Abstract
We have shown that the Sleeping Beauty (SB) transposon system can mediate stable expression of both reporter and therapeutic genes in human primary T cells and that trans delivery (i.e., transposon and transposase are on separate plasmids) is at least 3-fold more efficient than cis delivery. One concern about trans delivery is the potential for integration of the transposase-encoding sequence into the cell genome with the possibility of continued expression, transposon remobilization, and insertional mutagenesis. To address this concern, human peripheral blood lymphocytes were nucleofected with transposase plasmid and a DsRed transposon. Eighty-eight stable DsRed+ T cell clones were generated and found to be negative for the transposase-encoding sequence by PCR analysis of genomic DNA. Genomic PCR was positive for transposase in 5 of 15 bulk T cell populations that were similarly transfected and selected for transgene expression where copy numbers were unexpectedly high (0.007–0.047 per cell) by quantitative PCR. Transposase-positive bulk T cells lacked transposase plasmid demonstrated by Hirt (episomal) extracted DNA and showed no detectable transposase by Southern hybridization, Western blot, and quantitative RT-PCR analyses. Cytogenetic and array comparative genomic hybridization analyses of the only identified transposase-positive clone (O56; 0.867 copies per cell) showed no chromosomal abnormality or tumor formation in nude mice although transposon remobilization was detected. Our data suggest that SB delivery via plasmid in T cells should be carried out with caution because of unexpectedly high copy numbers of randomly integrated SB transposase.
The Sleeping Beauty (SB) transposon system is a nonviral method that can be used for integration and persistent transgene expression in a variety of somatic cell types. Here, Huang et al. report on the safety of these SB vectors; specifically, they examine the frequency of random integration by SB in human primary T cells. They also look at whether an SB+ T-cell clone triggers numeric or structural chromosome alterations and tumor growth in nude mice.
Introduction
The Sleeping Beauty (SB) transposon system provides a nonviral method of mediating stable transgene expression in mammalian cells and tissues (Ivics et al., 1997; Izsvak and Ivics, 2004). SB, a reconstructed member of the Tc1/mariner superfamily, consists of two components: the catalytic transposase and a transposon encoding any DNA cargo. Gene transfer, achieved by transposition, occurs by a “cut-and-paste” mechanism (Ivics et al., 1997). On codelivery to a target cell, the transposase recognizes and binds to the inverted/direct repeat (IR/DR) sequences designating the transposon ends, excises this sequence from the donor molecule, and inserts it into the cellular genome (Ivics et al., 1997). SB has been used successfully to mediate integration and persistent gene expression in a variety of somatic cell types, including human primary T cells (Huang et al., 2006, 2008, 2009; Singh et al., 2008; Peng et al., 2009) and CD34+ hematopoietic progenitor cells isolated from umbilical cord blood (Mátés et al., 2009; Sumiyoshi et al., 2009; Xue et al., 2009). These studies suggest the potential use of this nonviral vector system for ex vivo gene transfer applications, where one goal is to achieve life-long expression of the introduced gene sequences.
The primary expectation of gene delivery studies using the SB transposon system is to achieve genomic integration of transposon-encoded sequences. This is generally achieved by supplying an SB-encoding plasmid as the source of the transposase-encoding component of the system. Our previous work demonstrated that a trans delivery strategy appeared to be 3-fold more efficient than the SB cis vector for mediating stable gene transfer (Huang et al., 2006). This trans delivery method has proven to be effective in mediating both in vitro and in vivo transposition (reviewed by Iszvak and Ivics, 2004; Hackett et al., 2005). However, provision of the transposase-encoding sequence as a codeliverable DNA molecule may be problematic in that there exists the potential for undesired integration and persistent expression of the enzyme. Continued expression of transposase after the initial transposition event raises the possibility of subsequent transposon excision and reintegration, with associated risk of genotoxicity, a result previously observed for Cre and Fok-1 recombinases in mammalian cells (Loonstra et al., 2001; Pfeifer et al., 2001; Silver et al., 2001; Alwin et al., 2005).
To address this problem, modified viral vectors have been developed to limit the duration and intensity of nuclease expression (Pfeifer et al., 2001; Silver et al., 2001; Galla et al., 2004). Nonviral strategies have employed RNA as an alternative to DNA for short-term expression of Cre or FLP recombinases in cultured human cells (Van den Plas et al., 2003), single-cell mouse embryos (de Wit et al., 1998), and mouse embryonic stem cells (Ponsaerts et al., 2004), demonstrating precisely targeted sequence modification while limiting the potential for integration of the nuclease. In vitro-transcribed RNA as a source of SB transposase has been used successfully to mediate transposition in cultured cells and adult mice (Wilber et al., 2006, 2007).
Successful use of the SB transposon system in trans for stable genetic modification of human T cells and CD34+ hematopoietic progenitor cells prompted the studies reported here. These studies are intended to examine the frequency with which random integration of the transposase-encoding sequence may occur when the transposase and transposon are codelivered in trans to human peripheral blood T cells.
Materials and Methods
Human T cell gene transfer
Human T cell gene transfer with the SB transposon/transposase has been previously described (Huang et al., 2006, 2008, 2009). Peripheral blood was either purchased from Memorial Blood Centers (St. Paul, MN) or obtained from healthy donors with informed consent approved by the University of Minnesota Institutional Review Board. Mononuclear cells from peripheral blood (PBMNCs) were isolated with lymphocyte separation medium (Mediatech Cellgro, Herndon, VA) and washed with phosphate-buffered saline (PBS) (Invitrogen, Carlsbad, CA) containing 0.5% bovine serum albumin (BSA) (Sigma, St. Louis, MO). Viable cells (5 × 106), transposon plasmid (5 μg), and 0, 5, 10, 15, or 20 μg of transposase plasmid were resuspended in human T cell nucleofection solution and subsequently transferred into the supplied cuvette, and transfected (setting U-14; Lonza, Walkersville, MD). The cells were immediately resuspended in prewarmed growth medium and seeded into 24-well plates containing human T cell medium (RPMI 1640 [Invitrogen], 10% FCS [Hyclone, Logan, UT], 2 mM l-glutamine, penicillin [50 U/ml], streptomycin [50 μg/ml], 25 μM 2-mercaptoethanol) supplemented with recombinant human interleukin (rhIL)-2 (50 IU/ml; Chiron, Emeryville, CA) and rhIL-7 (10 ng/ml; National Cancer Institute Biological Resources Branch, Rockville, MD) and incubated at 37°C/5% CO2 overnight or for 2–4 hr.
Cells were activated with anti-CD3/CD28 beads (provided by B.L. Levine, University of Pennsylvania, Philadelphia, PA) at a target-to-bead ratio of 1:3 (Levine et al., 2002). Five days later, beads were removed and activated T cells were maintained in human T cell medium supplemented with rhIL-2 (50 IU/ml) and rhIL-7 (10 ng/ml) and restimulated every 10 to 14 days with OKT3, allogeneic PBMNCs, and Epstein–Barr virus (EBV)-transformed lymphoblastoid cell lines (LCLs) (Ortho Biotech, Raritan, NJ) (Riddell and Greenberg, 1990). Cells were maintained in culture over a period of 2–3 months and periodically harvested for determination of transgene expression by flow cytometry analysis, using a FACSCalibur (BD Biosciences, San Jose, CA).
Single cell-derived T cell clones were generated by FACS at 1 cell per well in 96-well plates from SB transposon-transfected T cells 3 weeks after nucleofection as previously described (Riddell and Greenberg, 1990; Huang et al., 2006, 2009). Previously described DsRed+ T cell clones (L6, L22, O9, O21, O56, and O69) and DsRed– clones (NO11 and NO71) were generated by limiting dilution 3 weeks after nucleofection of 5 × 106 PBMNCs with 5 μg of SB DsRed transposon and 5–10 μg of SB10 DNA (Huang et al., 2006). T cell clones were expanded in 24-well plates and flasks with OKT3 (Riddell and Greenberg, 1990).
Generation of SB-transfected bulk T cells
Eighteen bulk T cell populations (Table 1) were generated by nucleofection of PBLs or umbilical cord blood (UCB)-derived mononuclear cells with 8 different SB transposons (Table 1) plus either SB10 or SB11 DNA or SB11 mRNA, and were enriched for expression of the transposon-encoded transgene. The plasmid template for in vitro transcription of SB11 transposase was previously described (Wilber et al., 2006, 2007). Capped RNA transcripts (SB11 or enhanced green fluorescent protein [eGFP]) were generated with an mMESSAGE mMACHINE kit (Ambion, Austin, TX) and the RNA products were treated with DNase I to remove DNA template. RNA was purified with a MEGAclear kit (Ambion) and washed twice with 70% ethanol. PBMNCs were nucleofected with SB transposon pKT2/Cag-DsRed (5 μg) and SB11 or eGFP mRNA (10 or 15 μg) in the presence of RNasin RNase inhibitor (40 U; Promega, Madison, WI) using human T cell Nucleofector solution (Lonza). About 3 weeks after transfection, T cells were either sorted by FACS for DsRed, selected on the basis of drug resistance (e.g., Zeocin, 0.2 mg/ml; Invitrogen), or isolated by immunomagnetic selection with Miltenyi beads (e.g., anti-NGFR; Miltenyi Biotec, Bergisch Gladbach, Germany), and further cultured for 2–3 months. Transgene expression was confirmed by flow cytometric and functional analyses (e.g., CD19 chimeric antigen receptor [CAR], cytosine deaminase) as previously described (Huang et al., 2006, 2008).
Table 1.
Transposon and Transposase Combinations Used for Generation of Transgenic Bulk T Cell Populations
| Bulk T cells | T cell source | Transposon | Transgene(s) | Transposase |
|---|---|---|---|---|
| SB21 | PBLs | pT2/Cag-DsReda | DsRed | SB10 DNA |
| SB81 | PBLs | pT2/Cag-NGCDa | NGCD | SB10 DNA |
| SB82 | PBLs | pT2/Cag-NGCDa | NGCD | SB10 DNA |
| AP1 | PBLs | pT2/EF1-fLuc_PGK-GFP:Zeo | GFP-Zeo fusion/fLuc | SB10 DNA |
| AP2 | PBLs | pT2/EF1- fLuc_PGK-GFP:Bsd | GFP-Bsd fusion/fLuc | SB10 DNA |
| D2-NGCD | PBLs | pT2/Cag-NGCDa | NGCD | SB10 DNA |
| PBL15 | PBLs | pKT2/Ubc-19BB_mCMV-QBINGCD | CD19 CAR/NGCD | SB10 DNA |
| UCB15 | UCB | pKT2/Ubc-19BB_mCMV-QBINGCD | CD19 CAR/NGCD | SB10 DNA |
| UCB32 | UCB | pT2/PGK-19BB_mCMV-QBICD20b | CD19 CAR/CD20 | SB10 DNA |
| PBL32-1 | PBLs | pT2/PGK-19BB_mCMV-QBICD20b | CD19 CAR/CD20 | SB10 DNA |
| PBL32-2 | PBLs | pT2/PGK-19BB_mCMV-QBICD20b | CD19 CAR/CD20 | SB10 DNA |
| NGCD-gLuc | PBLs | pT2/EF1-NGCD_mCMV-gLuc | NGCD-gLuc | SB10 DNA |
| PBL9 | PBLs | pKT2/Cag-DsRedc | DsRed | SB11 DNA |
| UCB1 | UCB | pKT2/Cag-DsRedc | DsRed | SB11 DNA |
| PBL1m | PBLs | pKT2/Cag-DsRedc | DsRed | SB11 mRNAd |
| PBL1d | PBLs | pKT2/Cag-DsRedc | DsRed | SB11 DNA |
| PBL2m | PBLs | pKT2/Cag-DsRedc | DsRed | SB11 mRNAd |
| PBL2d | PBLs | pKT2/Cag-DsRedc | DsRed | SB11 DNA |
PBL, peripheral blood lymphocytes; UCB, umbilical cord blood; NGCD, truncated nerve growth factor receptor–cytosine deaminase fusion; EF1, human elongation factor-1α promoter; Cag, chimeric CMV-enhancer chicken β-actin promoter; PGK, human phosphoglycerate kinase promoter; GFP, green fluorescent protein; Zeo, Zeocin resistance gene; Bsd, blasticidin resistance gene; 19BB, CD19 CAR with 4-1BB signaling domain; QBINGCD, QBI translational enhancer element proceeding NGCD; QBICD20, QBI proceeding CD20; fLuc, firefly luciferase; gLuc, gaussia luciferase; CD19 CAR, chimeric antigen receptor for CD19; CD20, CD20 antigen.
Huang et al. (2006).
Huang et al. (2008).
Huang et al. (2010).
Wilber et al. (2006).
Genomic DNA polymerase chain reaction analysis
Genomic DNA (gDNA) was isolated from T cell clones and cell lines with a Gentra Systems Puregene DNA purification kit (Qiagen, Valencia, CA). Genomic DNA (250 ng) was used as template for a 25-μl PCR containing 1 × PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 μM primers, and 0.5 U of GoTaq Flexi DNA polymerase (Promega). SB10- and SB11-specific primer sequences were as follows: forward primer (5′-AGC CGT CAT ACC GCT CAG GA-3′) and reverse primer (5′-CCC ATG TGC AGT TGC AAA CC-3′). The β-actin gene-specific forward primer (5′-CGC CCT TTC TCA CTG GTT CT-3′) and reverse primer (5′-GTC ACA CTG GGG AAG CCA CT-3′) were used as a PCR internal control. The PCR was carried out by an initial denaturation at 94°C for 10 min, followed by 30 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 45 sec, and followed by extension at 72°C for 5 min. PCR products were separated by 1.7–2% agarose gel electrophoresis.
Analysis of transposase copy numbers by quantitative TaqMan PCR
The copy numbers of the transposase-encoding sequences in bulk T cell lines that were positive by genomic DNA PCR were further analyzed by TaqMan qPCR. One SB10/11– bulk T cell line (PBL1m), one mock T cell line, and clone O56 were used as negative and positive controls. The standard curve of SB10 copy numbers was generated by dilution of the SB10 plasmid ranging from 0, 0.004, 0.012, 0.037, 0.11, 0.33, 1, 3, to 9 copies according to the web site at http://www.uri.edu/research/gsc/resources/cndna.html. Genomic DNA isolated from nine pooled mock T cells was used to dilute SB10 plasmid when producing the standard curve. The sequences of primer pairs and probe specific for both SB10 and SB11 were designed with Primer Express software (Applied Biosystems) as follows: 5′-CAA AGC CCT GAC CTC AAT CCT A-3′ (forward primer), 5′-CTT GCT CGC ACA CGC TTT T-3′ (reverse primer), and 5′-AAA ATT TGT GGG CAG AAC T-3′ (FAM) (probe sequence). TaqMan copy number assays were performed according to the manufacturer's instructions (Applied Biosystems). Briefly, 10 μl of 2 × TaqMan universal PCR master mix, 2 μl of 20 × SB10/SB11 primer/probe set, and 2 μl of 20 × RNase P (VIC) TaqMan copy number reference were mixed with 20 ng of gDNA in a total 20-μl reaction. PCR conditions were 10 min at 95°C, 40 cycles of 15 sec at 95°C, and 1 min at 60°C. Experiments were performed in duplicate or triplicate for each sample. The copy numbers of clone O56 and bulk T cell lines were calculated from the standard curve of SB10 plasmid and further confirmed with CopyCaller v1.0 software (Applied Biosystems).
RT-PCR
Total RNA was purified with an RNeasy mini kit (Qiagen) and treated with DNase. Reverse transcription reactions were carried out with SuperScript III and random hexamer primers; a reaction without reverse transcription (no SuperScript III) was used to control for potential contaminating genomic DNA. First-strand cDNA was used as template for SB10 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PCR with SB10-specific primers and GAPDH-F (5′-GAA GGT GAA GGT CGG AGTC-3′) and GAPDH-R (5′-GAA GAT GGT GAT GGG ATT TC-3′). PCRs were carried out under the same conditions as described for genomic PCR and separated by 2% agarose gel electrophoresis.
Quantitative RT-PCR
Total RNA was isolated from T cell clones and bulk samples with TRIzol reagent, treated with DNase, and reverse transcribed into cDNA (Invitrogen). The sequences of primer pairs and probe specific for both SB10 and SB11 are described in the section Analysis of Transposase Copy Numbers by Quantitative TaqMan PCR. One microliter of cDNA was used as the template; 12.5 μl of 2 × TaqMan gene expression PCR master mix was mixed with the template, primers, and probe (Invitrogen). The total reaction volume was 25 μl. PCR conditions were 10 min at 95°C, 40 cycles of 15 sec at 95°C, and 1 min at 60°C. Experiments were performed in triplicate for each sample. The mRNA levels of SB10 and SB11 were normalized to the mRNA level of endogenous 18S mRNA control by subtracting the cycle threshold (Ct) value of 18S mRNA from the Ct value of the gene. The fold difference was calculated compared with the control (2–ΔΔCt).
PCR cloning of transposition sites
Linker-mediated PCR used to recover the genomic DNA sequences flanking transposon inserts has been described previously (Huang et al., 2006). Nested PCR products were sequenced directly and/or cloned into pCR2.1-TOPO vector (Invitrogen). DNA sequencing was performed at the BioMedical Genomics Center at the University of Minnesota (Minneapolis, MN). The sequence results were subjected to BlastN analysis against the human genome, using the University of California at Santa Cruz (UCSC) database. Cancer-related genes were identified using the Cancer Genes database at Memorial Sloan-Kettering Cancer Center (New York, NY; http://cbio.mskcc.org/CancerGenes/). Transcriptional start site (TSS) data were extracted from the UCSC database, originally from SwitchGear Genomics. The closest TSS on the same strand as the mapped region was used in the analysis.
Western blot
Two million cells from T cell clones, cell lines, and HeLa cells (used as a positive control after transfection with SB10 transposase plasmid) were harvested and lysed in 200 μl of ice-cold lysis buffer, which contained 1 × PBS, 1% sodium dodecyl sulfate (SDS), and 10% glycerol. The lysates were immediately boiled for 5 min. Total cell lysate supernatants were collected by centrifugation at 16,000 × g for 10 min at 4°C. Samples were denatured under reducing conditions and electrophoresed by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (10% SDS–PAGE). The samples were then transferred to polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA) and immunoblotted with mouse anti-SB10 (R&D Systems, Minneapolis, MN) or rabbit polyclonal anti-SB antibody (kindly provided by P. Hackett, University of Minnesota, Minneapolis, MN) and secondary horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit IgG. The signals were visualized with enhanced chemiluminescence (ECL) reagent (GE Healthcare, Piscataway, NJ).
Southern blot analysis
Southern hybridization was performed as previously described (Huang et al., 2006). Briefly, 10 μg of genomic DNA extracted from each T cell clone was digested with EcoRI and BamHI overnight at 37°C. Ten-microgram amounts of genomic DNA from T cells with or without 6 pg of transposase plasmid were used as positive and negative controls, respectively. Genomic DNA fragments were separated by electrophoresis through 0.9% agarose gels and transferred to a Hybond membrane (GE Healthcare). A 260-bp SB10-specific PCR fragment was generated, using SB10 plasmid as template and SB10-specific primers as described previously, isolated, purified, and 32P radiolabeled with a Prime-It II random primer labeling kit (Stratagene, La Jolla, CA). Probe hybridization was performed in 0.2 M NaHPO4 (pH 7.2)/1 mM EDTA/1% bovine serum albumin (BSA)/7% SDS at 65°C overnight. Blots were subsequently washed three times and exposed to Kodak X-ray film (Carestream Health, Rochester, NY).
Flow cytometric analysis
Flow cytometric analysis was carried out on a FACSCalibur, and the data were analyzed with FlowJo software (Tree Star, Portland, OR).
Hirt DNA isolation and analysis
Unintegrated low molecular weight DNA (Hirt DNA) extracted from human T cell populations was previously described (Hirt, 1967; Huang et al., 2006). Hirt DNA (500 ng) was used to transform 25 μl of ElectroMAX DH10B bacterial cells (Invitrogen), using a Bio-Rad electroporator (Bio-Rad). Colonies resulting from each bacterial transformation were quantified, and if present, plasmid DNA was extracted from 20 colonies of each group and further evaluated by restriction digest for the presence of pT2/DsRed- and pUb-SB10-encoding plasmids.
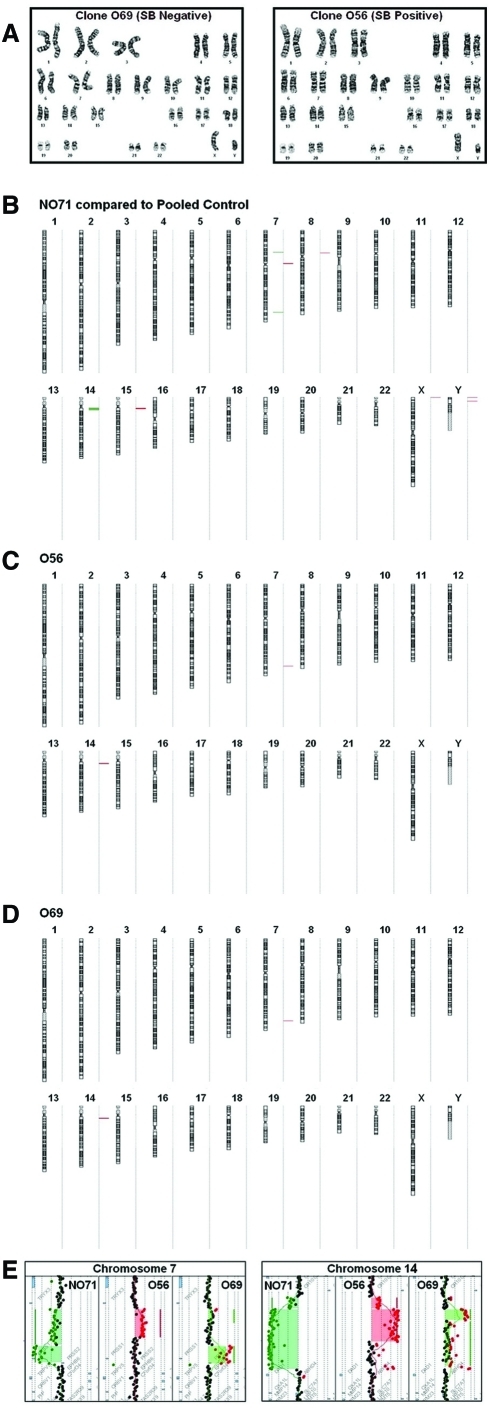
Cytogenetic analysis
O56 and O69 clones were maintained according to an OKT3 expansion protocol (Riddell and Greenberg, 1990). On day 7 after OKT3 stimulation, dead cells were removed with Ficoll-Hypaque (Mediatech Cellgro) and 10 × 106 cells were resuspended in 10 ml of human T cell culture medium. Cells were sent to the University of Minnesota Cytogenetics Core Laboratory for analysis. Briefly, cells were treated with colcemid for 3 hr and then harvested according to a standard cytogenetic protocol. Twenty metaphases were evaluated by G-banding at a 400- to 425-band level resolution.
Array comparative genomic hybridization analysis
Genomic DNA from the T cell control clone NO71 (DsRed transposon– and SB10–) (Huang et al., 2006) was isolated, restriction digested, and labeled with fluorochrome Cyanine 5, using random primers and exo-Klenow fragment DNA polymerase. For the control experiment, genomic DNA from peripheral blood of pooled sex-matched controls was labeled concurrently with fluorochrome Cyanine 3. For the analysis of T cell clones O56 and O69 derived from the same blood donor as clone NO71, their DNA was labeled with Cyanine 5 and run against DNA from the T cell control NO71, labeled with Cyanine 3. The Cyanine 3 and Cyanine 5 samples were combined and array comparative genomic hybridization (CGH) was performed with a microarray constructed by Agilent Technologies (Palo Alto, CA) that contains approximately 170,000 distinct biological oligonucleotides spaced at an average interval of 13 kb. The ratio of the samples (O56 and O69) to control DNA (NO71) for each oligo was calculated with Feature Extraction software 10.5 (Agilent Technologies). The abnormal threshold was applied using DNA Analytics 4.0.85. A minimum of three oligos that had a minimal absolute ratio value of 0.3 (based on a log2 ratio) was used as a criterion for a gain or loss of gene copy.
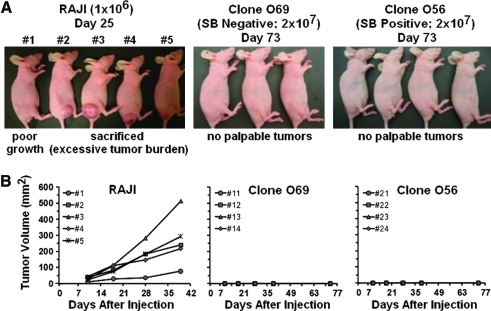
Determination of tumorigenicity of SB10+ O56 clone in nude mice
Eight-week-old female nude mice (BALB/cBy-Hfh11nu, cat. no. 000711) were purchased from Jackson Laboratory (Bar Harbor, ME). On day −1, mice were irradiated (4.0 Gy, cesium-137). On day 0, mice were subcutaneously injected via the right flank with either 2 × 107 SB10+ O56 clone cells, 2 × 107 SB10– O69 clone cells, or 1 × 106 Raji cells. Tumor growth was measured with calipers and computed as length × width (mm2). Growth was monitored up to 73 days. Mice were killed when tumors reached a size that caused them to be moribund. Digital images of mice were taken with a digital camera on days 25 and 73. These studies were conducted at the University of Minnesota and conformed to institutional guidelines and approval.
Results
Persistent expression of SB transposase can be observed in T cells after trans delivery
We previously reported on several single cell-derived clones of human peripheral blood T cells generated by cotransfection of a transposon encoding for expression of DsRed (pT2/DsRed) and the SB transposase (pUb-SB10) followed by limiting dilution (Huang et al., 2006). These clones were evaluated for the presence of transposase-encoded DNA sequences. For this, genomic DNA was prepared from all six T cell clones and evaluated by PCR using transposase-specific primers and β-actin as a control. The PCR assay revealed that one of these clones (O56) was positive for transposase-encoding DNA compared with O69 as a representative negative clone (Fig. 1A). To determine whether the integrated sequence in clone O56 conferred expression of the transposase protein, cell lysates were prepared from each clone and immunoblotting was performed with antibodies specific for the transposase or cellular tubulin. The immunoblot demonstrated that clone O56 was positive for expression of a 42-kDa protein equal in size to that observed for HeLa cells transiently transfected with the SB transposase expression plasmid used here as a positive control (Fig. 1B). Combined, these data show that the transposase-coding sequence was stably integrated into the cellular genome in a manner permissive for expression of the full-length gene product.
FIG. 1.
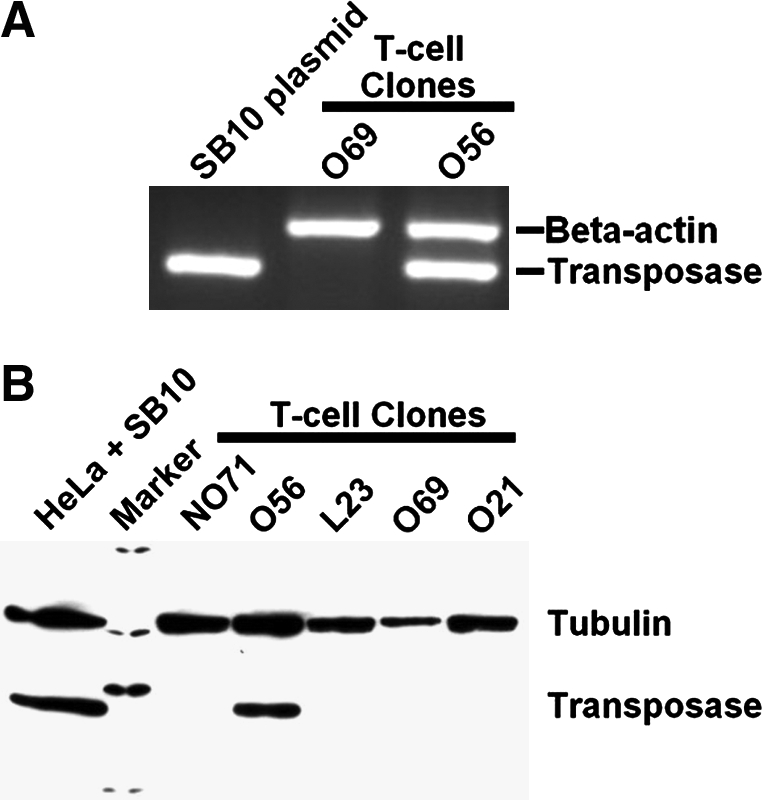
Confirmation of SB10 integration and expression in the O56 clone. (A) Genomic DNA analysis of SB10 expression in O56 and O69 T cell clones. SB10 plasmid DNA was used as a positive control. (B) Western blotting confirmation of SB10 expression in the O56 clone.
Eighty-eight additional human T cell clones generated in trans with SB transposase were negative for transposase sequence
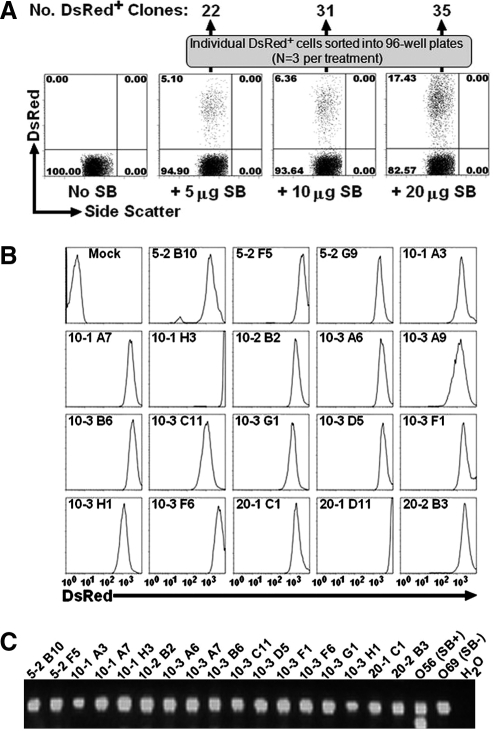
Our finding that one of six stable T cell clones demonstrated stable integration accompanied by persistent expression of the transposase suggested a surprisingly high incidence for this event (Fig. 1B, and data not shown). To more rigorously evaluate the potential for stable integration of the transposase, we tested an extensive number of transgenic T cell clones. For this, peripheral blood lymphocytes (PBLs) were isolated from two independent donors and nucleofected with transposon (pT2/DsRed; 5 μg) and various amounts of transposase (pUB-SB10; 0, 5, 10, or 20 μg). In each case, transfected cells were maintained in continuous culture for 4 weeks before flow cytometry analysis was performed to evaluate for expression of the DsRed reporter. At this time point, none of the T cells transfected with the transposon alone exhibited expression of DsRed whereas cells cotransfected with 5, 10, or 15 μg of transposase showed stable expression in 5.1, 6.4, and 17.4% of the cells (Fig. 2A), respectively. For each of these conditions, positive cells were enriched by individually sorting DsRed+ cells into wells of three independent 96-well tissue culture plates. After a period of expansion, 88 clones, consisting of 22, 31, and 35 clones from the 5-, 10-, and 20-μg doses of pUb-SB10, respectively, were isolated for subsequent analysis (Fig. 2A). All 88 of these single cell-derived clones were evaluated by flow cytometry for maintenance of DsRed expression, with representative examples presented in Fig. 2B. The persistent expression of DsRed observed for all single-cell clones is consistent with integration of the transposon-encoded sequence. Each of these 88 clones was tested for random integration of the transposase-encoding DNA sequence by PCR analysis of genomic DNA. When compared with clone O56, none of the 88 newly derived T cell clones was positive for transposase DNA; control reactions for β-actin confirmed sample integrity, and results for 17 clones are shown in Fig. 2C. These results suggest that the potential for integration of the transposase-encoding DNA at the single-cell level is rare, and we have observed 1 event out of 94 clones tested (a frequency of ≤1%).
FIG. 2.
Determination of SB10 integration in multiple T cell clones. (A) Generation of T cell clones by FACS sorting of DsRed+ cells derived from SB-transfected PBLs. (B) Flow cytometric analysis of DsRed expression in T cell clones. Nineteen representative clones of 88 are shown. Nomenclature of each T cell clone was assigned as follows: The first number indicates the amount of transposase used for transfection; the second number indicates the plate number; the third number indicates the well position. (C) Genomic PCR analysis of SB10 expression in T cell clones. Seventeen of 88 clones are shown.
Heterogeneous human T cell populations transfected in trans demonstrate the presence of high copy numbers of transposase sequences for which expression is undetectable
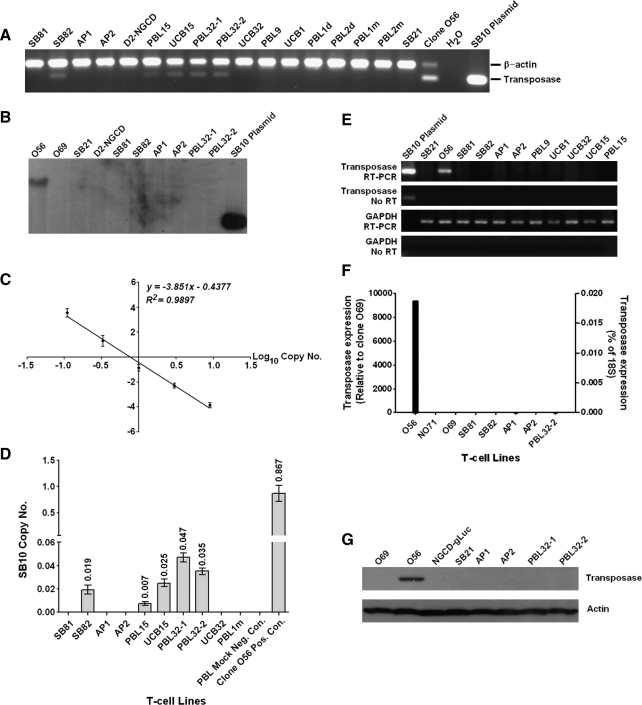
The studies described previously were performed with single T cell clones established from bulk populations that had been sorted and further expanded for weeks in culture. However, clinical application will likely require the use of a heterogeneous cell population that has not been subjected to persistent in vitro culture. Therefore, we tested for the presence of the transposase in bulk cell populations that had been cotransfected, enriched for transposon-encoded transgene-positive cells, and maintained in culture for 2–3 months. Human T cells were isolated from 18 independent sources of PBLs or UCB and cotransfected with SB transposons encoding 8 different transgenes (Table 1 and Fig. 3) and either pUb-SB10 or -SB11 DNA or SB11 mRNA (10 μg). PBLs from two donors transfected with SB transposon and SB11 mRNA (PBL1m and PBL2m) showed 26 and 4% DsRed+ cells compared with the same PBLs transfected with SB11 DNA (PBL1d and PBL2d) showing 13 and 9% DsRed+ cells on day 33 after transfection. Both PBLs cotransfected with DsRed transposon and control eGFP mRNA had 0.11 and 0.09% DsRed+ populations (data not shown), indicating that SB11 mRNA is as efficient as SB10 or SB11 DNA in mediating stable transgene expression in primary human T cells. Transfected bulk T cells were selected on the basis of >90% purity of transgene expression 3–4 weeks after gene transfer and further expanded in culture. Here, genomic PCR analysis identified 5 of 15 SB10 or SB11 DNA-transfected cell populations that were positive for transposase-encoding sequences (Fig. 4A). Alternatively, Southern blot analysis of several samples testing positive for transposase showed complete absence of the transposase-encoded sequence in bulk T cells, whereas clone O56 was positive for hybridization as demonstrated by the appearance of an ∼9-kb fragment (Fig. 4B). As a result, the copy number of transposase-encoding sequences in bulk cell populations and SB10+ clone O56 was determined by quantitative TaqMan PCR relative to a standard curve consisting of gDNA isolated from a pool of mock-treated T cells supplemented with increasing quantities of SB10 plasmid (Fig. 4C). On the basis of this standard curve, the copy number of the transposase in clone O56 was approximately 0.867 per cell. Unexpectedly, the copy numbers of the transposase in the five bulk T cell lines testing positive by gDNA PCR were high, ranging from 0.007 to 0.047 when compared with reported frequencies for random integration of plasmid ranging from 0.00001 to 0.000001 per cell (Fig. 4D) (Doetschman et al., 1988).
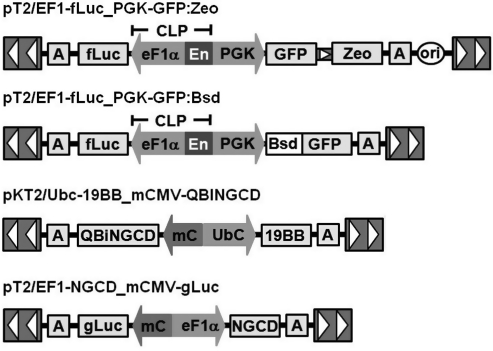
FIG. 3.
The SB transposon vectors used in this study. SB transposons contain inverted repeat/direct repeat sequences (IR/DR, indicated by arrowheads) flanking the gene of interest. CLP, CpG-less promoter; EF1α, a human elongation factor-1α promoter; PGK, human phosphoglycerate kinase promoter; Ubc, human ubiquitin C promoter; mC, minimal CMV promoter; Zeo, Zeocin resistance gene; GFP, green fluorescent protein; Bsd, blasticidin resistance gene; fLuc, firefly luciferase; NGCD, truncated human nerve growth factor receptor and cytosine deaminase fusion gene; 19BB, CD19 chimeric antigen receptor (CAR) with CD3ζ and 4-1BB signaling domain; gLuc, Gaussia luciferase; A, polyadenylation signal.
FIG. 4.
Molecular analysis of SB10 integration and expression in SB-transfected bulk T cells. (A) Genomic PCR showing that 5 of 15 bulk T cell populations transfected with SB10 or SB11 DNA plasmid were positive for transposase-encoding sequences. (B) Southern hybridization showing no detectable genomic integration of SB10 or SB11. (C) Standard curve of SB10 plasmid copy numbers by qPCR, where each point represents the mean ± SEM (n = 6 wells) of three independent assays. The standard curve for SB10 copy numbers was generated by dilution of the SB10 plasmid, ranging from 0, 0.004, 0.012, 0.037, 0.11, 0.33, 1, 3, to 9 copies. SB10 plasmid copy numbers less than 0.012 were considered not detectable and the copy number of 0.037 was not present on the linear line. Thus, the line equation established for the standard curve was based on the copy numbers ranging from 0.11, 0.33, 1, 3, to 9. (D) Copy numbers of the SB transposase in SB10+ or SB11+ bulk T cell lines and SB10+ clone O56. Data are presented as means ± SEM (n = 6 wells) of three independent assays. (E) RT-PCR showing no mRNA transcripts in genomic PCR-positive bulk T cells. (F) Quantitative RT-PCR showing undetectable mRNA transcript in SB10+ or SB11+ bulk T cells and an approximately 9000-fold higher expression level in O56 compared with O69. One of two representative results is shown. (G) Western blotting showing no SB10 or SB11 protein in genomic PCR-positive bulk T cells.
Six representative bulk cell populations were further examined to determine whether the quantitative PCR result was reflective of genomic integration or the persistence of the transposase-encoding plasmid as an episome (Table 2). For this, low molecular weight DNA (Hirt DNA) was isolated from bulk human T cell populations and electroporated into competent bacteria before plating on selective growth medium. After overnight incubation, bacterial colonies, if present, were cultured for characterization by restriction digest as the transposon, transposase (pUb-SB10), or unknown recombinant. Transformation efficiency was controlled by using pUC19 DNA, and T cells transfected with pT2/DsRed plus pUB-SB10 followed by only 7 days of culture served as a positive control for episomal DNA material. Under these conditions, no positive colonies for any transposon or pUb-SB10 were recovered from Hirt DNA isolated from SB10+ bulk human T cell populations. This finding indicates that the PCR results were a reflection of genomic integration of the transposase rather than persistence of episomal plasmid.
Table 2.
Identity of Recovered Plasmids for Cytoplasmic Hirt DNA Preparationsa
| |
|
|
|
Identity of recovered plasmids |
|
|
|---|---|---|---|---|---|---|
| T cells | Genomic PCR for SB10 | Total colonies | Number evaluated | Transposon | Transposase | Unknown |
| PBL pT2/DsRed+pUb-SB10 day 7 | n.d. | 3000 | 20 | 14 | 3 | 3 |
| SB82 | + | 4 | 4 | 0 | 0 | 4 |
| NGCD-gLuc | n.d. | 4 | 4 | 0 | 0 | 4 |
| AP1 | + | 5 | 5 | 0 | 0 | 5 |
| AP2 | + | 0 | 0 | 0 | 0 | 0 |
| PBL32-1 | n.d. | 1 | 1 | 0 | 0 | 1 |
| PBL32-2 | + | 0 | 0 | 0 | 0 | 0 |
| Clone O56 | + | 1 | 1 | 0 | 0 | 1 |
| pUC19 control | 4.65 × 1010 | n.d. | n.d. | n.d. | n.d. | |
n.d., not done.
Shown is a representative example of two independent transformations using Hirt DNA.
Cell populations found to be genomic PCR-positive for the transposase were further evaluated for expression of the RNA transcripts. RT-PCR was performed with primers specific for the transposase or GAPDH as a loading control and amplification of the RNA template as a test for contaminating genomic DNA. Figure 4E demonstrates that all samples except the plasmid control had equivalent amplification of GAPDH whereas only clone O56 produced an amplicon of the expected size for the transposase compared with the plasmid control. The TaqMan quantitative RT-PCR assay revealed that SB10+ clone O56 expressed a nearly 9000-fold higher level of SB10 than did SB10– clone O69. Background levels of SB10 or SB11 expression were detected in five bulk T cell samples, the DsRed–SB10– NO71 clone, and O69 (Fig. 4F). Likewise, transposase protein was undetectable for SB10+ bulk T cell samples where equivalent loading was confirmed with antibodies specific for cellular β-actin (Fig. 4G). These molecular assays demonstrate that stable SB10 integration in PCR-positive bulk T cells occurs with a frequency of 0.007–0.047 copies per cell level, which is insufficient to produce detectable bands on Southern blot, mRNA, and protein, further suggesting that random integration of SB transposase is unexpectedly high, but the frequency of continued expression of SB transposase is a rare event in T cells.
Human T cells with stable expression of SB transposase do not show abnormal chromosome alterations and fail to produce tumors in immunodeficient mice
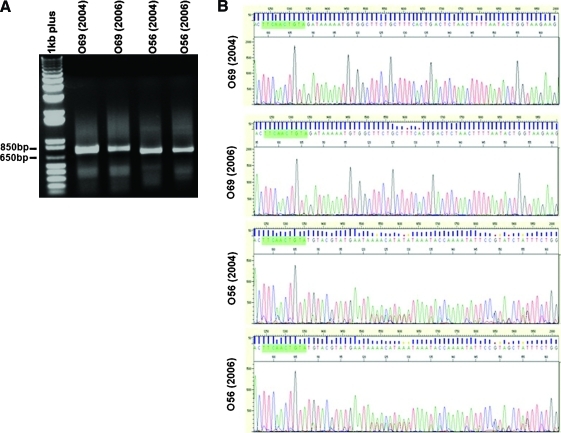
Although random integration of the transposase is evident in heterogeneous T cell populations, we have identified only a single incidence in which this resulted in persistent expression of the gene product. Interestingly, this event (clone O56) represents what could be categorized as a worst case scenario for genetic modification of primary human T cells when using the SB transposon system (i.e., every cell maintains expression of the transposase). To demonstrate the functionality of integrated transposase in clone O56, we studied transposon remobilization in clone O56 and clone O69 after in vitro culture in the years 2004 and 2006. Linker-mediated PCR resulted in a single band in both clones O56 and O69 cultured over these times (Fig. 5A). Sequencing analysis of PCR products revealed that clone O69 had only a single amplified PCR sequence whereas clone O56 had multiple integrations (Fig. 5B). Therefore, PCR products from O56 were further subject to TOPO cloning and resequencing. Table 3 shows that clone O56 cultured in the years 2004 and 2006 had two and five different integration sites on chromosome 11, respectively. However, Southern blot analysis, using an enzyme that cuts once within the transposon-encoded sequence, showed no difference in banding patterns for clone O56 cultured over time (data not shown). These data suggest that new integration events detected by linker-mediated PCR represent mobilization events that occurred in only a minority of the cells. Nevertheless, we conclude that SB10 transposase in clone O56 actively mediates transposon remobilization.
FIG. 5.
Characterization of transposon integrations in clones O56 and O69 cultured over time. (A) Linker-mediated PCR showing single bands in clone O56 and clone O69 cultured in the years 2004 and 2006. (B) Sequencing of PCR products showing mixed integrants in clone O56 but not in clone O69. Green highlights indicate the transposon sequence. Color images available online at www.liebertonline.com/hum.
Table 3.
Characterization of Transposon Integration Events
| T cell clone | Year cultured | Chromosome location: hit from: | Located gene | Gene symbol | Gene orientation | Cancer-related gene | Proximal TSS | Distance to TSS (bp)a | Integration frequency |
|---|---|---|---|---|---|---|---|---|---|
| O69 | 2004 | chr13:55526482 | Intergenic | PRR20A | (+) | Yes | chr13:56613053 | −1086572 | N/A |
| 2006 | chr13:55526482 | Intergenic | PRR20A | (+) | Yes | chr13:56613053 | −1086572 | N/A | |
| 2004 | chr11:34925030 | Intronic | PDHX | (+) | Yes | chr11:34894252 | +30777 | 21/24 | |
| 2004 | chr11:34925570 | Intronic | PDHX | (+) | Yes | chr11:34894252 | +30777 | 3/24 | |
| 2006 | chr11:34925025 | Intronic | PDHX | (+) | Yes | chr11:34894252 | +30777 | 1/24 | |
| O56 | 2006 | chr11:34925030 | Intronic | PDHX | (+) | Yes | chr11:34894252 | +30777 | 20/24 |
| 2006 | chr11:34925570 | Intronic | PDHX | (+) | Yes | chr11:34894252 | +30777 | 2/24 | |
| 2006 | chr11:34925572 | Intronic | PDHX | (+) | Yes | chr11:34894252 | +30777 | 1/24 | |
| 2006 | chr11:34874627 | Intronic | APIP | (–) | No | chr11:34894515 | +20112 | 1/24 |
N/A, not applicable because of pure PCR products, which were not subject to TOPO cloning; TSS, transcriptional start site; PRR20A, proline-rich 20A; PDHX, pyruvate dehydrogenase complex component X; APIP, APAF1-interacting protein.
+, intragenic; –, upstream of TSS.
Next, we examined whether the presence of active transposase in clonal T cells may lead to genome instability and cell transformation. G-banded karyotype analysis revealed no numerical or structural chromosome aberrations in SB10+ O56 or SB10– O69 (Fig. 6A). Because G-banded karyotype analysis detects only gross chromosomal alterations, array CGH analysis was also performed. The array CGH showed that the only copy number changes in clones O56 and O69, relative to the control clone NO71 (DsRed transposon–SB10–), were in the 7q34 and 14q11.2 regions that contain the known polymorphic T cell receptor genes. As the control clone NO71 had copy number losses in these regions (compared with DNA from pooled male controls), the gains for portions of these regions observed in O56 and O69 when hybridized against NO71 would indicate no loss of the regions relative to pooled controls. Those portions of the 7q and 14q regions that showed no loss in O56 and O69 relative to NO71 (e.g., the distal portions of 7q34 and 14q11.2 in clone O56) would represent the same loss for this region (compared with pooled controls) as was harbored by NO71 (Fig. 6B–E). Importantly, the array CGH results demonstrate that there is no significant difference between SB10+ O56 and SB10– O69 clones and no evidence of significant copy number changes other than those related to T cell clonality. In addition, animals implanted with either the SB10+ O56 clone or SB10– O69 clone showed no evidence of tumor growth for up to 10 weeks, whereas animals implanted with Raji cells developed tumors and became moribund after 5 weeks (Fig. 7A and B). These data demonstrate that human T cells with SB10 stably integrated remain karyotypically normal and do not form tumors in mice.
FIG. 6.
Cytogenetic and array CGH analyses of O56 clone. (A) G-band analysis of SB10+ O56 and SB10– O69 clones showing a normal 46,XY male karyotype. (B) Array CGH showing losses at 7q34 and 14q11.2 in the control clone NO71 (compared with pooled male control specimens) and gains in the same regions in clones O56 and O69 (compared with NO71). (C) Array CGH showing copy number gains against control clone NO71 at 7q34 and 14q11.2 regions in clone O56. (D) Array CGH showing copy number gains against NO71 at 7q34 and 14q11.2 regions in clone O69. (E) The control NO71, compared with the pooled control DNA, showed two regions of loss encompassing approximately 346 and 155 kb within 7q34, respectively. The start point for the loss of the 346-kb region was estimated at bp 141663456 and the stop point at bp 142009059. The ratio value of −1 was consistent with a deletion of this region on one chromosome 7 allele. This region contains TCRBV genes. The start point for the loss of the 155-kb region was estimated at 142021348 and the stop point at 142176133. The ratio for this region fell between −2 and −4, consistent with a homozygous loss. Mapped to this region are other TCRB-related genes. Two regions of loss within 14q11.2, encompassing 128 and 633 kb, respectively, were noted. The start point for the 128-kb region was estimated at 21285126 and the stop point at 2142207. Mapped to this region are TCRAV-related genes. The start point for the 633-kb region was estimated at 21420105 and the stop point at 22052858. Mapped to this region are also TCRAV-related genes. Clone O56 shows a ratio of 1.0 for the proximal part of 7q34 and a ratio of 0 for the remaining portion of this 7q34 region. As the control clone NO71 had deletion in these regions, this would indicate that O56 had no loss for the proximal region, and had the same loss as the control NO71 for the distal portion of this region. Clone O69 had a small gain for a portion of the 7q34 region, likely indicating a mixture of cells that had the same loss as the control NO71 and those that had no loss. Clone O69 had a ratio of approximately 2.0 for the distal portion of 7q34, indicating no loss for this region. For the 14q11.2 region, the ratios for O56 and O69 were generated using the control T cell clone NO71, which showed a ratio of −3 for this region. Thus, the +0.3 ratio for O56 indicates that there is no loss for the proximal portion of 14q11.2 region in these cells; the remainder of this region shows a profile consistent with the presence of some cells with loss, and others not. The findings are similar for O69, which shows no loss for the very proximal region, and some cells with loss for the more distal portion of the region. Of note, for NO71 compared with pooled controls, additional gains were seen in the near centromeric regions of chromosomes 7, 8, and 15, and in the pseudoautosomal regions of Xp and Yp. These gains are among the benign copy number variants that are well documented in published databases to occur in healthy controls, and are interpreted as germline variants of no relevance to the present experiments. Color images available online at www.liebertonline.com/hum.
FIG. 7.
The O56 clone did not induce tumor formation in nude mice. (A) Tumor growth in nude mice; mice injected with the SB10+ O56 clone showed no tumor growth on day 73. In Raji cell-injected mice, one mouse (#1) showed initial tumor growth and then the tumor regressed spontaneously. (B) Tumor volume versus days after injection. Color images available online at www.liebertonline.com/hum.
Discussion
Three important observations concerning random integration of SB transposase on trans delivery in human primary T cells appear in this paper. First, the frequency of random integration of SB transposase resulting in expression of SB10 or SB11 protein accounts for about 1% (1 of 94 T cell clones analyzed). Second, although random integration of the SB transposase was observed in approximately 33% of bulk T cells and the copy numbers were unexpectedly high (5 of 15 bulk cells tested with 0.007–0.047 copies of the SB transposase), no transposase mRNA transcript or protein could be detected. Third, an SB10+ T cell clone (O56; 0.867 copies) with stable transposon encoded transgene expression shows active transposon remobilization but no apparent numeric or structural chromosome alterations and no tumor growth in nude mice. Thus, our results suggest that random integration of SB transposase is high but that stable integration of functional SB transposase-encoding sequences is low in human T cells, warranting caution in the use of SB transposase DNA for human T cell gene therapy.
Transient expression of the transposase is a requirement of the transposition process. However, persistent expression suggests a situation in which the transposon could become remobilized after the initial insertion event. This method has proven quite effective for discovering novel cancer-causing genes in both normal and tumor-prone mice (Carlson et al., 2005; Collier et al., 2005; Dupuy et al., 2005; Su et al., 2008; Keng et al., 2009; Rahrmann et al., 2009; Starr et al., 2009). In these studies, mice were engineered for constitutive or tissue-restricted expression of the transposase to mobilize transposons from a resident concatemer. Some of the new integration events induce expression of an oncogene or inactivation of a tumor suppressor gene. Common to several of these studies is the use of animals demonstrating constitutive or induced expression of the transposase as a control cohort for transposase-mediated mutagenesis (Carlson et al., 2005; Collier et al., 2005; Dupuy et al., 2005; Su et al., 2008; Keng et al., 2009; Rahrmann et al., 2009; Starr et al., 2009). Only when animals are also predisposed to cancer, that is, deficient in the tumor suppressor genes encoding p19Arf or p53, has there been demonstrated morbidity (Collier et al., 2005; Keng et al., 2009). This was not observed for normal mice, where the most extensive studies were carried out for more than 1 year (Collier et al., 2005; Keng et al., 2009).
Additional studies intending to model defined somatic tumorigenesis have coinfused normal mice (Wiesner et al., 2009) or tumor-predisposed mice (Carlson et al., 2005) with transposons harboring an activated oncogene and transposase under delivery conditions that can target a specific tissue. After SB-mediated gene transfer of an activated version of the N-RAS oncogene to the liver of Arf-deficient mice, Carlson and colleagues identified three independent tumors where the codelivered transposase had become randomly integrated. Western blot confirmed that one of these tumors demonstrated constitutive expression of the transposase.
Using nucleofection as a method for achieving ex vivo gene transfer into human T cells, we detected SB10 functional integration in one of the first 6 clones but not in the additional 88 clones. In each case, we used similar ratios of transposon- and transposase-encoding plasmids. The only difference between these two experiments is the method used to generate T cell clones, with limiting dilution and FACS-based cell sorting used in the first and second experiments, respectively. It is possible but highly unlikely that cell sorting disfavors the growth of clones with integrated transposase compared with limiting dilution.
We also detected SB transposase integration in 5 of 15 bulk T cell populations stably expressing transposon-encoded transgenes, using genomic PCR. Unexpectedly, five genomic PCR-positive bulk T cell populations demonstrated approximately 700–4700 times higher frequencies of random transposase integration than previously documented levels for plasmid random integration (0.7–4.7 vs. 0.001–0.0001%) (Doetschman et al., 1988). This higher than expected level of random integration of SB transposase could be attributed to the following: First, all bulk T cells were enriched for transposon-encoded transgenes by FACS, immunomagnetic isolation, or drug selection (Table 1). Thus, transposase expression in every cell is required to achieve transposition. Second, the amount of SB transposase for trans delivery (10–15 μg of transposase and 5 μg of transposon per 5 × 106 T cells) was high. Third, one of the intrinsic properties of the SB transposase is to “cut” DNA, suggesting a potential for increased frequency of random integration. Fourth, the method of nucleofection used here to achieve T cell gene transfer may have an increased potential compared with alternative gene transfer methods (e.g., calcium phosphate coprecipitation or liposome-mediated transfection) for promoting random integration. Fifth, the frequency of random integration is cell type specific. For example, McIvor and colleagues reported that NIH3T3 cells had a six times higher stable baseline transfection frequency relative to HeLa cells (0.15 vs. 0.025%, respectively) as defined by transfection with pT2/neo (SB transposon encoding the neomycin resistance gene) alone (Converse et al., 2005). In similar studies using an unrelated nonviral integrating vector system, the Calos group also demonstrated an ∼31-fold higher integration frequency in NIH3T3 cells than in 293 cells (0.25 vs. 0.008%) as defined by transfection with bacteriophage ΦC31 cargo-containing vector without integrase (Thyagarajan et al., 2010). By comparison, the frequency of SB transposase random integration in human T cells appears similar (0.7–0.47 vs. 0.15–0.25%) to these results reported for NIH3T3 cells.
Even with these unexpectedly high levels of random integration of transposase-encoding sequences, all bulk T cells failed to express detectable levels of SB10 or SB11 mRNA and protein. This result can be explained in that random integration of the codelivered transposase occurs in only a small percentage of cells that can be detected only by sensitive PCR and TaqMan qPCR amplification, not by Southern hybridization. Random integration of the transposase in this manner may not favor the production of detectable transcripts and protein. Our data clearly demonstrate that SB10 or SB11 functional integration is a low-frequency event. With respect to the tumorigenic potential of transposase-positive T cells demonstrating constitutive expression of a transposon-encoded transgene (e.g., the O56 clone), we observed no tumor formation in nude mice 73 days after cell implantation. However, we cannot exclude the possibility that nude mice may not be the best model for assessing human T cell transformation. In addition, it may be necessary to monitor the potential for tumor development for a longer period of time to ensure tumor negativity because it is possible that tumor formation may occur at a much later time point.
From our results, it can be derived that the frequency of functional SB transposase integration is a low but viable possibility. However, this does not exclude the potential use of DNA as a source for transposase. Although we were able to identify a single clone that was positive for SB10 transposase integration with relatively low copy number (0.867 copies), protein production, and active transposon remobilization, additional studies confirmed that there was no resultant tumor growth in mouse models or alterations to the chromosomal structure of the genomic DNA, even by array CGH analysis. It is evident that integration of the transposase can lead to local transposon “hopping” as shown in only a minority of clone O56 populations. The transposase used in previous studies for eliciting insertional mutagenesis was engineered for induced activity and may not represent a risk when using the SB transposon system to mediate gene transfer.
The advantages of a DNA transposase system are plentiful when compared with mRNA as a source of SB transposase in human T cell engineering: low cost, simple production, similar efficiency, and easy handling in transfection. However, it may be better to use mRNA as a source of SB transposase or DNA with a coexpressed suicide gene for hematopoietic stem cell gene transfer as these cells are highly subject to insertional mutagenesis (Hacein-Bey-Abina et al., 2003; Mátés et al., 2009; Sumiyoshi et al., 2009; Xue et al., 2009). Supporting this alternative, we observed similar efficiencies of stable gene transfer for two independent donor T cell populations when in vitro-transcribed transposase-encoding mRNA was compared with plasmid. Ultimately, our results warn of potential risks involved in the mediation of gene transfer by the SB transposon system, which was approved in 2008 for the “first-in-human” gene therapy trial by the Recombinant DNA Advisory Committee (RAC) (VandenDriessche and Chuah, 2009). In this trial, T cells genetically engineered by the SB transposon system will be adoptively transferred into patients with CD19+ B cell malignancies (Singh et al., 2008). Because of the high frequency of transposase random integration in human T cells as demonstrated in this study, use of SB transposase mRNA or DNA along with a suicide gene should be considered.
Acknowledgments
The authors thank Dr. David Largaespada (University of Minnesota, Minneapolis, MN) for helpful discussion about array CGH data and local “hopping,” Dr. R. Scott McIvor (University of Minnesota) for critical reading of this manuscript, Dr. Perry Hackett (University of Minnesota) for independent confirmation of copy number calculations, Dr. Betsy Hirsch (University of Minnesota) for interpretation of the G-banding and array CGH data, Dr. Mingqiang Ren (Medical College of Georgia, Augusta, GA) for helpful discussion about copy number determination by qPCR and standard curve, and Dr. Nikunj Somia (University of Minnesota, Minneapolis, MN) for providing the blasticidin–GFP fusion sequence. The authors also thank Dr. San Ming Wang and Dr. Yeong C. Kim (Northwestern University, Evanston, IL), and Dr. Zheng Jin Tu (University of Minnesota, Minneapolis, MN) for help in bioinformatic analyses of integration sites. K.H. was a recipient of an Undergraduate Research Opportunity Program (UROP) Award, University of Minnesota, and of an American Society of Hematology Research Trainee Award. This work was supported by grants from the Alliance for Cancer Gene Therapy, the Gabrielle's Angel Foundation for Cancer Research, the Sidney Kimmel Foundation for Cancer Research Kimmel Scholar Program, the University of Minnesota (an AHC Translational Research grant), the University of Minnesota Medical School Dean's Commitment, and in part by the Children's Cancer Research Fund in Minneapolis (X.Z.). The cytogenetic and array CGH analyses were performed in the Cytogenetics Core Laboratory at the University of Minnesota with support from the Comprehensive Cancer Center (NIH Grant P30 CA077598-09). K.H.'s current address: Medical College of Wisconsin, 8701 Watertown Park Road, Milwaukee, WI 53226.
Contribution Statement
X.H. designed, performed, and supervised the research, analyzed the data, and wrote the paper. K.H. performed the research, analyzed the data, and wrote the paper. M.W. performed the research, analyzed the data, and wrote the paper. H.G. and C.L. performed the research and analyzed the data. A.W. provided critical reagents and wrote the paper. X.Z. oversaw the research and wrote the paper.
Author Disclosure Statement
The authors declare no competitive financial interests.
References
- Alwin S. Gere M.B. Guhl E. Effertz K. Barbas C.F., III Segal D.J. Weitzman M.D. Cathomen T. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- Carlson C.M. Frandsen J.L. Kirchhof N. McIvor R.S. Largaespada D.A. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17059–17064. doi: 10.1073/pnas.0502974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier L.S. Carlson C.M. Ravimohan S. Dupuy A.J. Largaespada D.A. Cancer gene discovery in solid tumors using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- Converse A.D. Belur L.R. Gori J. Liu G. Amaya F. Aguilar-Cordova E. Hackett P.B. McIvor R.S. Counterselection and co-delivery of transposon and transposase functions for Sleeping Beauty-mediated transposition in cultured mammalian cells. Biosci. Rep. 2005;24:577–594. doi: 10.1007/s10540-005-2793-9. [DOI] [PubMed] [Google Scholar]
- de Wit T. Drabek D. Grosveld F. Microinjection of Cre recombinase RNA induces site-specific recombination of a transgene in mouse oocytes. Nucleic Acids Res. 1998;26:676–678. doi: 10.1093/nar/26.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman T. Maeda N. Smithies O. Targeted mutation of the Hprt gene in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8583–8587. doi: 10.1073/pnas.85.22.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy A.J. Akagi K. Largaespada D.A. Copeland N.G. Jenkins N.A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Galla M. Will E. Kraunus J. Chen L. Baum C. Retroviral pseudotransduction for targeted cell manipulation. Mol. Cell. 2004;16:309–315. doi: 10.1016/j.molcel.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Von Kalle C. Schmidt M. McCormack M.P. Wulffraat N. Leboulch P. Lim A. Osborne C.S. Pawliuk R. Morillon E. Sorensen R. Forster A. Fraser P. Cohen J.I. de Saint Basile G. Alexander I. Wintergerst U. Frebourg T. Aurias A. Stoppa-Lyonnet D. Romana S. Radford-Weiss I. Gross F. Valensi F. Delabesse E. Macintyre E. Sigaux F. Soulier J. Leiva L.E. Wissler M. Prinz C. Rabbitts T.H. Le Deist F. Fischer A. Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hackett P.B. Ekker S.C. Largaespada D.A. McIvor R.S. Sleeping Beauty transposon-mediated gene therapy for prolonged expression. Adv. Genet. 2005;54:189–232. doi: 10.1016/S0065-2660(05)54009-4. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Huang X. Wilber A.C. Bao L. Tuong D. Tolar J. Orchard P.J. Levine B.L. June C.H. McIvor R.S. Blazar B.R. Zhou X. Stable gene transfer and expression in human primary T cells by the Sleeping Beauty transposon system. Blood. 2006;107:483–491. doi: 10.1182/blood-2005-05-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Guo H. Kang J. Choi S. Zhou T.C. Tammana S. Lees C.J. Li Z.Z. Milone M. Levine B.L. Tolar J. June C.H. McIvor R.S. Wagner J.E. Blazar B.R. Zhou X. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19+ lymphoid malignancies. Mol. Ther. 2008;16:580–589. doi: 10.1038/sj.mt.6300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Wilber A. McIvor R.S. Zhou X. DNA transposons for modification of human primary T lymphocytes. Methods Mol. Biol. 2009;506:115–126. doi: 10.1007/978-1-59745-409-4_9. [DOI] [PubMed] [Google Scholar]
- Huang X. Guo H. Tammana S. Jung Y.C. Mellgren E. Bassi P. Cao Q. Tu Z.J. Kim Y.C. Ekker S.C. Wu X. Wang S.M. Zhou X. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and PiggyBac transposons in human primary T cells. Mol. Ther. 2010;18:1803–1813. doi: 10.1038/mt.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z. Hackett P.B. Plsterk R.H. Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Izsvak Z. Ivics Z. Sleeping Beauty transposon: Biology and applications for molecular therapy. Mol. Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Keng V.W. Villanueva A. Chiang D.Y. Dupuy A.J. Ryan B.J. Matise I. Silverstein K.A.T. Sarver A. Starr T.K. Akagi E. Tessarollo L. Collier L.S. Powers S. Lowe S.W. Jenkins N.A. Copeland N.G. Llovet J.M. Largaespada D.A. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat. Biotechnol. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B.L. Bernstein W.B. Aronson N.E. Schlienger K. Cotte J. Perfetto S. Humphries M.J. Ratto-Kim S. Birx D.L. Steffens C. Landay A. Carroll R.G. June C.H. Adoptive transfer of costimulated CD4+ T cells induces expansion of peripheral T cells and decreased CCR5 expression in HIV infection. Nat. Med. 2002;8:47–53. doi: 10.1038/nm0102-47. [DOI] [PubMed] [Google Scholar]
- Loonstra A. Vooijs M. Beverloo H.B. Allak B.A. van Drunen E. Kanaar R. Berns A. Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátés L. Chuah M.K. Belay E. Jerchow B. Manoj N. Acosta-Sanchez A. Grzela D.P. Schmitt A. Becker K. Matrai J. Ma L. Samara-Kuko E. Gysemans C. Pryputniewicz D. Miskey C. Fletcher B. Vandendriessche T. Ivics Z. Izsvák Z. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- Pfeifer A. Brandon E.P. Kootstra N. Gage F.H. Verma I.M. Delivery of the Cre recombinase by a self-deleting lentiviral vector: Efficient gene targeting in vivo. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11450–11455. doi: 10.1073/pnas.201415498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsaerts P. Brown J.P. Van den Plas D. Van den Eeden L. Van Bockstaele D.R. Jorens P.G. Van Tendeloo V.F. Merregaert J. Singh P.B. Berneman Z.N. Messenger RNA electroporation is highly efficient in mouse embryonic stem cells: Successful FLPe- and Cre-mediated recombination. Gene Ther. 2004;11:1606–1610. doi: 10.1038/sj.gt.3302342. [DOI] [PubMed] [Google Scholar]
- Rahrmann E.P. Collier L.S. Knutson T.P. Doyal M.E. Kuslak S.L. Green L.E. Malinowski R.L. Roethe L. Akagi K. Waknitz M. Huang W. Largaespada D.A. Marker P.C. Identification of PDE4D as a proliferation promoting factor in prostate cancer using a Sleeping Beauty transposon-based somatic mutagenesis screen. Cancer Res. 2009;69:4388–4397. doi: 10.1158/0008-5472.CAN-08-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D.P. Livingston D.M. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- Starr T.K. Allaei R. Silverstein K.A. Staggs R.A. Sarver A.L. Bergemann T.L. Gupta M. O'Sullivan M.G. Matise I. Dupuy A.J. Collier L.S. Powers S. Oberg A.L. Asmann Y.W. Thibodeau S.N. Tessarollo L. Copeland N.G. Jenkins N.A. Cormier R.T. Largaespada D.A. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q. Prosser H.M. Campos L.S. Ortiz M. Nakamura T. Warren M. Dupuy A.J. Jenkins N.A. Copeland N.G. Bradley A. Liu P. A DNA transposon-based approach to validate oncogenic mutations in the mouse. Proc. Natl., Acad. Sci. U.S.A. 2008;105:19904–19909. doi: 10.1073/pnas.0807785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi T. Holt N.G. Hollis R.P. Ge S. Cannon P.M. Crooks G.M. Kohn D.B. Stable transgene expression in primitive human CD34+ hematopoietic stem/progenitor cells, using the Sleeping Beauty transposon system. Hum. Gene Ther. 2009;20:1607–1626. doi: 10.1089/hum.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B. Olivares E.C. Hollis P. Ginsburg D.S. Calos M.P. Site-specific genomic integration in mammalian cells mediated by phage ΦC31 integrase. Mol. Cell. Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenDriessche T. Chuah M.K.L. Moving gene therapy forward with mobile DNA. Hum. Gene Ther. 2009;20:1559–1561. doi: 10.1089/hum.2009.1109. [DOI] [PubMed] [Google Scholar]
- Van den Plas D. Ponsaerts P. Van Tendeloo V. Van Bockstaele D.R. Berneman Z.N. Merregaert J. Efficient removal of LoxP-flanked genes by electroporation of Cre-recombinase mRNA. Biochem. Biophys. Res. Commun. 2003;305:10–15. doi: 10.1016/s0006-291x(03)00669-7. [DOI] [PubMed] [Google Scholar]
- Wiesner S.M. Decker S.A. Larson J.D. Erickson K. Forster C. Gallardo J.L. Long C. Demorest Z.L. Zamora E.A. Low W.C. SantaCruz K. Largaespada D.A. Ohlfest J.R. De novo induction of genetically engineered brain tumors in mice using plasmid DNA. Cancer Res. 2009;15:431–439. doi: 10.1158/0008-5472.CAN-08-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilber A. Frandsen J.L. Geurts J.L. Largaespada D.A. Hackett P.B. McIvor R.S. RNA as a source of transposase for Sleeping Beauty-mediated gene insertion and expression in somatic cells and tissues. Mol. Ther. 2006;13:625–630. doi: 10.1016/j.ymthe.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Wilber A. Wangensteen K.J. Chen Y. Zhuo L. Frandsen J.L. Bell J.B. Chen Z.J. Ekker S.C. McIvor R.S. Wang X. Messenger RNA as a source of transposase for Sleeping Beauty transposon-mediated correction of hereditary tyrosinemia type I. Mol. Ther. 2007;15:1280–1287. doi: 10.1038/sj.mt.6300160. [DOI] [PubMed] [Google Scholar]
- Xue X. Huang X. Nodland S.E. Mátés L. Ma L. Ivics Z. Izsvák Z. Lebien T.W. McIvor R.S. Wagner J.E. Zhou X. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood. 2009;114:1319–1330. doi: 10.1182/blood-2009-03-210005. [DOI] [PubMed] [Google Scholar]