreased cell number and [3H]Thymidine incorporation (P < 0.01). In conclusion, 1) GH caused acute changes in several novel genes, suggesting that many GH-induced signaling pathways and target genes remain to be discovered; 2) because Tbx3 expression is regulated in osteoblasts and blockage of Tbx3 expression decreased cell number and DNA synthesis, we propose that Tbx3 is an important determinant of osteoblast cell number.
Keywords: insulin-like growth factor I, lit/lit mouse, signaling pathways
Growth hormone (GH) is important in the development and maintenance of bone. This has been demonstrated in several experiments utilizing human and animal models that are GH deficient and/or treated with GH. In GH-deficient adults, decreased bone mineral density (BMD) can be corrected with exogenous GH treatment (20, 38). In addition, GH treatment increases bone formation markers as well as serum concentrations of IGF-I (14, 19, 38). We (27) and others (42) have demonstrated that mice deficient in GH production or action exhibit significant deficits in body weight, femur length, and volumetric BMD. It is well known that GH may act directly on specific tissues or by binding to its receptors and stimulating the release of IGF-I. When GH binds to its receptors, JAK is phosphorylated and several pathways, including STAT, MAPK, and phosphatidylinositol 3-kinase, are activated (16, 31). Through these pathways, GH regulates the transcription of specific genes, such as IGF-I. Recent evidence has demonstrated that GH may also act independently of IGF-I in bone. In a previous study (27), we reported that the rate of gain in femur length and periosteal circumference during the postpubertal growth period are impaired to a greater extent in GH-deficient lit/lit mice compared with IGF-I knockout mice. Furthermore, previous published work has suggested an involvement of IGF-independent effects of GH in stimulating growth (21, 35, 42). However, the molecular pathways by which GH exerts its anabolic effects in bone remain to be established. Therefore, our first objective was to identify genes in bone that are acutely regulated by GH treatment through the use of whole genome microarray analysis. To meet our objective, we evaluated the effect of a single effective dose of GH using the GH-deficient lit/lit mouse model. This model is characterized as a dwarf mouse with a missense mutation of the GH-releasing hormone receptor (GHRHR) and no detectable concentrations of GH (3, 9, 13, 17, 27); therefore, it provides a sensitive model for examining the effects of GH on target genes in bone. Our second objective was to further characterize specific genes identified by microarray analysis in mediating GH effects in bone. Specifically, we examined the role of a novel transcription factor, T-box (Tbx)3, in regulating osteoblast proliferation in vitro. In general, we determined that several thousand genes are acutely regulated by a single injection of GH in bone and that Tbx3 is an important regulator of osteoblast proliferation.
MATERIALS AND METHODS
Animals
C57BL/6J-Ghrhrlit [GHRHR gene disruption, allele name little (lit/lit)] mice were purchased from Jackson Laboratories (Bar Harbor, ME). The lit/lit mouse is a dwarf mouse strain characterized by no detectable concentrations of circulating GH and, as a consequence, low serum IGF-I concentrations caused by a missense mutation in the GHRHR gene that abolishes the function of the receptor (9, 17). At 4 wk of age, 24 mice were treated with a single injection of human (h)GH [4 mg/kg body wt; a kind gift from Biosidus (Buenos Aires, Argentina); n = 12] or PBS (n = 12) by subcutaneous injection. The dose was selected on the basis of our previous findings that daily injections of GH (4 mg/kg body wt) in lit/lit mice induced circulating concentrations of IGF-I to levels similar to those of wild-type mice (23). At 6 h (n = 6 GH injected; n = 6 PBS injected) or 24 h (n = 6 GH injected; n = 6 PBS injected) after the single injection of GH, mice were euthanized by CO2 inhalation followed by decapitation. Femur, muscle, tibia, and liver were removed and immediately stored in liquid nitrogen and then at −70°C until analyzed.
The experimental procedures performed in this study were in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Animal Studies Subcommittee at the Jerry L. Pettis Memorial Veteran Affairs Medical Center.
RNA Extraction
RNA was extracted from the bone, muscle, and liver of each mouse (n = 6/group) using TRIzol (Invitrogen, Carlsbad, CA) and an RNeasy Mini Kit (Qiagen, Valencia, CA). Briefly, frozen tissues samples were ground with a mortar and pestle with 1 ml of TRIzol until a thin powder was visible. RNA was then extracted using the RNeasy Mini Kit according to the manufacturer’s instructions. Following extraction, up to 10 µg of RNA were DNase treated with a DNA-free kit (Ambion, Austin, TX) to remove any residual DNA. RNA quality was determined using a Bioanalyzer 2100 (Agilent, Palo Alto, CA), and RNA was quantified using a NanoDrop Spectrophotometer (Wilmington, DE).
Microarray Analysis
Within each treatment group, RNA from two animals was pooled together for a total of three replicates per treatment group. Total bone RNA (500 ng) from each pooled sample (n = 12) was reverse transcribed and linear amplified using a primer containing oligo(dT) and a T7 RNA polymerase promoter. After synthesis of the first and second strands of cDNA, the product was used in an in vitro transcription reaction to generate cRNA in the presence of cyanine 3- or cyanine 5-labeled UTP. The labeled cRNA was purified using RNeasy spin columns (Qiagen) to remove free nucleotides. Two micrograms of fragmented cyanine 3-labeled cRNA of control sample were mixed with equal amounts of cyanine 5-labeled cRNA of GH-treated sample, and the mixtures were hybridized to 22K mouse development oligo microarrays (Agilent, Palo Alto, CA) for 17 h at 60°C with constant rotation. The slide was then washed in sodium saline citrate (SSC) buffer and dried with pressurized nitrogen. All slides were scanned using an Agilent DNA microarray scanner, and the images were processed and analyzed using Agilent feature extraction software. Expression analysis of the data was analyzed using GeneSpring 7.0 (Silicon Genetics, Redwood City, CA).
Gene Expression Analysis
Quantitative real-time reverse transcriptase (RT)-PCR analysis was used to verify the microarray data as well as to further examine the regulation of specific genes that were differentially expressed. For analysis of samples used in the microarray, gene expression was determined for each individual animal (n = 6/treatment group). For in vitro experiments, gene expression was determined for four to six replicates per treatment group. Specifically, RNA samples were normalized to 300 ng per sample and reverse transcribed with oligo(dT) primers and SuperScript II (Invitrogen) to produce cDNA at a total volume of 20 µl. Real-time RT-PCR analysis was performed using a Stratagene Brilliant SYBRGreen Master Mix (Stratagene, La Jolla, CA) and an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). Briefly, a total volume of 25 µl was loaded into a 96-well plate (1 µl of cDNA product, 0.125 µl of 10 µM of forward and reverse primer, and 12.5 µl of 1× SYBR Green Master Mix). The PCR conditions were 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 1 min, 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s peptidylprolyl isomerase A. RNA were used as endogenous controls. ΔCT values were determined (CT value for gene of interest minus CT value for control gene), and comparisons of the CT values were used for relative quantification of gene expression (11). Gene-specific primers were used to amplify IGF-I (forward: 5′-CATCTCCAGTCTCCTCAGATC-3′, reverse: 5′-GTCCACACACGAACTGAAGAGC-3′), Tbx3 (forward: 5′-TTCCTACCTCACCGGGCG-3′, reverse 5′-CCGTTGGGAGGCAGCGT-3′), and PPIA (forward: 5′-TCCTGGACCCAAAACGCTCC-3′, reverse 5′-CCATGGCAAATGCTGGACCA-3′).
Tissue Culture Analysis
Tissue culture reagents
α-Minimal essential medium (α-MEM), Dulbecco’s modified Eagle’s medium (DMEM), penicillin-streptomycin suspension, and calf serum (CS) were purchased from GIBCO-BRL (Gaithersburg, MD). Bovine serum albumin (BSA) was purchased from Fluka (Buchs, Switzerland). The recombinant hGH was a gift from Biosidus (Buones Aires, Argentina). hIGF-I was purchased from GroPep (Adelaide, SA, Australia). Human bone morphogenic protein (BMP)-7 was a gift from Creative Biomolecules (Hopkinton, MA). Recombinant mouse tumor necrosis factor (TNF)-α was purchased from PeproTech (Rocky Hill, NJ). Wingless-related MMTV integration site 3A (Wnt3a) was purchased from R&D Systems (Minneapolis, MN). Human IGF-binding protein (IGFBP)-4 was purified as previously described (33). All other chemicals were purchased from Fisher Scientific (Tustin, CA) or Sigma (St. Louis, MO).
Osteoblast differentiation
Mouse bone marrow cells were isolated, counted with a hemocytometer, and plated as 20 × 106 cells per 90-mm petri dish. Bone marrow cells were cultured with α-MEM containing 10% CS, 100 U/ml penicillin, and 100 µg/ml streptomycin. Culture media were changed every 2 days until 75% confluence was reached (after ~6 days), and then 50 µg/ml ascorbic acid and 10 mM β-glycerophosphate were added to the media to induce differentiation into osteoblast cells, which was confirmed by nodule formation. One-half of the culture media was changed every 3 days for 24 days. Cells were extracted with TRIzol at days 0, 6, 12, 18, and 24 and stored at −70°C until RNA extraction was performed using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol.
Regulation of Tbx3 expression in MC3T3-E1 cells
For all in vitro experiments, MC3T3-E1 cells, a mouse osteoblast-like cell line, were cultured in DMEM or α-MEM containing 10% CS or fetal bovine serum (FBS) until 70% confluence. Media were changed to serum-free two times over a 24-h period. Cells were treated with GH (100 ng/ml), IGF-I (30 ng/ml), IGFBP-4 (300 ng/ml), TNF-α (20 ng/ml), BMP-7 (30 ng/ml), or Wnt3a (1 nM) for 24 h. RNA was extracted from the cells using an RNeasy Mini Kit according to the manufacturer’s protocol.
Transfection of cells with short interfering RNA
Short interfering (si)RNA for mouse Tbx3 (siRNA-1 and siRNA-2) and a scrambled negative control (siRNA-NC) were purchased from Dharmacon (cat. nos. D-064487-01, D-064487-02, D-001210-01-05). An RNAi human/mouse control kit was purchased from Qiagen, which was used to optimize transfection conditions. MC3T3-E1 cells were transfected with siRNA-1, -2, and -NC by use of RNAiFect (Qiagen) according to the manufacturer’s directions. Briefly, cells were plated in α- or DMEM containing 10% CS or FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin until 70% confluent. Cells were passaged with 1× trypsin and plated at a density of 3,000 cells/well for 96-well plates and 50,000 cells/well for 24-well plates. Cells were allowed to adhere to the plate (24 h) and transfected with 100 nM siRNA plus RNAiFect transfection reagent (1 or 4 µl/well for 96- and 24-well plates, respectively) for 24 h. This dose was chosen on the basis of preliminary data as well as the manufacturers’ recommendations (Dharmacon and Qiagen). In addition, cells were transfected with RNAiFect alone, siRNA-1 alone, or siRNA-NC, which served as negative controls. After transfection, media were changed to serum-free media (α- or DMEM and 0.1% BSA) for 24 h. Cells were then treated with appropriate growth factors for the desired length of time. To determine knockdown of Tbx3 expression by siRNA, RNA was extracted from 24-well plates, 24 h after transfection, using an RNeasy Mini Kit according to the manufacturer’s protocol, and expression was determined by real-time RT-PCR.
Determination of cell number
Cells were plated at 3,000 cells/well in a 96-well plate. After 24 h, cells were transfected with siRNA for 24 h, deprived of serum for another 24 h, and then treated with 1% CS. Cell number was determined by the alamarBlue assay 48 h after treatment. Specifically, cells were washed with PBS, and media were replaced with 100 µl of 10% alamarBlue (BioSource International, Camarillo, CA) diluted in phenol red-free DMEM. The fluorescence was determined 2 h later using a fluorescent plate reader (Fluorolite 1000; Dynex Technologies, Chantilly, VA). DNA synthesis was determined by a [3H]thymidine incorporation assay. Specifically, 18 h after treatment, 0.25 µCi of [3H]thymidine was added to each well and incubated for 6–8 h. Cells were washed with PBS and removed by swabbing with 10% trichloroacetic acid (TCA). Swabs were washed with TCA and 95% EtOH, and counts were determined using a scintillation counter (26).
Apoptosis
MC3T3-E1 cells were transfected and grown as described for the alamarBlue assay. Apoptosis was determined using an Apo-ONE Homogeneous Caspase-3/7 Assay (Promega, Madison, WI). Total protein was determined by the Bradford assay (Bio-Rad, Hercules, CA).
Statistical Analysis
Microarray data were analyzed using GeneSpring 7.0 (Silicon Genetics). Local background-subtracted median signal intensities were used as intensity measurements, and the data were normalized using LOWESS normalization (46). The genes that had a raw signal of 100 or greater and passed with flag value “present” were included in the analysis. To minimize the false positive/negative error rate in our microarray, we used a combination of confidence (P ≤ 0.05) and fold change (i.e., ≥1.0-, 1.5-, or 2.0-fold) to restrict our gene lists. For Gene Ontology (GO) analysis we used MappFinder (10). In this analysis, we used the genes that were significantly upregulated (fold change >1, P < 0.05) and significantly downregulated (fold change <1, P < 0.05) based on GeneSpring analysis. The Z-score was set at >0 and the permutated P at <0.05. To eliminate redundant minor GO categories, we set both the minimum number of changed genes and the minimum number of measured genes to three. Up- or downregulation of GH/IGF axis factors was used to confirm the effectiveness of the single GH injection to alter gene expression identified by microarray analysis (Table 2). The current experimental design and analysis are compliant with current MIAME guidelines (4), and the data are available in the NCBI database (acc. no. GSE3689). Real-time RT-PCR and cell number data were analyzed using Student’s t-test. Osteoblast time course data were analyzed using ANOVA (Statistica Software). A significant difference was determined at P ≤ 0.05.
Table 2.
GH-, IGF-, Wnt-, and BMP-associated genes acutely regulated by GH in bone (P ≤ 0.05)
| Common Name | 6 h | 24 h | Description | GenBank |
|---|---|---|---|---|
| GH-IGF axis | ||||
| Igf1 | 2.60 | 2.29 | Insulin-like growth factor I (Igf1), mRNA | AK038119 |
| Igf2 | 2.01 | Insulin-like growth factor II (Igf2), mRNA | NM_010514 | |
| Igfbp2 | 0.75 | Insulin-like growth factor-binding protein 2 (Igfbp2), mRNA | AK011784 | |
| Igfbp3 | 1.37 | 1.54 | Insulin-like growth factor-binding protein 3 (Igfbp3), mRNA | AK077477 |
| Igf2bp1 (IMP1) | 2.11 | 2.55 | Insulin-like growth factor II-binding protein 1 (Igf2bp1), mRNA | AK013940 |
| Igf2bp3 (IMP3) | 0.87 | Insulin-like growth factor II-binding protein 3 (Igf2bp3), mRNA | AK011689 | |
| Ghitm | 0.72 | Growth hormone-inducible transmembrane protein (Ghitm), mRNA | AK005636 | |
| Stat1 | 0.44 | Signal transducer and activator of transcription 1 (Stat1), mRNA | AK018544 | |
| Stat4 | 0.40 | Signal transducer and activator of transcription 4 (Stat4), mRNA | AK037598 | |
| Stat5a | 0.60 | Signal transducer and activator of transcription 5A (Stat5a), mRNA | AK090254 | |
| Stat6 | 1.42 | Signal transducer and activator of transcription 6 (Stat6), mRNA | NM_009284 | |
| Wnt signaling | ||||
| Wnt5a | 1.87 | 2.03 | Wingless-related MMTV integration site 5A | AK031512 |
| Dvl2 | 0.83 | 0.81 | Dishevelled 2, dsh homolog (Drosophila) (Dvl2), mRNA | NM_007888 |
| Axin | 0.94 | Axin 1 (axis inhibition protein 1) (fused protein) (fragment). | XM_128515 | |
| Wif1 | 1.33 | Wnt inhibitory factor 1 (Wif1), mRNA | AK077698 | |
| Fbxw4 | 1.43 | F-box and WD-40 domain protein 4 (Fbxw4), mRNA | AK004414 | |
| Csnk2a2 | 0.53 | Casein kinase II, alpha 2, polypeptide (Csnk2a2), mRNA | AK002927 | |
| Frzb | 0.55 | Frizzled-related protein (Frzb), mRNA | AK019093 | |
| Tle2 | 0.69 | Transducin-like enhancer of split 2, homolog (Drosophila) E(spl) (Tle2), mRNA | NM_019725 | |
| Apc | 1.05 | Adenomatosis polyposis coli (Apc), mRNA | AK042021 | |
| Csnk1a1 | 2.31 | Casein kinase 1, alpha 1 (Csnk1a1), mRNA | AK012299 | |
| BMP-associated genes | ||||
| Bmp2 | 0.74 | 0.78 | Bone morphogenetic protein 2 (Bmp2), mRNA | NM_007553 |
| Bmp4 | 0.86 | Bone morphogenetic protein 4 (Bmp4), mRNA | NM_007554 | |
| Bmp7 | 0.51 | Bone morphogenetic protein 7 (Bmp7), mRNA | AK020411 | |
| Madh5 | 2.77 | MAD homolog 5 (Drosophila) (Madh5), mRNA | AK018077 | |
RESULTS
Microarray Analysis
We identified 19,081 genes on the microarray chips that were expressed at an intensity of 100 or greater. Thirty-two percent (6,160) of these genes were differentially expressed at P ≤ 0.05 at 6 and/or 24 h (Supplemental Table S1; see AJP Endocrinol Metab web site),1 and the number of genes identified at each time point that fit these criteria are listed in Table 1. Overall, a greater number of genes were upregulated than downregulated following a single injection of GH, and more genes were differentially expressed at 24 than at 6 h. Interestingly, 17% (1,034) of the 6,160 genes were differentially expressed at both 6 and 24 h.
Table 1.
Number of genes differentially expressed (P ≤ 0.05) in bones of GH-deficient (lit/lit) mice following GH treatment
| 6 h | 24 h | 6 and 24 h | ||||
|---|---|---|---|---|---|---|
| Fold Change | Increase | Decrease | Increase | Decrease | Increase | Decrease |
| >2.0 | 417 | 152 | 592 | 561 | 282 | 44 |
| >1.5 | 842 | 445 | 1558 | 1122 | 504 | 110 |
| <1.5 | 691 | 835 | 938 | 763 | 212 | 208 |
| Total* | 1,533 | 1,280 | 2,496 | 1,885 | 716 | 318 |
GH, growth hormone.
Of the 6,160 genes differentially expressed at P ≤ 0.05, 1,034 genes were expressed at both 6 and 24 h.
To more closely evaluate the large number of genes, we used three main approaches to organize the data. First, we used GO analysis to categorize genes that were significantly up- or downregulated (P < 0.05). We identified 395 genes that fell into 49 GO categories (Supplemental Table S2). Interestingly, more than half of the genes differentially expressed by GH were ESTs and therefore could not be included in the GO analysis.
Second, to confirm the effectiveness of the single injection of GH, we identified genes associated with the GH-IGF axis that were up- or downregulated by GH treatment at P ≤ 0.05 (Table 2). Specifically, IGF-I and IGFBP-3 were upregulated more than 2- and 1.5-fold, respectively, in bone at 6 and 24 h. In addition, STAT1, -4, and -5a were downregulated and STAT6 was upregulated at 24 h. These data indicate that our single dose of GH was effective within 6 h and triggered negative feedback mechanisms by 24 h.
Third, we examined all genes to determine whether major pathways involved in the regulation of bone were acutely regulated by GH. Using the criteria of genes differentially expressed at P ≤ 0.05, we identified several Wnt-signaling genes (Table 2). Specifically, Wnt5a was upregulated twofold at both 6 and 24 h after GH treatment. In addition, several genes downstream of Wnt were differentially expressed, including Dishevelled (Dvl) 2, Wnt inhibitory factor 1 (Wif1), frizzled-related protein (Frzb), axis inhibition protein (Axin) 1, adenomatosis polyposis coli (Apc), and casein kinase (Csnk)1α1 and Csnk2α2. Surprisingly, we determined that BMP-2, -4, and -7 were downregulated 1.2- to 2-fold by GH treatment at 6 and/or 24 h, but Mad homolog (h) 5 was upregulated 2.8-fold at 24 h (Table 2).
Real-Time RT-PCR Analysis
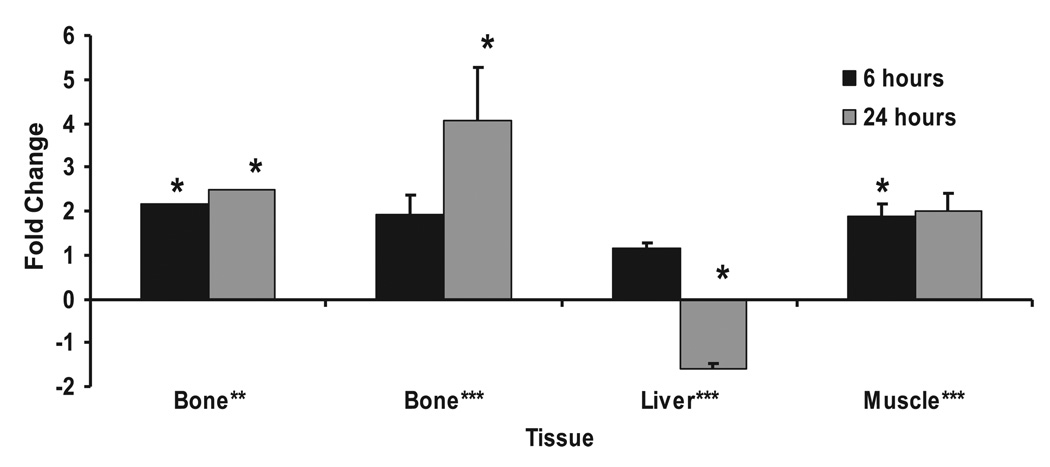
To further validate our microarray data, we examined the gene expression of IGF-I in the bones of lit/lit mice with real-time RT-PCR. Similar to the microarray data, IGF-I expression increased (3.4-fold, P ≤ 0.01) at 6 h following GH treatment (data not shown). For subsequent studies, we focused on Tbx3, a novel transcription factor of which little is known, which was upregulated 2.0- and 2.5-fold (P ≤ 0.05) at 6 and 24 h, respectively, in the microarray analysis. It is known that Tbx3 plays an important role in limb development, but to our knowledge no information is available about its role in mediating GH actions. Real-time RT-PCR analysis demonstrated that Tbx3 is expressed in bone, muscle, and liver (Fig. 1). Following GH treatment in the lit/lit mice, Tbx3 expression increased 4.1-fold (P ≤ 0.05) at 24 h in bone and 1.9-fold (P ≤ 0.05) at 6 h in muscle. In the liver, Tbx3 expression decreased 1.6-fold (P = 0.05) at 24 h.
Fig. 1.
T-box (Tbx)3 mRNA expression increases in bones of growth hormone (GH)-deficient (lit/lit) mice after a single injection of GH. Data are presented as fold change from control mice (means ± SE). *Significant difference compared with controls at P ≤ 0.05. **Data from microarray analysis (n = 3). ***Data from real-time RT-PCR analysis (n = 6).
Regulation of Tbx3 mRNA Expression in Mouse Preosteoblast Cells
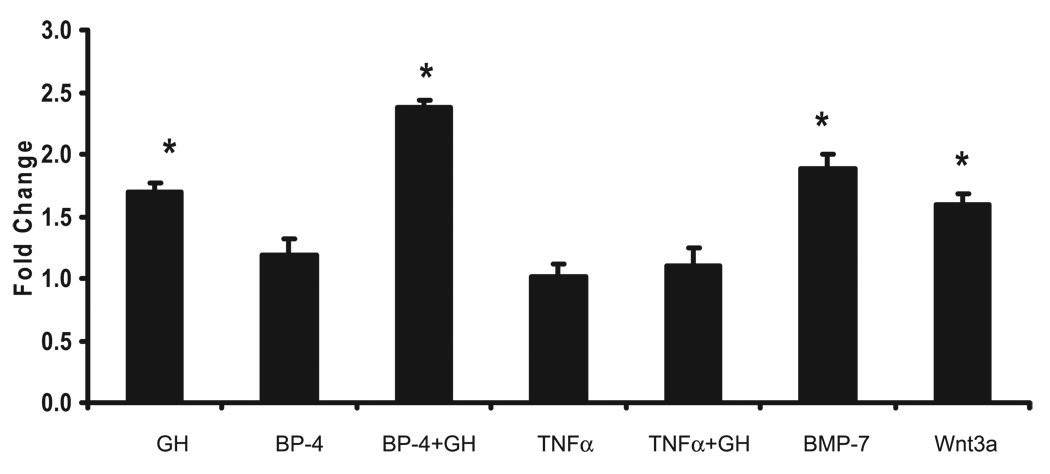
In MC3T3-E1 cells, GH induced (P ≤ 0.05) Tbx3 expression 1.6-fold (Fig. 2), and pretreatment with IGFBP-4, a known inhibitor of IGF-I, did not block the GH-induced increase in Tbx3 expression (Fig. 2). In addition, pretreatment of cells with TNF-α, a known inhibitor of STAT5b, abolished the GH-induced expression of Tbx3 (Fig. 2).
Fig. 2.
Tbx3 mRNA expression in response to GH, IGF-binding protein (IGFBP)-4, TNF-α, bone morphogenic protein (BMP)-7, and wingless-related MMTV integration site 3A (Wnt3a) treatment in a mouse preosteoblast cell line (MC3T3-E1). For each experiment, n = 6/treatment. GH = 100 ng/ml; IGFBP-4 = 300 ng/ml; TNF-α = 20 ng/ml; BMP-7 = 30 ng/ml; Wnt3a = 1 nM. Data are presented as fold change from control group (means ± SE). *Significant increase in expression vs. Control group at P ≤ 0.05.
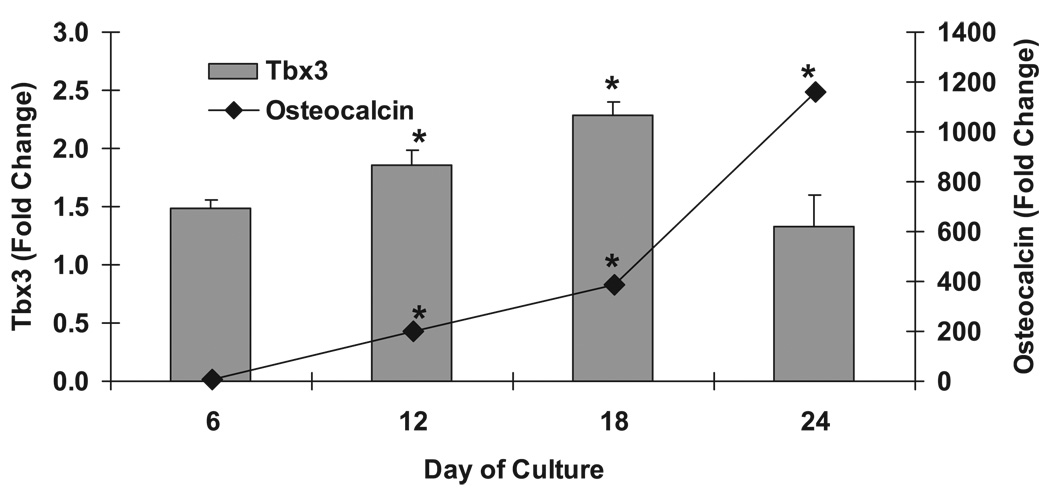
To further examine the role of Tbx3 in bone, mouse bone marrow cells were cultured in media to induce differentiation into osteoblasts and nodule formation. During the 24 days of cell culture, the cells differentiated into osteoblast cells, as confirmed by increased osteocalcin expression (Fig. 3). Similar to osteocalcin, Tbx3 expression increased 1.8- and 2.3-fold at days 12 and 18, respectively, compared with day 0 of cell culture (P ≤ 0.05). In addition, in MC3T3-E1 cells, BMP-7 and Wnt3a, known stimulators of osteoblast differentiation, increased Tbx3 expression 1.9- and 1.6-fold, respectively (P ≤ 0.05; Fig. 2).
Fig. 3.
Tbx3 mRNA expression increases during differentiation of mouse bone marrow stromal cells into osteoblast cells. Data are presented as fold change from day 0 (means ± SE); n = 6/day. *Significant increase in Tbx3 or osteocalcin expression vs. day 0 at P ≤ 0.05.
Knockdown of Tbx3 Expression Using siRNA
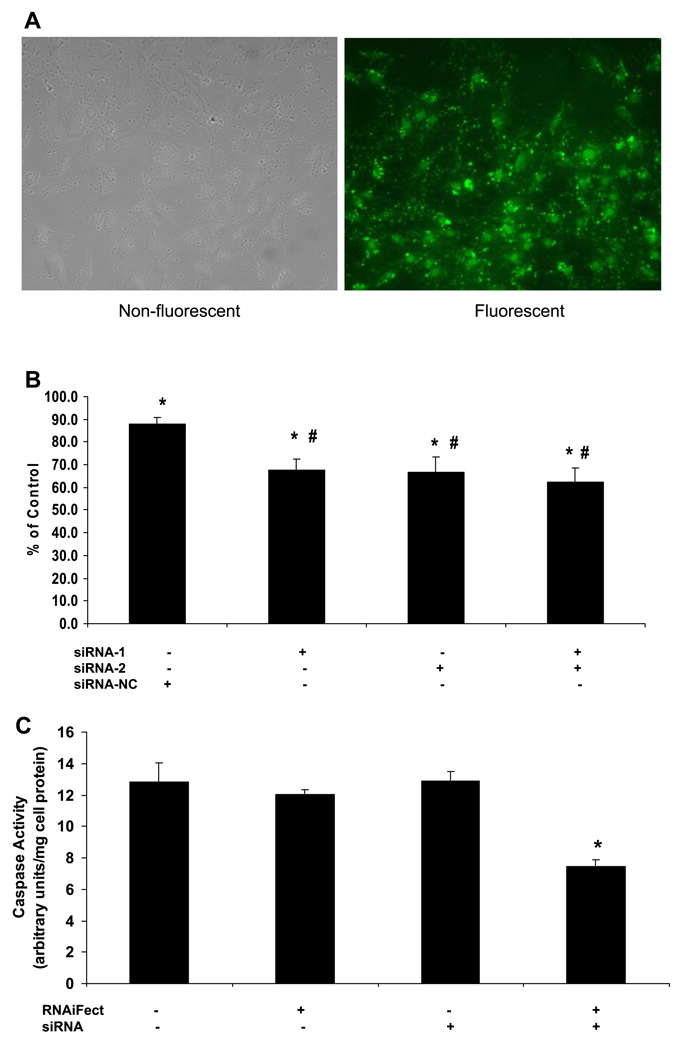
To determine the optimal conditions and transfection efficiency, cells were transfected with various amounts of a negative control, fluorescently labeled siRNA, and RNAiFect (purchased from Qiagen), for 6 h. Through comparison of the number of cells transfected (fluorescent) vs. total number of cells (nonfluorescent) by use of a fluorescent microscope, we determined that ~60–70% of the cells were successfully transfected (Fig. 4A). On the basis of these data (Fig. 4A) and the manufacturers’ recommendations, we transfected cells with 100 nM siRNA and 1 µl/100 µl reaction of RNAifect for our experiments.
Fig. 4.
Tbx3 short interfering (si)RNA reduces osteoblast cell number but does not induce apoptosis in MC3T3-E1 cells. A: transfection efficiency of MC3T3-E1 cells using Alexa fluorlabeled nonsilencing siRNA (200 nM). Cells were incubated for 6 h, and transfection efficiency was determined by number of nonfluorescent vs. fluorescent cells. About 60–70% of cells were successfully transfected. Each image is representation of 2 replicates. B: effect of Tbx3 siRNA on cell number. siRNA-1 and -2, Tbx3-specific siRNA; siRNA-NC, scrambled negative control siRNA. Total siRNA concentrations for each treatment were 100 nM. Data are expressed as %control (RNAiFect alone) and presented as means ± SE; n = 6/treatment. *P < 0.05 vs. Control. #P < 0.05 vs. siRNA-NC. C: effect of Tbx3 siRNA on apoptosis. Tbx3 siRNA-1 = 100 nM. RNAiFect, transfection reagent. Data are expressed as means ± SE; n = 6/treatment. *P < 0.001 vs. Control.
To determine the efficiency of knockdown of Tbx3 expression, cells were transfected with 100 nM Tbx3 siRNA-1, siRNA-2, and the combination of siRNA-1 and -2. On the basis of real-time RT-PCR analysis, we determined that Tbx3 expression was reduced 31 ± 5, 22 ± 5, and 31 ± 2% with siRNA-1, siRNA-2, and the combination, respectively (P < 0.01), compared with RNAiFect alone (data not shown).
Tbx3 Is an Important Mediator of Osteoblast Proliferation
First, to determine the specificity of the Tbx3 siRNA, we determined, by alamarBlue assay, the response of osteoblast cell number to transfection with the two Tbx3 siRNA (siRNA-1 and -2) and the siRNA-NC. A slight reduction (12%) in cell number was observed with the siRNA-NC; however, a greater reduction (32–38%) was observed with each Tbx3 siRNA as well as with the combination (P < 0.05; Fig. 4B). Therefore, we concluded that the Tbx3 siRNAs were specific to the Tbx3 gene. For further experiments, we used Tbx3 siRNA-1. Tbx3 siRNA-1 significantly reduced cell number (8–38%) in the presence and absence of serum in multiple experiments. The variability observed in the reduction may be due to variation in the transfection efficiency in each experiment. To determine whether the reduction in cell number was associated with a change in DNA synthesis, incorporation of [3H]thymidine was determined. A similar response was observed, such that knockdown of Tbx3 reduced [3H]thymidine incorporation into the DNA (17 to 20%) in the presence and absence of serum (P < 0.01; data not shown). To determine whether the decrease in cell number and DNA synthesis was due to increased apoptosis, we next determined caspase activity. We determined that Tbx3 siRNA did not increase caspase activity, however, it does appear to reduce the basal caspase activity in MC3T3-E1 cells (P < 0.001; Fig. 4C).
DISCUSSION
Microarray Analysis
The IGF-dependent and -independent pathways by which GH acts in bone are not well known. Through the use of microarray analysis in a model that is very sensitive to a single injection of GH, we have identified several genes induced by GH of which little information is known. In addition, we have provided evidence that several genes associated with known signaling pathways (i.e., Wnt and BMP) are acutely regulated by GH in bone. Interestingly, we identified a novel transcription factor, Tbx3, which is acutely regulated by GH as well as other growth factors in bone and is an important regulator of osteoblast proliferation.
We have provided the first report of a global analysis of gene expression in bone in response to GH administration in a GH-sensitive model. As our microarray chips included 22,000 genes and we used the GH-deficient lit/lit mouse model, we were not surprised that we identified over 6,100 genes that were acutely regulated by a single injection of GH in bone. Because more than half of the genes were of an unknown function, or expressed sequence tags (ESTs), we conclude that there are many genes involved in mediating GH actions that have not been identified. Interestingly, several of the ESTs identified include a zinc finger motif, which indicates a potential role for these genes as transcription factors. In addition, proteins encoded by several ESTs contained a proline-rich region, which is similar to the GH receptor in which a proline-rich region is required for phosphorylation of JAK (32). Future studies are needed to establish whether one or more of these ESTs are important mediators of GH effects in bone.
It is not surprising that more genes were upregulated than downregulated at both time points, since it has been well established that GH has anabolic effects on several tissues, including bone (29). For example, GH treatment increases proliferation of osteoblast cells in vitro and increases bone formation in vivo (29). In addition, many of the effects of GH are mediated by IGF-I. Therefore, the greater number of genes differentially expressed at 24 vs. 6 h was most likely due to activation of genes downstream of GH, such as IGF-I, which in turn stimulates additional genes. However, further analysis is needed to determine which of the genes differentially expressed at 6 h in the bones of GH-treated mice compared with vehicle-treated control mice are the result of direct actions of GH independent of IGF-I. It should be mentioned that, because we used hGH, there is a possibility that some of the differentially expressed genes may be the result of GH acting through the prolactin receptor (43). However, we evaluated several known downstream targets of prolactin (FGF-2, IRF-1, Bax, Jak2, Tec) (7, 30, 37, 39) and found no significant change in their expression. On the basis of these findings, and the findings that expression levels of many genes associated with the GH/IGF axis showed the anticipated change after a single injection of GH, we are confident that the changes observed are in response to GH acting through the GH receptor.
As expected, several genes associated with the GH-IGF axis were acutely regulated by GH. Interestingly, we identified two novel proteins, IGF-II mRNA-binding protein [(IMP)1 and -3], which are members of a family of RNA-binding proteins that play a role in posttranscriptional processes (18). Very little is known about these proteins, and only a few target genes have been identified (18). We observed an increase in IMP1 and a decrease in IMP3 following a single injection of GH. These data are consistent with previous reports that IMP1 is essential for normal embryonic and postnatal development (18), whereas IMP3 is implicated in the suppression of growth (28). On the basis of our data and previous reports, we hypothesize that these proteins may be important mediators of GH action in bone.
Examination of all genes differentially expressed at P ≤ 0.05 revealed several signaling genes about which little is known concerning their role in mediating GH effects in any tissue. Interestingly, several of these genes are from two major pathways, Wnt and BMP, which are important regulators of osteoblast differentiation. On the basis of our microarray data, it appears that these pathways may be involved in cross talk with the GH-IGF axis. We observed upregulation of Wnt5a in bone, which is supported by recent evidence that Wnt5a is produced by osteoblasts (44). In addition, several genes downstream of Wnt (β-catenin, Dvl2, Axin, Csnk2α2, Apc, and Csnk1α1) were up- or downregulated by GH, providing the first evidence for a role for Wnt-signaling genes (canonical and noncanonical pathways) in mediating GH actions in bone. Interestingly, three genes associated with the BMP pathway, an important regulator of bone formation (25), were downregulated by a single injection of GH. This is in contrast to a previous report in which several injections of GH stimulated BMP-2 and -4 in rats, as well as IGF-I (24). Although the different observations may be due to the different duration of GH treatment in the two experiments, the significance and mechanisms involved in the decreased expression in BMPs in the present study needs further evaluation. Interestingly, the Madh5 protein, involved in BMP signaling, was upregulated by GH treatment more than twofold at 24 h. Our data, combined with recent evidence that interactions between IGF, BMP, and Wnt-signaling genes occur (2, 41), supports the possibility that cross talk may occur between these major signaling pathways in mediating GH effects in bone. However, since some of the genes identified by these criteria (P ≤ 0.05) changed only by <1.5-fold, further confirmation of changes in the expression of these genes is needed to confirm the role of these genes in mediating GH action in bone.
Comparison of GH-responsive genes identified in bone in this study with those identified in other tissues in previous reports reveals similarities and dissimilarities. Genes with functions related to receptor signaling, membrane trafficking, apoptosis, and metabolism have been reported to be regulated by GH in previous microarray experiments using liver tissue (1, 15), as well as in our study. Specific genes related to these categories that were altered in response to GH in both liver and bone include translocase, IGFBP-2, tubulin-γ, transferrin, Bcl-like apoptosis facilitator, fatty acid-CoA ligase, and ribosomal proteins. There are a number of potential explanations for the limited similarity in GH-responsive genes between our study and previous studies in the liver. First, there may be species-and tissue-specific differences in the genes that are regulated by GH. Second, GH’s effect on target genes may be dependent on the age of the animals. Third, we administered only a single injection of GH and examined the genes that responded 6–24 h after treatment, whereas in previous studies multiple injections of GH were administered. Further microarray studies using multiple GH-responsive tissues from the same animals after a single injection are required to determine whether GH regulates similar target genes in different tissues.
Regulation of Tbx3 Expression
Another interesting finding in this study relates to upregulation of Tbx3 (a novel transcription factor) at 6 and 24 h after a single injection of GH, which was confirmed by real-time RT-PCR. Because we were interested in identifying molecular pathways involved in mediating GH action, our initial focus was on the transcription factors. We chose to further investigate Tbx3 because, to our knowledge, these are the first data that suggest an involvement of Tbx3 in mediating GH effects in bone. In addition, we demonstrated that Tbx3 is expressed in several tissues, its response to GH is tissue specific, and very little information is known about Tbx3 except that 1) it can act as a regulator of transcription, 2) a mutation of the gene results in ulnar mammary syndrome in humans, and 3) a homozygous mutation of Tbx3 in mice is embryonically lethal (6, 8).
Through further in vitro analysis, we demonstrated that GH effects on Tbx3 expression in bone may be independent of IGF-I. In this regard, we found that pretreatment of MC3T3-E1 cells with IGFBP-4, a known inhibitor of IGF-I, did not block GH-induced Tbx3 expression. In addition, the abolished GH induction of Tbx3 expression by pretreatment with TNF-α, a known inhibitor of STAT5b, indicates that GH may stimulate Tbx3 through the JAK-STAT pathway. However, further experiments are needed to confirm that this is the primary pathway by which GH regulates Tbx3 expression in bone. In contrast to bone and muscle, GH treatment decreased Tbx3 expression in liver; however, the significance of the differential expression of Tbx3 in bone vs. liver remains to be established.
Previous experiments have determined that a mutation of the Tbx3 gene in humans results in human ulnar mammary syndrome, which is characterized by limb malformation (5). In addition, a homozygous mutation of the Tbx3 gene in mice results in severe limb defects, similar to ulnar mammary syndrome, and is embryonically lethal (8). Consistent with these data, we demonstrated in vitro that Tbx3 is an important regulator of osteoblast proliferation. Specifically, two siRNA, specific for Tbx3, knocked down expression of Tbx3, and Tbx3 siRNA reduced basal- and serum-induced osteoblast cell proliferation but did not increase cell apoptosis. Similar findings have been observed in rat bladder carcinoma cells (22). On the basis of these data, we predict that Tbx3 may play an important role in the regulation of osteoblast cell number. Additionally, the increased expression of Tbx3 during osteoblast differentiation and in response to Wnt3a and BMP-7 treatment suggests that Tbx3 may also be involved in regulating osteoblast differentiation. This is supported by previous findings in chick limb development, which demonstrated that Tbx3 is dependent on sonic hedgehog (Shh), regulates BMP signaling, and is induced by BMP-4 (40). In addition, our results are similar to a recent report that Wnt signaling is needed to induce and maintain Tbx3 expression in early mammary gland development (12). However, the mechanisms by which Tbx3 regulates osteoblast proliferation and differentiation are not known. On the basis of the knowledge that Tbx proteins contain a DNA-binding domain and act as transcriptional activators and/or repressors by binding to transcription factors such as Hox, dHand, and Gli3 (34, 36, 45), we propose that Tbx3 may alter osteoblast proliferation and differentiation by interacting with other transcription modulators. We are currently in the process of further defining the role of Tbx3 in bone formation as well as the molecular mechanisms by which Tbx3 regulates osteoblasts. In conclusion, we have demonstrated that Tbx3 is an important regulator of osteoblast proliferation and may play a role in regulating osteoblast differentiation. Experiments are currently underway to determine the role of Tbx3 in osteoblast differentiation and in regulating GH action in bone.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sean Belcher for secretarial assistance and Joe Rung-Aroon and Erica Winter for technical assistance.
GRANTS
This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grants AR-048139 and AR-31062.
Footnotes
The Supplemental Material for this article (Supplemental Tables S1 and S2) is available online at http://ajpendo.physiology.org/cgi/content/full/00592.2005/DC1.
REFERENCES
- 1.Ahluwalia A, Clodfelter KH, Waxman DJ. Sexual dimorphism of rat liver gene expression: regulatory role of growth hormone revealed by deoxyribonucleic Acid microarray analysis. Mol Endocrinol. 2004;18:747–760. doi: 10.1210/me.2003-0138. [DOI] [PubMed] [Google Scholar]
- 2.Attisano L, Labbe E. TGFbeta and Wnt pathway cross-talk. Cancer Metastasis Rev. 2004;23:53–61. doi: 10.1023/a:1025811012690. [DOI] [PubMed] [Google Scholar]
- 3.Beamer WH, Eicher EM. Stimulation of growth in the little mouse. J Endocrinol. 1976;71:37–45. doi: 10.1677/joe.0.0710037. [DOI] [PubMed] [Google Scholar]
- 4.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp TR, Kortlever RM, Lingbeek M, Trettel F, Mac-Donald ME, van Lohuizen M, Bernards R. TBX-3, the gene mutated in Ulnar-Mammary Syndrome, is a negative regulator of p19ARF and inhibits senescence. J Biol Chem. 2002;277:6567–6572. doi: 10.1074/jbc.M110492200. [DOI] [PubMed] [Google Scholar]
- 6.Carlson H, Ota S, Campbell CE, Hurlin PJ. A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome. Hum Mol Genet. 2001;10:2403–2413. doi: 10.1093/hmg/10.21.2403. [DOI] [PubMed] [Google Scholar]
- 7.Clevenger CV. Role of prolactin/prolactin receptor signaling in human breast cancer. Breast Dis. 2003;18:75–86. doi: 10.3233/bd-2003-18108. [DOI] [PubMed] [Google Scholar]
- 8.Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- 9.Donahue LR, Beamer WG. Growth hormone deficiency in “little” mice results in aberrant body composition, reduced insulin-like growth factor-I and insulin-like growth factor-binding protein-3 (IGFBP-3), but does not affect IGFBP-2, -1 or -4. J Endocrinol. 1993;136:91–104. doi: 10.1677/joe.0.1360091. [DOI] [PubMed] [Google Scholar]
- 10.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorak MT. REAL-TIME PCR. 2005 [online at tripod.com/genetics/realtime.html” LOCATOR-TYPE=”URL”> http://dorakmt.tripod.com/genetics/realtime.html]
- 12.Eblaghie MC, Song SJ, Kim JY, Akita K, Tickle C, Jung HS. Interactions between FGF and Wnt signals and Tbx3 gene expression in mammary gland initiation in mouse embryos. J Anat. 2004;205:1–13. doi: 10.1111/j.0021-8782.2004.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eicher EM, Beamer WG. Inherited ateliotic dwarfism in mice. Characteristics of the mutation, little, on chromosome 6. J Hered. 1976;67:87–91. doi: 10.1093/oxfordjournals.jhered.a108682. [DOI] [PubMed] [Google Scholar]
- 14.Fernholm R, Bramnert M, Hagg E, Hilding A, Baylink DJ, Mohan S, Thoren M. Growth hormone replacement therapy improves body composition and increases bone metabolism in elderly patients with pituitary disease. J Clin Endocrinol Metab. 2000;85:4104–4112. doi: 10.1210/jcem.85.11.6949. [DOI] [PubMed] [Google Scholar]
- 15.Flores-Morales A, Stahlberg N, Tollet-Egnell P, Lundeberg J, Malek RL, Quackenbush J, Lee NH, Norstedt G. Microarray analysis of the in vivo effects of hypophysectomy and growth hormone treatment on gene expression in the rat. Endocrinology. 2001;142:3163–3176. doi: 10.1210/endo.142.7.8235. [DOI] [PubMed] [Google Scholar]
- 16.Frank SJ. Growth hormone signalling and its regulation: preventing too much of a good thing. Growth Horm IGF Res. 2001;11:201–212. doi: 10.1054/ghir.2001.0237. [DOI] [PubMed] [Google Scholar]
- 17.Godfrey P, Rahal JO, Beamer WG, Copeland NG, Jenkins NA, Mayo KE. GHRH receptor of little mice contains a missense mutation in the extracellular domain that disrupts receptor function. Nat Genet. 1993;4:227–232. doi: 10.1038/ng0793-227. [DOI] [PubMed] [Google Scholar]
- 18.Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedstrom M, Saaf M, Brosjo E, Hurtig C, Sjoberg K, Wesslau A, Dalen N. Positive effects of short-term growth hormone treatment on lean body mass and BMC after a hip fracture: a double-blind placebo-controlled pilot study in 20 patients. Acta Orthop Scand. 2004;75:394–401. doi: 10.1080/00016470410001141-1. [DOI] [PubMed] [Google Scholar]
- 20.Holmes SJ, Economou G, Whitehouse RW, Adams JE, Shalet SM. Reduced bone mineral density in patients with adult onset growth hormone deficiency. J Clin Endocrinol Metab. 1994;78:669–674. doi: 10.1210/jcem.78.3.8126140. [DOI] [PubMed] [Google Scholar]
- 21.Isaksson OG, Jansson JO, Gause IA. Growth hormone stimulates longitudinal bone growth directly. Science. 1982;216:1237–1239. doi: 10.1126/science.7079756. [DOI] [PubMed] [Google Scholar]
- 22.Ito A, Asamoto M, Hokaiwado N, Takahashi S, Shirai T. Tbx3 expression is related to apoptosis and cell proliferation in rat bladder both hyperplastic epithelial cells and carcinoma cells. Cancer Lett. 2005;219:105–112. doi: 10.1016/j.canlet.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 23.Kasukawa Y, Baylink DJ, Guo R, Mohan S. Evidence that sensitivity to growth hormone (GH) is growth period and tissue type dependent: studies in GH-deficient lit/lit mice. Endocrinology. 2003;144:3950–3957. doi: 10.1210/en.2002-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Bartold PM, Young WG, Xiao Y, Waters MJ. Growth hormone induces bone morphogenetic proteins and bone-related proteins in the developing rat periodontium. J Bone Miner Res. 2001;16:1068–1076. doi: 10.1359/jbmr.2001.16.6.1068. [DOI] [PubMed] [Google Scholar]
- 25.Linkhart TA, Mohan S, Baylink DJ. Growth factors for bone growth and repair: IGF, TGF beta and BMP. Bone. 1996;19:1S–12S. doi: 10.1016/s8756-3282(96)00138-x. [DOI] [PubMed] [Google Scholar]
- 26.Mohan S, Linkhart T, Jennings J, Baylink D. Chemical and biological characterization of low-molecular-weight human skeletal growth factor. Biochim Biophys Acta. 1986;884:243–250. doi: 10.1016/0304-4165(86)90169-8. [DOI] [PubMed] [Google Scholar]
- 27.Mohan S, Richman C, Guo R, Amaar Y, Donahue LR, Wergedal J, Baylink DJ. Insulin-like growth factor regulates peak bone mineral density in mice by both growth hormone-dependent and -independent mechanisms. Endocrinology. 2003;144:929–936. doi: 10.1210/en.2002-220948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monk D, Bentley L, Beechey C, Hitchins M, Peters J, Preece MA, Stanier P, Moore GE. Characterisation of the growth regulating gene IMP3, a candidate for Silver-Russell syndrome. J Med Genet. 2002;39:575–581. doi: 10.1136/jmg.39.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohlsson C, Bengtsson BA, Isaksson OG, Andreassen TT, Slootweg MC. Growth hormone and bone. Endocr Rev. 1998;19:55–79. doi: 10.1210/edrv.19.1.0324. [DOI] [PubMed] [Google Scholar]
- 30.Peirce SK, Chen WY. Human prolactin and its antagonist, hPRL-G129R, regulate bax and bcl-2 gene expression in human breast cancer cells and transgenic mice. Oncogene. 2004;23:1248–1255. doi: 10.1038/sj.onc.1207245. [DOI] [PubMed] [Google Scholar]
- 31.Piwien-Pilipuk G, Huo JS, Schwartz J. Growth hormone signal transduction. J Pediatr Endocrinol Metab. 2002;15:771–786. doi: 10.1515/jpem.2002.15.6.771. [DOI] [PubMed] [Google Scholar]
- 32.Postel-Vinay MC, Kelly PA. Growth hormone receptor signalling. Baillieres Clin Endocrinol Metab. 1996;10:323–336. doi: 10.1016/s0950-351x(96)80455-1. [DOI] [PubMed] [Google Scholar]
- 33.Qin X, Strong DD, Baylink DJ, Mohan S. Structure-function analysis of the human insulin-like growth factor binding protein-4. J Biol Chem. 1998;273:23509–23516. doi: 10.1074/jbc.273.36.23509. [DOI] [PubMed] [Google Scholar]
- 34.Rallis C, Del Buono J, Logan MP. Tbx3 can alter limb position along the rostrocaudal axis of the developing embryo. Development. 2005;132:1961–1970. doi: 10.1242/dev.01787. [DOI] [PubMed] [Google Scholar]
- 35.Sanders EJ, Harvey S. Growth hormone as an early embryonic growth and differentiation factor. Anat Embryol (Berl) 2004;209:1–9. doi: 10.1007/s00429-004-0422-1. [DOI] [PubMed] [Google Scholar]
- 36.Smith J. T-box genes: what they do and how they do it. Trends Genet. 1999;15:154–158. doi: 10.1016/s0168-9525(99)01693-5. [DOI] [PubMed] [Google Scholar]
- 37.Stevens AM, Yu-Lee LY. Multiple prolactin-responsive elements mediate G1 and S phase expression of the interferon regulatory factor-1 gene. Mol Endocrinol. 1994;8:345–355. doi: 10.1210/mend.8.3.8015552. [DOI] [PubMed] [Google Scholar]
- 38.Sugimoto T, Kaji H, Nakaoka D, Yamauchi M, Yano S, Sugishita T, Baylink DJ, Mohan S, Chihara K. Effect of low-dose of recombinant human growth hormone on bone metabolism in elderly women with osteoporosis. Eur J Endocrinol. 2002;147:339–348. doi: 10.1530/eje.0.1470339. [DOI] [PubMed] [Google Scholar]
- 39.Too CK, Knee R, Pinette AL, Li AW, Murphy PR. Prolactin induces expression of FGF-2 and a novel FGF-responsive NonO/p54nrbrelated mRNA in rat lymphoma cells. Mol Cell Endocrinol. 1998;137:187–195. doi: 10.1016/s0303-7207(97)00240-2. [DOI] [PubMed] [Google Scholar]
- 40.Tumpel S, Sanz-Ezquerro JJ, Isaac A, Eblaghie MC, Dobson J, Tickle C. Regulation of Tbx3 expression by anteroposterior signalling in vertebrate limb development. Dev Biol. 2002;250:251–262. [PubMed] [Google Scholar]
- 41.Verras M, Sun Z. β-Catenin is involved in insulin-like growth factor 1-mediated transactivation of the androgen receptor. Mol Endocrinol. 2005;19:391–398. doi: 10.1210/me.2004-0208. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Zhou J, Cheng CM, Kopchick JJ, Bondy CA. Evidence supporting dual, IGF-I-independent and IGF-I-dependent, roles for GH in promoting longitudinal bone growth. J Endocrinol. 2004;180:247–255. doi: 10.1677/joe.0.1800247. [DOI] [PubMed] [Google Scholar]
- 43.Wennbo H, Gebre-Medhin M, Gritli-Linde A, Ohlsson C, Isaksson OG, Tornell J. Activation of the prolactin receptor but not the growth hormone receptor is important for induction of mammary tumors in transgenic mice. J Clin Invest. 1997;100:2744–2751. doi: 10.1172/JCI119820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 45.Wilson V, Conlon FL. The T-box family. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-6-reviews3008. REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0048. research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.