Abstract
The CD1 family of antigen-presenting molecules is able to display lipids to T cells by binding them within a hydrophobic groove connected to the protein surface. In particular, the CD1b isotype is capable of binding ligands with greatly varying alkyl chain lengths through a complex network of interconnected hydrophobic pockets. Interestingly, mycobacterial lipids, such as glucose monomycolate (GMM) exclusively bind to CD1b. We determined the crystal structure of one of the three expressed bovine CD1b proteins, CD1b3, in complex with endogenous ligands, identified by mass spectrometry as a mixture of phosphatidylcholine and phosphatidylethanolamine, and analyzed the ability of the protein to bind glycolipids in vitro. The structure reveals a complex binding groove architecture, similar to the human ortholog but with consequential differences. Intriguingly, in bovine CD1b3 only the A’, C’ and F’ pockets are present while the T’ pocket previously described in human CD1b is closed. This different pocket conformation could affect the ability of boCD1b3 to recognize lipids with long acyl chains such as GMM. However, even in the absence of a T’ tunnel, bovine CD1b3 is able to bind mycolates from Rhodococcus ruber in vitro.
Keywords: CD1b, Bos taurus, lipid antigens, mycolic acid
Introduction
Tuberculosis, the disease caused in humans by infection with Mycobacterium tuberculosis, represents a major cause of mortality worldwide, with increasing reports of multidrug-resistant strains appearing in several countries (1). In addition, bovine tuberculosis, caused by M. bovis, is of economical and zoonotic concern. Unfortunately, the attenuated bacillus Calmette-Guerin (BCG) vaccine currently available is of little protective efficacy in humans and animals (2), highlighting the need of alternative approaches for the development of an effective vaccine against tuberculosis. In recent years, mycobacterial glycolipid and lipid antigens have been the focus of much attention in this context due to the T cell stimulatory activity of these compounds, the stable chemical structure of the lipid backbone due to the fact that lipids are not directly encoded by genes, and the low level of polymorphism exhibited by the CD1 family of antigen-presenting molecules that display these antigens to T cells (3). During mycobacterial infection in humans and animals, CD1-restricted and glycolipid-specific T cell responses can be detected ex vivo, suggesting a physiologically relevant role for this mode of antigen presentation. Despite the fact that all these features would make CD1-presented lipids excellent vaccine subunit candidates, the CD1 system is often seen as static, not subject to evolutionary pressure exerted by the changing environmental challenges of an evolving species, and often classified as part of the innate immune system.
CD1 molecules, similar to MHC Class I molecules, exist on the surface of antigen-presenting cells as non-covalent heterodimers formed by a CD1 heavy chain and a beta-2-microglobulin “light” chain (β2m)3 (4). While the β2m chain consists of a single immunoglobulin-like domain, the CD1 heavy chain is anchored to the membrane via a single-pass transmembrane domain and contains a short intracellular cytoplasmic tail and three extracellular domains (denominated α1, α2 and α3). The C-terminal cytoplasmic tail usually contains trafficking motifs responsible for the intracellular localization of the protein (5). The α1 and α2 ectodomains form the antigen-binding groove responsible for the recognition of self and non-self antigens, whereas the α3 domain non-covalently associates with β2m. The CD1 binding groove is highly hydrophobic and defined by two alpha helices that sit above an antiparallel beta-sheet platform. Antigens are generally bound with the alkyl chains buried in the deep hydrophobic groove while the glycosidic or polar portion of the molecule is exposed at the surface for recognition by a T cell receptor or TCR (4). However, while the overall architecture of these antigen-presenting molecules is conserved, the size, shape and number of individual pockets differ between the various CD1 isotypes.
In humans, all group 1 CD1 proteins (CD1a, CD1b, and CD1c) are highly expressed on immature thymocytes and immature and mature dendritic cells while group 2 CD1 (CD1d) is expressed at lower levels on many cell types (3). A fifth CD1 molecule, CD1e, is not expressed on the cell surface but is found to be involved in facilitating lipid loading onto CD1b (6). CD1d is the restricting element for NKT cells, a cell population with a limited T cell repertoire that can quickly release large amounts of IFN-γ and other cytokines (7). Group 1 CD1 proteins are known for their ability to present lipid antigens to T cells with a diverse T cell repertoire (8, 9). Each of the five human CD1 isotypes possesses typical characteristics in terms of size and shape of the antigen binding groove as well as its sub-cellular location. This has led to the general idea that human group 1 CD1 represents all possible options to present an optimal variety of lipid antigens to T cells: CD1b, with the biggest antigen binding groove, is the only isotype that can bind mycolates and uses lysosomal factors to present exogenous antigens; CD1a has the smallest groove and functions without intersecting with the lysosome, while CD1c is intermediate in both regards (10, 11). Because all CD1 molecules except CD1d have been deleted in muroid rodents, the human family of CD1 proteins is often considered to be complete and representing all CD1 proteins that exist. However, mammals other than primates lack some isotypes and/or express multiple variants of certain isotypes (12–14). In particular, cattle possess three functional CD1B genes, called CD1B1, CD1B3 and CD1B5 that differ in sequence mainly in their α1 and α2 domains and in their cytoplasmic tail (13). In humans, the single CD1b protein has been found to present mammalian gangliosides (15), and mycobacterial antigens such as sulfoglycolipids (16), phosphatidylinositol mannosides, and glucose monomycolate (GMM)(17). The crystal structure of the human CD1b antigen-binding groove identified the presence of four interlinked hydrophobic channels (A’, C’, F’ and T’) providing the potential to accommodate up to 60 carbon atoms (18). However, an open F’ pocket and the presence of a unique C’ portal under the α2 helix which connects the inner binding groove to the outer solvent allowed for the binding of longer chains such as mycobacterial monomycolates (up to C90) that would normally exceed the size of the binding groove (18, 19). The crystallographic findings are well supported by in vitro-derived data using the GMM-reactive, CD1b-restricted human T cell line LDN5. GMM with short (C30–36 excluding the hexose ring), intermediate (C50–58), and long (C74–82) mycolic acid backbones are all presented by CD1b to LDN5 cells (20, 21).
In an attempt to better understand the functional differences between the bovine CD1b proteins including their antigen-binding specificity we determined the crystal structure of the first sequenced bovine CD1b gene product, CD1b3 (boCD1b3), and analyzed its binding capacity towards glycolipids, especially mycobacterial mycolates, in vitro.
Materials and Methods
Molecular cloning of bovine CD1b transcripts
Full length CD1b3 mRNA had been cloned previously (22). A full length CD1b1 mRNA was present in the EST database (EH164202), available at www.ncbi.nlm.nih.gov. No full length CD1b5 transcript was present in the databases. Based on the genomic sequence of CD1b5, primers for CD1b5 were designed: CD1b5FullLengthFor: 5’-AGTTCTACTTCCCATTGAAATGCTGCTTCTG-3’; CD1b5FullLengthRev: 5’-GTAATTGCTCTAAATGGGAAAGAAGACACG-3’. PCR was performed on bovine thymic cDNA with PFU Turbo polymerase (Stratagene, Agilent Technologies) according to manufacturers suggestions under the following cycling conditions: an initial denaturation of 7 min. at 95°C, followed by 35 cycles of 30 sec. at 95°C, 45 sec. at 58°C, 1 min. at 72°C, followed by a final elongation step of 7 min. at 72°C. PCR products were excised from an agarose gel, purified, and ligated in a Topo4blunt vector for sequencing. The CD1b5 mRNA sequence was deposited into the GenBank database (accession number GU325785). Sequence alignments figures were prepared with the software Espript (23) using similarity scores assigned with the Risler matrix (24).
Protein expression and purification
The recombinant soluble bovine CD1b3-β2m heterodimeric molecule was produced using the baculovirus expression system as previously described for mouse CD1d (25). Briefly, the protein expressed in insect cells was purified by Nickel affinity chromatography (HisTrap FF, GE Healthcare) and eluted with a linear gradient of 20–250 mM imidazole, followed by anion-exchange chromatography on a MonoQ 5/50 column (GE Healthcare) equilibrated with 10 mM Tris-HCl pH 8.0 and elution using a linear gradient of 0–350 mM NaCl. The fractions containing the boCD1b3-β2m heterodimer were pooled together and further purified by gel filtration on a Superdex200 10/300 column (GE Healthcare) equilibrated in 50 mM HEPES pH 7.5, 150 mM NaCl. The fractions containing the complex were then concentrated to 10 mg/ml in 10 mM HEPES pH 7.5, 30 mM NaCl for subsequent crystallization and analysis.
Mass spectrometry analysis of boCD1b3
The intact non-covalent complex of boCD1b3 with the lipids was analyzed with native nano ESI mass spectrometry using borosilicate capillaries to introduce the sample. Before measurement the sample was buffer exchanged to 50 mM ammonium acetate pH 6.8 with 5kD centrifuge filters (Millipore). The MS and fragment ion MS/MS spectra were recorded with a high mass modified QToF instrument (26), the parent ion and neutral loss MS/MS analysis were performed with Quattro Ultima triple quadrupole instrument both from Waters Manchester, UK. The desolvation energies were arranged by setting the voltage on the entrance of the mass spectrometer 'sample cone voltage' to 125V (referred to as high desolvation energy in the text) or 50V (low desolvation energy).
Protein crystallization
Initial crystallization trials were performed by a nanoliter dispensing liquid handling robot (Phoenix, Art Robbins) using commercially available crystallization screens (PEG/Ion I & II, Wizard I & III, Hampton Research and Emerald Biosciences). Promising conditions were optimized manually and diffracting crystals were grown in 100 mM sodium cacodylate pH 6.5, 2 M ammonium sulfate by sitting drop vapor diffusion at 4°C, mixing 1 µl of protein at 10 mg/ml with 1 µl of precipitant.
Structure determination and refinement
Crystals were flash-cooled at 100K in crystallization solution containing 20% glycerol. Data collection was performed at beamline 7.1 at the Stanford Synchrotron Radiation Laboratory (SSRL, Stanford, CA) and processed to a resolution of 2.3 Å with the HKL2000 software (27). The protein crystallized in space group P21212 with cell parameters a = 137.53 Å, b = 139.97 Å, c = 111.95 Å, α = β = γ = 90° and with four CD1-lipid complex in the asymmetric unit (estimated solvent content of 56.2%).
Molecular replacement was performed using Molrep (28, 29), and the crystal structure of the human CD1b (PDB ID 2H26) (30) with the ligand removed as a search model. The initial phases derived from this model were refined using maximum-likelihood restrained refinement coupled with TLS refinement in Refmac (29, 31). Three TLS groups per CD1b-β2m complex were defined, containing the α1, α2 domains and the glycolipid ligand (group 1), the α3 domain (group 2) and the β2m chain (group 3), respectively. A set of 5% of the reflections were set aside for the calculation of Rfree to monitor refinement progress. Tight NCS constraints between the four molecules present in the asymmetric unit were imposed and progressively released towards the final stages of refinement. 2Fo-Fc and Fo-Fc maps were used in the program Coot (32) for modeling and manual building. The quality of the final model was assessed with Molprobity (33) and the validation tools available in Coot. Data collection and refinement statistics are presented in Table I. The architecture and size of the binding groove pockets were analyzed using Hollow (34) and CastP (35). Residues pKs were determined with the multiconformation continuum electrostatics approach implemented in the software MCCE 2.0 (36), as previously described for the human CD1b structure (37). Shake-omit maps were generated by removing the ligands from the structure and randomly perturbating the coordinates, occupancy and B-factor of each atom by 0.2 Å, 0.05 units and 20 Å2 respectively with the software Moleman2 (38). The resulting structure was then refined with the software Refmac as described earlier. Molecular representations were generated with the programs Pymol (39) and APBS (40).
Table I.
Data collection and refinement statistics.
| Data collection statistics | |
|---|---|
| Space group | P21212 |
| Cell dimension | |
| a, b, c, (Å) | 137.53, 139.97, 111.95 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution range (Å) [outer shell] | 50-2.3 [2.36-2.30] |
| No. reflections | 96251 |
| Rmerge (%) | 8.0 [62.0] |
| Multiplicity | 5.0 [4.9] |
| Average I/σI | 21.5 [2.4] |
| Completeness (%) | 99.5 [99.8] |
| Refinement statistics | |
| No. atoms | 12721 |
| Protein | 11632 |
| Ligand (PC) | 108 |
| Ligand (PE) | 102 |
| Carbohydrate | 317 |
| Waters | 506 |
| Other ligands and ions | 56 |
| R/Rfree | 0.201/0.248 |
| Ramachandran plot (%) | |
| Favored | 97.9 |
| Allowed | 100 |
| R.m.s. deviations | |
| Bonds (Å) | 0.012 |
| Angles (°) | 1.457 |
| B-factors (Å2) | |
| Protein | 43.1 |
| Ligand PC | 59.8 |
| Ligand PE | 59.0 |
| Carbohydrate | 58.6 |
| Waters | 21.2 |
| Other ligands and ions | 53.2 |
In vitro loading of lipid antigens
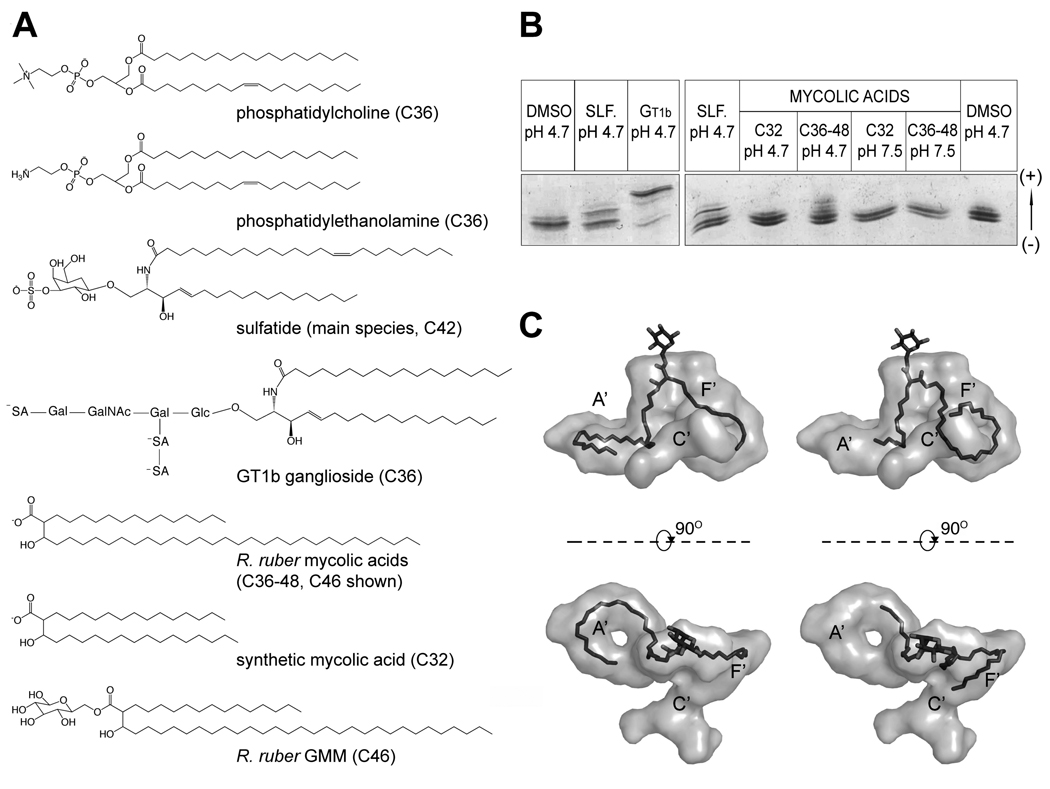
Brain porcine sulfatide extract (Avanti Polar Lipids Inc) were used together with a purified extract of Rhodococccus ruber mycolic acids (C36–C48), a synthetic version of a short mycolic acid (C32), purified trisialoganglioside GT1b from bovine brain (minimum purity 96%, Sigma Aldrich) and purified extract of mycolic acids from Mycobacterium tuberculosis (average size C80, minimum purity 98%, Sigma-Aldrich) to assess in vitro loading of antigens by boCD1b3. The R. ruber mycolic acids were generated by KOH hydrolysis of purified GMMs, followed by extraction into diethyl ether. Analysis by thin layer chromatography showed mycolic acids as the only lipids present in the sample (data not shown). Aliquots of 10 µg of purified boCD1b3 at a concentration of 20 µM were loaded overnight at room temperature in the presence of 6 times molar excess of each ligand (in DMSO, final concentration 3–5%, or in the presence of a final concentration of 0.05% Tween 20). Loading was performed in the presence of 100 mM TrisHCl pH 7 or with protein previously buffer-exchanged against 50 mM Na acetate pH 4.7, 100 mM NaCl. The binding of charged lipids was assessed by isoelectric focusing on a PhastGel IEF 3–9 using a PhastSystem (GE Healthcare) followed by staining with Coomassie dye.
Modeling of R. ruber GMM in the boCD1b3 binding groove
A C46 (C16 α chain and a C30 meromycolate chain) glucose monomycolate model representative of the Rhodococccus ruber mycolates was generated and manually modeled in the binding groove of boCD1b3 by following the electron density of the endogenously bound ligand, using the program Coot (32). The chain lengths were consistent with the reported sizes of alpha and meromycolate chains in mycolic acids of the genus Rhodococcus (α chain C12–18, C34–48 in total)(41). The lipid was modeled in two different orientations, corresponding to the two conformations observed for the ligands in the non-crystallographic boCD1b3 dimer. The alkyl chains were real-space refined against the electron density present in the groove while the polar glucose residue was placed in a similar orientation to the one observed for this portion of the GMM molecule in the huCD1b/GMM structure (19).
LDN5 T cell assay
C1R cells were electroporated with bovine CD1b3 full-length cDNA, cloned into pcDNA3.1. After growing for 6 weeks on medium containing 0.2 mg/ml G418 (Sigma-Aldrich), cells that stained positively with the BCD1b.3 antibody were FACS sorted. LDN5 T cells and human CD1b transfected C1R cells were previously reported (21). Before the assay, C1R cells were grown for a week in medium without G418. Expression of CD1b was confirmed by flowcytometry on the same day as the T cell assay. 7.5 *104 T cells were cultured with 5.0 *104 C1R cells for 24 hours, in the presence or absence of serial dilutions of GMM isolated from M. phei, N. farcinica, and R, equi, representing GMM with long, intermediate, and short chain mycolic acids (21). 50 µl of supernatant was tested for the presence of IFN-γ by a sandwich ELISA.
Results
BoCD1b3-boβ2m structure
The boCD1b3-boβ2m crystal structure was determined to a resolution of 2.3 Å in space group P21212 with four CD1-β2m heterodimers in the asymmetric unit, which arrange in two CD1-β2m pairs. Superposition of the eight individual molecules present in the asymmetric unit shows RMSD values ranging from 0.27 to 0.41 Å (Cα atoms) for the CD1b heavy chains and RMSD values ranging from 0.20 to 0.25 Å (Cα atoms) for the β2m chains. Given the high similarity observed between the chains in the asymmetric unit, and for the purpose of clarity, only one pair of structurally distinct boCD1b3-boβ2m heterodimers (composed of chains A, B and C, D) will be described through the text, unless otherwise specified.
The crystal structure of boCD1b3 reveals the expected domain architecture observed for other CD1 molecules, with the heavy chain non-covalently associated with the shorter β2m chain (Figure 1) (4, 42). The heavy chain is composed of three domains (α1, α2 and α3) with α1 and α2 constituting the antigen-binding superdomain. The boCD1b3 α1-α2 superdomain shares 72% sequence identity (ClustalW) (43) with human CD1b, for which four crystal structures in complex with different ligands have been previously determined (PDB ID 1GZQ in complex with phosphatidylinositol, 1GZP with the ganglioside GM2, 1UQS with GMM and 2H26 with PC and a long hydrophobic spacer) (18, 19, 30). Superposition of the α1-α2 superdomains (residues 8–180) of boCD1b3 and huCD1b yielded RMSD values of 0.76 Å (PDB ID 1GZQ), 0.73 Å (1GZP), 0.82 Å (1UQS) and 0.77 Å (2H26) showing a highly conserved topology in this region of the molecule.
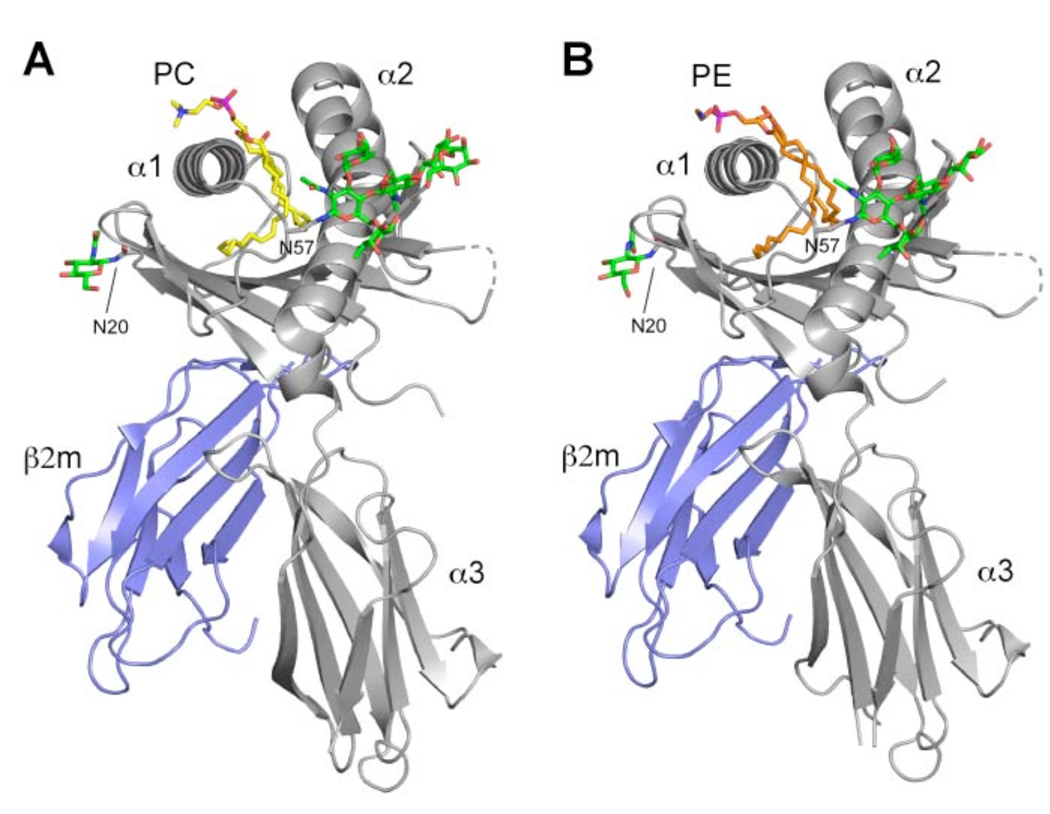
Figure 1.
Overview of the boCD1b3-PC/PE structure. CD1 heavy chain is in grey with the α1, α2 and α3 domains indicated and β2m is in blue. The bound PC molecule (A) is shown in yellow and PE in orange (B), while N-linked oligosaccharides at positions Asn20 and Asn57 are shown in green. The disordered loop comprising residues 106–109 of the heavy chain is shown as a dashed line.
Electron density for N-linked oligosaccharides is observed at residues Asn20, 57 and 128 of the heavy chain. The most well-ordered oligosaccharide is attached to Asn57 and interestingly shows the presence of an uncommon α1–3 fucose residue on the proximal GlcNAc, in addition to the more common α1–6 linked core fucose (Figure 1). Although uncommon, similarly linked carbohydrates have previously been described as the result of α1–3 fucosyltransferase activity in Drosophila melanogaster and other insect cells (44, 45).
Binding pocket shape
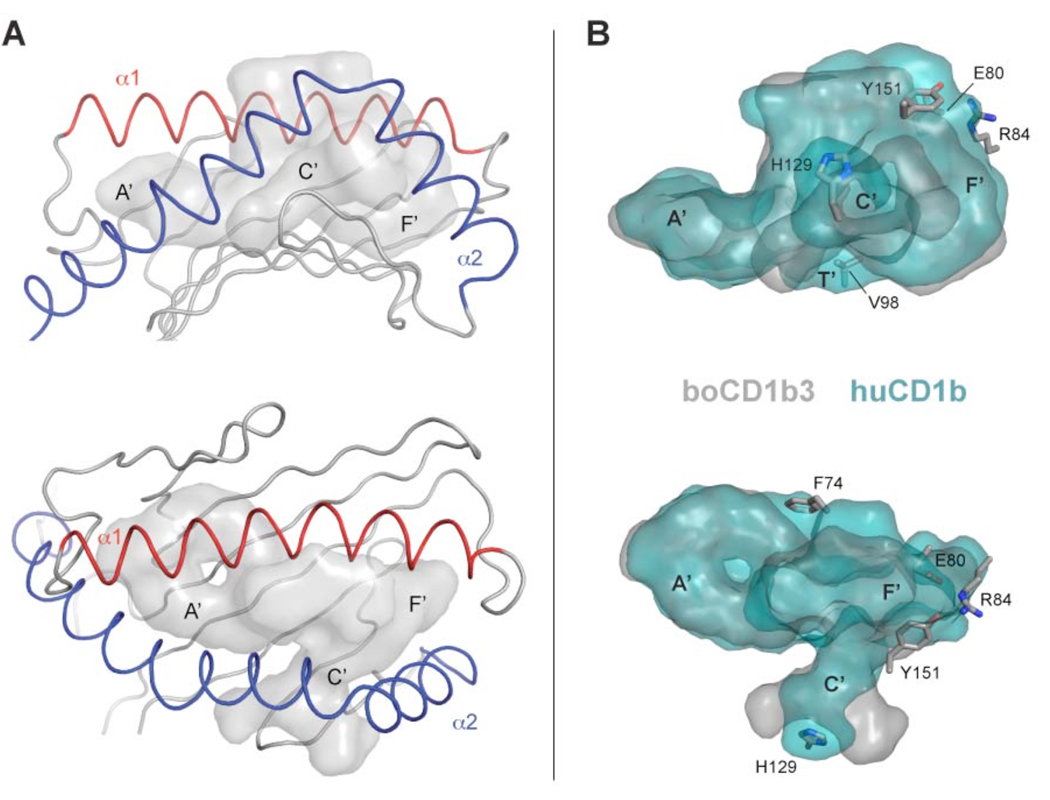
BoCD1b3 has been predicted by sequence homology to possess a binding pocket similar to the one observed in human CD1b (22). Comparison of the binding grooves between bovine CD1b3 and human CD1b reveals an almost identical pocket volume (2396 Å3 vs 2402 Å3, as calculated with CASTp) (35) and a similar topology (Figure 2A and 2B). In boCD1b3, the A’, C’ and F’ pockets appear to be retained, although with minor differences, while the T’ tunnel is interrupted by Val98 (glycine in huCD1b). This results in the separation of the two main pockets. The A’ pocket is buried under the N-terminal half of the α1 helix and laterally closed by the C-terminal portion of the α2 helix, in a position similar to huCD1b. The A’ pocket shows the typical “doughnut” shape built around the central pole defined by Leu12 and Phe70. The F’ pocket is located at the opposite side of the binding groove, and pinned in between the conserved residues Phe77 and Phe144. However, in boCD1b3 the F’ pocket appear to be closed at its top, mainly due to the presence of an arginine residue at position 84 of the α1-helix (a phenylalanine in huCD1b). This residue is involved in several polar and ionic interactions with Glu80 and Tyr151, creating a F’ roof that “caps” the opening of the pocket (Figures 2B and S1). As a result, the F’ roof prevents the alkyl chains to egress from the pocket towards the CD1-TCR interface, contrarily to what has been suggested for huCD1b (18). However, it is possible that the protonation of Glu80 at the acidic pH values observed in late endosomes and lysosomes (reported to be in the range of 4.5–5.5 for dendritic cells) (46) could result in the disruption of the network with Arg84 and Tyr151 and increased loading of long chain lipids with a mechanism similar to the one described for huCD1b (37). In human CD1b, two residues located at one end of the α1 helix, Asp60 and Glu62, are found to tether the rigid α1 helix with a flexible area of the α2 helix and the loop containing residues 50–60. The protonation of these two residues at the acidic pH values observed in late endosomes was shown to result in the disruption of these tethering interactions and in increased loading of long chain lipids. The two residues Asp60 and Glu62 are conserved in boCD1b3, together with their respective ionic partners Lys55 and Arg168. As the pK of charged residues can be heavily influenced by the local environment, the multiconformation continuum electrostatics approach was used to estimate the pK of Glu80, as previously described for the human CD1b structure (36, 37). The obtained pK value of 4.15 suggests that, within the range of pH observed in the lysosomes, a substantial proportion of the Glu80 sidechains are protonated, consistent with the hypothesis of a pH-dependent mechanism for the opening of the F’ roof. Moreover, boCD1b3 could also be able to bind antigens with long acyl chains owing to the presence of an open C’ portal extending under the α2 helix (Figure 2A). This portal, previously described in the structures of huCD1b in complex with PI, GM2 (18) and GMM (19) (but closed in the structure containing endogenous ligands) (30), is also present in boCD1b3, being defined by the residues 112–113, 126–131 and 145–163 of the heavy chain, together with a conserved disulfide bond (131–145) unique to CD1b molecules. Interestingly, the C’ portal in boCD1b3 appears to adopt a novel conformation, due to the differences in several residues in this area. The conformation of the boCD1b3 residue His129 (Ala129 in huCD1b) results in the blocking of the main exit of the C’ portal. However, the bulky side chains Met155 and Tyr163 of huCD1b are much smaller (Ser155 and Ser163) in boCD1b3 and open two alternative escape routes through the C’ portal on the side of the loop containing residues 126–131 (Figure S2). Both openings appear large enough to allow for the passage of alkyl chains from the C’ tunnel to the exterior of the protein.
Figure 2.
Architecture of the binding groove of boCD1b3 and comparison with huCD1b. A, The antigen-binding domain of boCD1b3, side and top view, is shown with the α1 and α2 helices indicated in red and blue respectively. The surface of the transparent antigen-binding groove is rendered in grey with the main pockets A’, F’ and the C’ portal labeled. B, Side and top view of the superposed huCD1b (PDB ID 1UQS, cyan) and boCD1b3 (grey) binding pocket. The boCD1b3 residues, which limit the pocket shape compared to huCD1b are shown in sticks representation. Side views are shown in the top row and top views are below.
Mass spectrometry analysis of boCD1b3 and its endogenous ligands
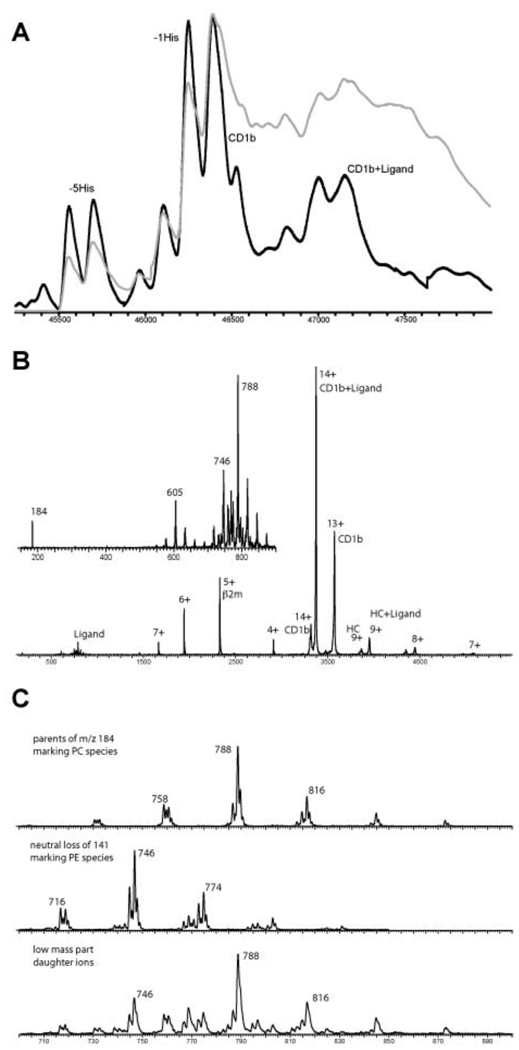
Since the protein was expressed in insect cells, an endogenous ligand was expected to be present in the hydrophobic groove of the molecule, as observed for other CD1 molecules expressed in the same system (30, 47). To determine the nature of the ligands bound to the boCD1b3 molecules, the purified protein was analyzed by native ESI-MS (48). Introducing the protein complex at high desolvation energy the recorded spectrum showed a charge envelope of sharp and intense peaks for the empty boCD1b3 and a set of broad and low intense peaks for the boCD1b3 ligand complex. For both series of peaks the 14+ is the most intense, being m/z 3313.6 for the empty boCD1b3 and m/z 3368.6 for the boCD1b3 ligand complex (data not shown). From this mass difference it was calculated that the average mass of the ligand is approximately 770 Da. The charge envelope of the empty boCD1b3 is showing that the glycosylation is uniform with a total molecular weight of approximately 46376 dalton which means that the three glycosylation sites provide an additional mass of around 3140 Da. Decreasing the desolvation energy is giving a mass spectrum with very broad peaks for the intact boCD1b3-ligand complex showing that this is the prevalent species in solution. Deconvolution of the charge envelopes in the spectra recorded at high and low desolvation energies are presented in Figure 3A (black line from high energy and grey line for low energy). The mass spectrum of boCD1b3-ligand complex recorded in the negative ion mode is only showing PE ions as PC cannot form negative ions. Subjecting the selected 14+ of the total complex to collision induced dissociation the recorded fragment ion MS/MS spectrum is showing a neutral loss and charged loss of ca. 770 Da, some peaks corresponding with β2m and further signals for the heavy chain+ligand and the empty heavy chain (Figure 3B). The inset of this figure is showing the low mass range with the ligand signals. To identify the exact nature of the ligands, two MS/MS scan methods were used to probe the headgroup fragmentation of the ligands. First a parent ion scan of m/z 184 (cholinephosphate) revealing the molecular ions of the phosphatidylcholine species, and second a neutral loss scan of 141 Da (ethanolaminephosphate) giving the molecular ions of the phosphatidylethanolamine species (Figure 3C). In this way two endogenous diacyl glycerolipids could be unambiguously identified by ESI-MS/MS namely phosphatidylcholine (PC) and phosphatidylethanolamine (PE) with 36 carbons in total (C18:1 and C18:0) as the major species (approximately 35% of the total fatty acids).
Figure 3.
Mass analysis of the lipid ligands in boCD1b3. A, Deconvoluted mass spectra at high (black) and low (grey) desolvation energy. B, Tandem MS spectrum of the selected 14+ precursor ions of the intact non-covalent complex of boCD1b3 and the lipid ligands (CD1b+Ligand). The low mass range inset of this tandem MS spectrum is showing m/z 184, phosphatidylcholine (PC) marker ion and m/z 788 as his most intense parent ion and further m/z 746 and his 141 Da neutral loss at m/z 605 from phosphatidylethanolamine (PE). C, The panel presents, respectively, the parent ion scan of m/z 184 giving the PC species, the neutral loss scan of 141 Da giving the PE species and for the same mass range the fragment ion spectrum of the complex of boCD1b3 and the lipids.
Modality of binding of PC and PE in the boCD1b3 binding groove
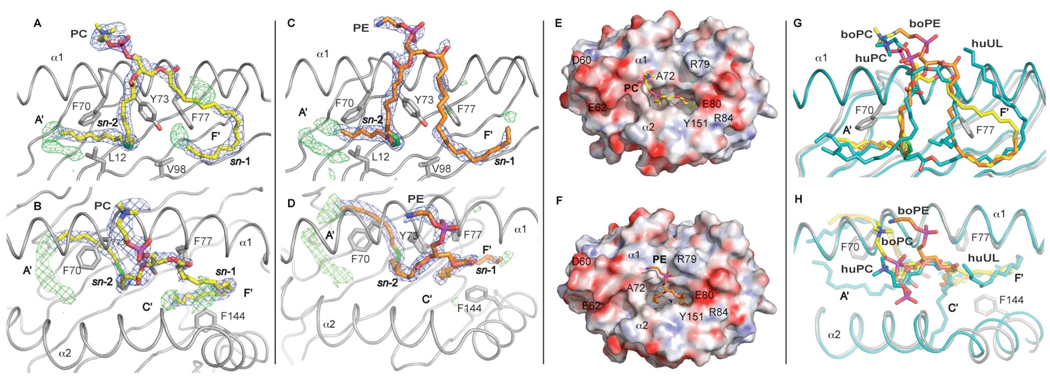
Clear electron density present in the binding groove confirmed the presence of the endogenous ligands. The electron density is well defined in the A’ pocket and illustrates that the endogenous ligand chain is fully encircling the central A’ pole in an anti-clockwise orientation. Electron density in the F’ pocket appeared to be less defined and to differ between the two molecules composing the crystallographic dimer. The main species of PC and PE as identified by ESI-MS (C36 with C18:1 at position sn-2 and C18:0 at position sn-1) were built into the binding groove during the final stages of refinement (Figure 4). In each of the two non-crystallographic dimers present in the asymmetric unit, one molecule was modeled with bound PC and one molecule with PE, to distinguish the slightly different electron densities observed in correspondence of the F’ pocket and the polar head of the ligand. It must be noted that, due to the similarity of the chemical composition of PC and PE (Figure 5A), this differences may not reflect two different chemical species but different conformations of the same compound. The PC/PE molecules were best modeled with the sn-2 chain bound to the A’ pocket and the sn-1 chain in the F’ pocket while the polar head is exposed outside the binding groove for potential recognition by a T cell receptor molecule. The density in the A’ pocket appears to be partially filled by the 18:1 acyl chain of PC/PE. The unsaturation between carbons 9 and 10 on the sn-2-linked acyl chain sits nicely at the junction between the inner cavity of the pocket and the channel connecting it to the surface as observed for other diacylglycerol ligands (49). Refinement of the longer but less common species of PC/PE with a C24:1 chain at the sn-2 position accounts for most, but not all, of the remaining electron density in the A’ pocket. This suggests that in the majority of CD1b-PC/PE complexes, when the C18:1 PC/PE is bound, a short chain lipid (possibly C8) could potentially act as a spacer. Spacer lipids with identical size have recently been described for the F’ pocket of mouse CD1d (50). The density in the F’ pocket appears less defined, possibly an indication of high flexibility in this region. However, lowering the sigma levels of the 2Fo-Fc electron density map to 0.6–0.8 (data not shown) suggests that the ligands could adopt alternative conformations, as illustrated by the two orientations of the modeled endogenous ligands. In case of the lipid binding mode observed for PC (Figure 4A and S3) the alkyl chain tail end is tucked underneath the ligand in the F’ pocket while for PE the alkyl chain fully encircles the F’ pocket in a counter-clockwise manner (Figure 4C and S3) and points towards the C’ portal.
Figure 4.
Endogenous ligands PC (A, B, E) and PE (C, D, F) bound to boCD1b3. A side view is presented with the α2 helix removed for clarity (A, C, G) together with the corresponding top view (B, D, E, F, H). A, B, C, D, The 2Fo-Fc electron density for the ligand, contoured at 1σ, is shown as a blue mesh. Some of the residues involved in the binding of the ligand and in the formation of the hydrophobic pocket are also depicted. The alkyl chain at the sn-1 position is inserted in the F’ pocket while alkyl chain at the sn-2 position, with the unsaturation shown in green, partially fills the A’ pocket. Positive difference density (Fo-Fc map contoured at 3σ and shown as green mesh) is visible at the bottom of the A’ pocket and, to a lesser extent, in the F’ pocket, possibly indicating lower occupancy species, such as ligands with a C24:1 fatty acid present in the binding groove, or possibly a C8 spacer ligand. E and F, Top view of the boCD1b3 binding pocket with the protein shown as a molecular surface with electrostatic potential (electronegative in red and electropositive in blue from −30 kT/e to 30 kT/e). The PC and PE ligands are shown in yellow and orange respectively. G and H, Top and side views of the superposed boCD1b3 (in grey) and human CD1b (PDB ID 2H26, in cyan) binding grooves in cartoon representation. The boCD1b3 ligands boPC and boPE (yellow and orange, respectively) are compared to the huPC and long spacer (huUL, human Unidentified Ligand, both in cyan) observed in the huCD1b structure.
Figure 5.
IEF analysis of boCD1b3 lipid binding and GMM models. A, Chemical structures of the lipids used in this study. The two endogenous ligands PC and PE are shown together with the lipids used in the in vitro binding assay, as well as the GMM species (C46) used for modeling. B, IEF gel illustrating loading of boCD1b3 with different lipids. DMSO pH 4.7: Loading control with DMSO. SLF pH 4.7: sulfatide, pH 4.7. GT1b pH 4.7: ganglioside GT1b, pH 4.7. C32 pH 4.7: C32 synthetic mycolic acid, pH 4.7. C36–C48 pH 4.7: R. ruber extract, pH 4.7. C32 pH 7.5: C32 synthetic mycolic acid, pH 7.5. C36–C48 pH 7.5: R. ruber extract, pH 7.5. C, Hypothetical model of a C46 GMM bound to the boCD1b3 binding groove (rendered as a grey surface). On the left side, side and top view of the binding pocket with the C30 meromycolate chain of the GMM molecule modeled in the A’ pocket. On the right side, side and top view of the binding pocket with the C30 chain of the GMM molecule modeled in the F’ pocket and extending toward the C’ portal.
The polar moieties of both PC and PE project outside the binding groove towards the α1 helix (Figure 4). PC and PE adopt different conformations of this portion of the molecule above residue Ala72, which acts as a “pole” around which the extended choline and ethanolamine groups can wrap themselves (Figure 4E and 4F). Interestingly, the crystal packing of the boCD1b3 crystals is such that each boCD1b3-PC complex is packed with the top of the antigen-binding groove against the antigen-binding groove of another, boCD1b3-PE complex. In particular the PC molecule contacts several residues from the α1 helix of the boCD1b3-PE complex (Glu65, Asp68, Leu69 and the PE molecule, cutoff 3.5 Å), while each PE molecule contacts residues from the α1 and α2 helices (Glu65, Leu69, Ile157, Arg160) of the boCD1b3-PC complex together with the corresponding PC ligand (Figure S4). This results in several interactions between the α1 and α2 helices of the two molecules and between the two ligands, which could affect their overall conformation.
Superposition of the two boCD1b3 molecules onto the human CD1b structure in complex with the endogenous ligand PC and a C41 hydrophobic spacer (PDB ID 2H26) (30) was used to compare the position and orientation of the ligand moieties in the binding groove (Figure 4G and H). In the huCD1b-PC-UL (UL, Unidentified Ligand) structure, the PC molecule binds, in a similar orientation to what has been observed for PC and PE in boCD1b3, with the sn-2 chain in the A’ pocket, while the sn-1 chain is bound in the C’ pocket. However, in huCD1b the polar phosphocholine headgroup is more closely located towards the α2 helix, while in case of boCD1b3 the polar headgroup is either situated close to the entrance of the groove above the A’ pocket (PC) or in the center of the groove, extending over the α1-helix (PE) between Ala72 and Arg79 (Figure 4 B, D, E, F). Therefore, boCD1b3 exhibits greater flexibility in binding polar headgroups of lipid antigens than the human CD1b ortholog.
In vitro loading of lipid antigens
The binding groove architecture and the ability of boCD1b3 to bind dual alkyl chain ligands such as PC and PE encouraged us to test in vitro the ability of boCD1b3 to bind different charged lipid antigens. Lipid loading can be directly visualized by native isoelectric focusing gel electrophoresis (IEF). Several charged ligands (Figure 5A) were incubated with boCD1b3 and successful loading was visualized by IEF (Figure 5B). Loading of a charged lipid can be monitored by observing a shift in the protein band on the gel, due to the change in isoelectric point caused by the additional lipid charge. Insect cell-expressed boCD1b3 migrates on the gel as two main species (Figure 5B, DMSO pH 4.7), possibly as a result of heterogeneity of the boCD1b3 protein, which also has been observed for mouse CD1d (51). A sphingolipid with a single charge such as sulfatide (3-sulfated β-D-galactosylceramide) can partially replace the endogenous ligands present in the binding groove, as demonstrated by the band shift observed in Figure 5B, lane 2 and 4 (labeled as SLF. pH 4.7). Almost complete loading was observed with the ganglioside GT1b, which carries three negative charges (labeled as GT1b pH 4.7). The incubation with this lipid resulted in a shift of the two bands of boCD1b3 towards the anode, further suggesting that heterogeneity of the protein, rather than the binding of lipids with different charge values lead to the observed double band. No detectable band shift is observed after incubation of the protein with a synthetic C32 mycolic acid (Figure 5B, lane 5 and 7), or with long (average size C80, purified extract from Mycobacterium tuberculosis) chain mycolic acid (data not shown). However, incubation of boCD1b3 with mycolic acid from Rhodococcus ruber (C36–48) clearly shows some degree of loading, as demonstrated by the appearance of a band at the same height of the main band observed for sulfatide-loaded boCD1b3 (compare Figure 5B, lane 4 with 6). Binding of this lipid is observed at acidic conditions (pH 4.7, lane 6) but not close to neutrality (pH 7.5, lane 8), consistent with the hypothesis proposed for the pH dependent antigen-capture of huCD1b (37) and suggesting a late-endosomal localization for this isoform of bovine CD1b. The inability of C32 and long mycolic acids to bind under these experimental conditions could relate to several factors such as an inability to displace the endogenous ligand already present in the binding groove by a shorter ligand (C32 mycolic acid) or the low solubility of the ligands, in particular the long form, in aqueous buffers. In vitro loading of CD1 with lipids of low solubility appears to be generally very insufficient (data not shown) and is not an indication of the inability of a particular lipid to bind to boCD1b3 in vivo.
GMM model
The ability of boCD1b3 to bind mycolic acids derived from Rhodococcus ruber prompted us to model the conformation that such ligands would assume in the binding pocket of boCD1b3 (Figure 5C). Two different modalities of binding of a C46 (C16 α chain, C30 meromycolate chain, Figure 5A) mycolic acid representative of the species present in R. ruber were modeled. In both conformations the glucose moiety is exposed on the surface of the binding groove, as observed in the huCD1b-GMM crystal structure (19). The two conformations differ in the relative position of the two alkyl chains in the main pockets of the binding groove. In one conformation, based on the modeled PC molecule, the C30 meromycolate chain fully occupies the A’ pocket while the shorter C16 alpha chain is inserted in the F’ pocket (Figure 5C, left side). Alternatively (Figure 5C, right side), the ligand was modeled with the C16 α chain partially occupying the A’ cavity and the longer C30 meromycolate chain in the F’ pocket. In the latter conformation the orientation of this longer chain follows the density observed for the PE molecule in the same region and suggests a potential modality of binding for longer chains that could exit the binding groove through the C’ portal. However, in the first model the size of the mycolate that can bind would be limited to C46–C48, as longer meromycolate chains (>C30) can not be bound in the A’ pocket.
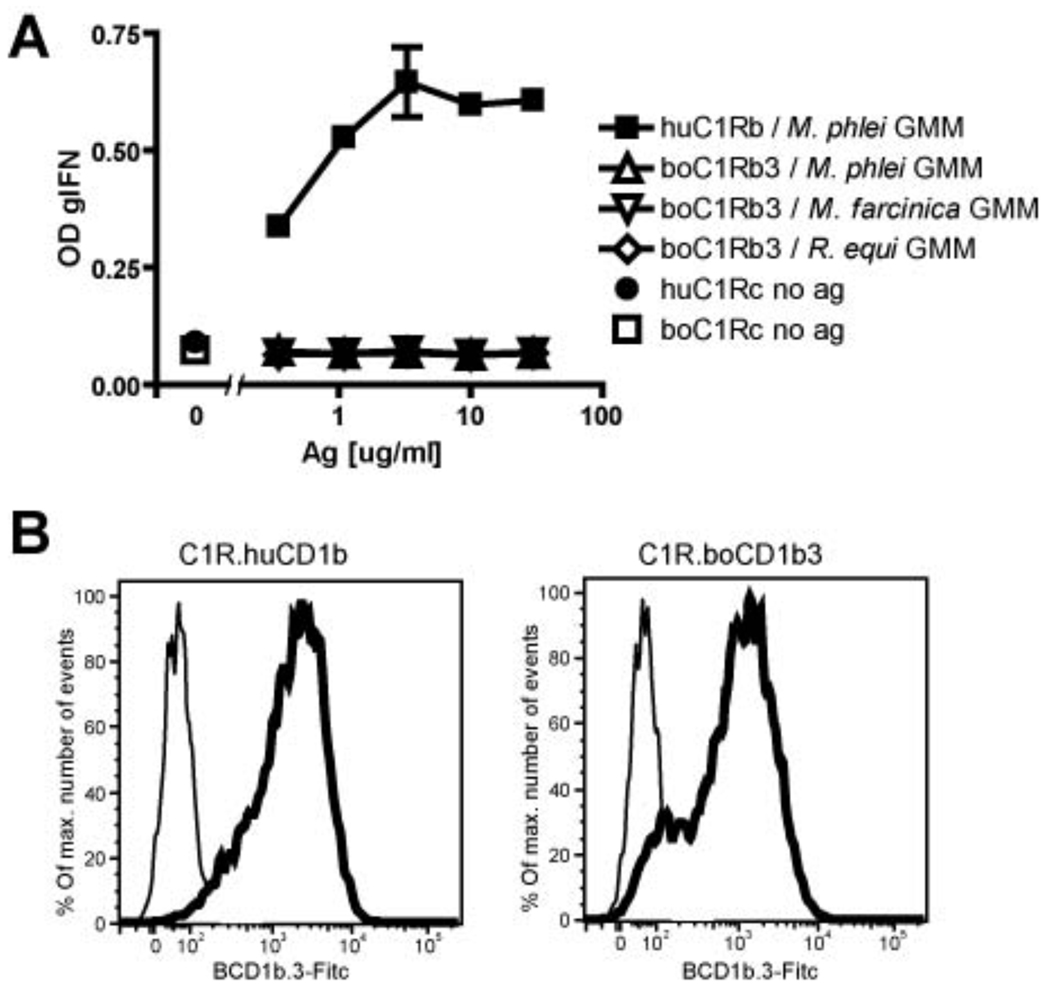
Lack of cross species presentation of GMM by CD1b
One of the remarkable features of the CD1d/α-galactosylceramide complex is its cross-species reactivity between humans and mice. Human NKT cells recognize the murine complex and vice versa. While no GMM-reactive bovine T cell line has been so far described, we tested whether bovine CD1b3 can present antigen to LDN5 T cells, which recognizes GMM of any chain length presented by human CD1b. The human lymphoblastoid cell line C1R transfected with bovine CD1b3 is not able to stimulate LDN5 cells, whereas C1R cells transfected with human CD1b do (Figure 6A). The expression of bovine and human CD1b on the surface of C1R cells was confirmed by staining with the antibody BCD1b.3 (Figure 6B). Consistent with these findings, previous studies revealed that mutation of only two residues on huCD1b (Glu80 and Thr157 to alanine among a panel of residues in the α1-α2 superdomain) resulted in a >1-log reduction in presentation of GMM to the LDN5 cell line, compared to wild-type huCD1b (52). This data suggests that these two residues mediate important contacts with the LDN5 TCR. Because in boCD1b3 Glu80 is in a different position (shifted by 2Å) and Thr157 is replaced by hydrophobic and bulky Ile157 (Figure S5), those TCR contacts may not be formed and are likely, at least in part, to be responsible for the lack of cross-species presentation between human and bovine CD1b molecules.
Figure 6.
Recognition of bovine and human CD1b by LDN5 cells. A, The presence of IFN-γ was measured in supernatants of co-cultures of LDN5 cells with human CD1b or bovine CD1b3-transfected C1R cells, and GMM. B, The expression level of CD1b was determined by flowcytometry, using the BCD1b.3 antibody (thick line), or an isotype control (thin line), followed by goat anti mouse FITC.
Comparison of boCD1b3 with boCD1b1 and boCD1b5
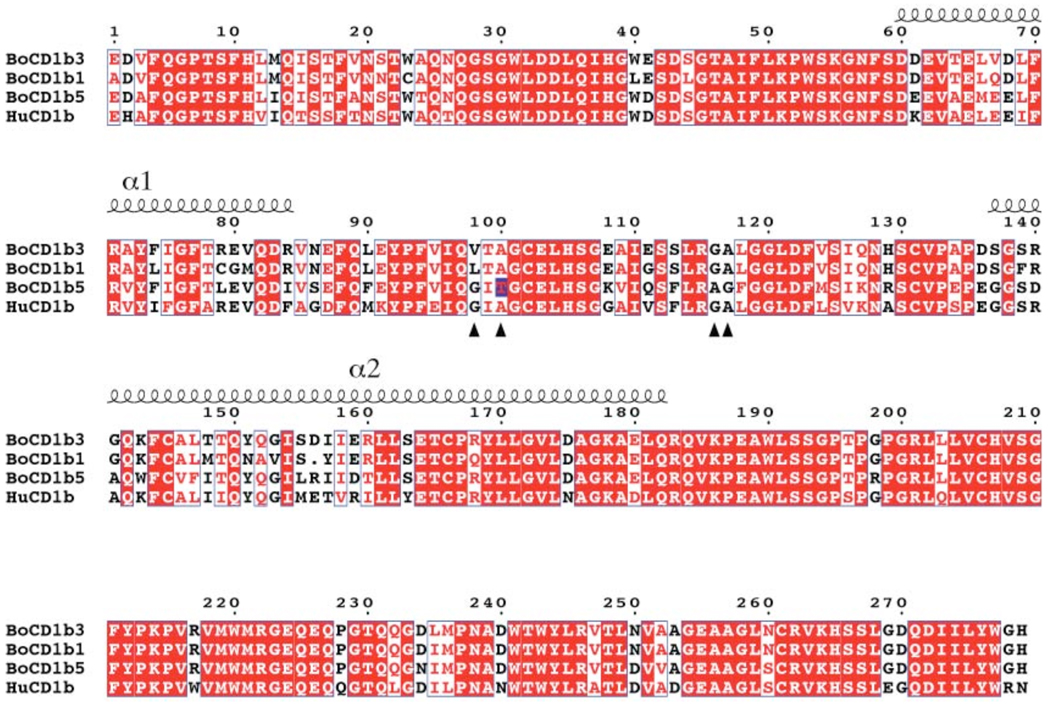
Cattle have been found to possess three potentially expressed CD1b proteins, boCD1b1, boCD1b3, boCD1b5 (22). Full length transcripts of CD1B3 and CD1B1 genes has been reported previously, and in this study we confirmed the expression of a full length transcript of the CD1B5 gene using a PCR-based approach (GenBank accession number GU325785). The three genes are highly conserved (sequence identity in the α1-α2 superdomain region for boCD1b3-boCD1b1 89%, boCD1b3–boCD1b5 76%, boCD1b1–boCD1b5 69%, Figure 7) and it can be expected that the general architecture of the binding groove with two main pockets, A’ and F’, and a C’ portal is maintained among these three proteins. However, based on the structural data presented here and the other available human and murine CD1 structures (18, 19, 47), some minor but potentially interesting differences can be predicted. Due to different residues at positions 80 and 84 (glycine/arginine in boCD1b1, glutamate/isoleucine in boCD1b5) the top of the F’ pocket could resemble huCD1b in the boCD1b5 structure and have an altogether different conformation in boCD1b1. On the other hand the C’ portal, due to the presence of bulky residues (histidine and arginine) at position 129 and relatively small side chains at positions 155 and 163 should show a similar conformation to the one observed in the boCD1b3 structure (Figure S1). Interestingly, due to the presence of a leucine at position 98, boCD1b1 is not expected to have a T’ tunnel, while boCD1b5 (with a glycine in the same position) should maintain this structural feature unique to huCD1b. However, we identified a potential polymorphism Ala/Thr at position 100 (evident by comparing the PCR product obtained as described in the Experimental Procedures with the genomic sequence publicly available at http://www.ensembl.org/Bos_taurus) that, together with the substitution of a glycine residue with an alanine at position 116, could affect and partially block the T’ tunnel in boCD1b5. The presence of a threonine in position 100 in the PCR product as described here is corroborated by other, partial bovine CD1b5 sequences available in EST databases (e.g. EH160235).
Figure 7.
Sequence alignments of the mature ectodomain of the three CD1b molecules present in Bos taurus (boCD1b1, boCD1b3, boCD1b5) with the human CD1b sequence. Identical amino acids are boxed in red. Red letters on white background indicate a similarity score among all possible pairs of residues per columns above 0.7, calculated according to the Risler matrix. The positions of the α1 and α2 helices are shown above the sequence while arrows indicate the residues determining the presence and conformation of the T’ tunnel. The polymorphism at position 100 of the boCD1b5 sequence is marked by a blue box.
Discussion
Three different isoforms of the CD1b molecule are expressed in Bos taurus, raising interesting questions about their structural and functional similarity to the human ortholog and the reason for their coexistence in the bovine genome. Despite the high sequence identity between the bovine and human proteins and a conserved overall structure, the binding groove architecture of the boCD1b3 structure described here indicates several differences compared to huCD1b. While the A’ pocket appears to have a highly preserved shape among CD1 molecules, differences with potential functional and immunological implications are observed for all the other pockets (C’, F’, T’) composing the binding groove.
Due to the presence of an arginine at position 84 (a phenylalanine in the human protein) the F’ pocket of boCD1b3 is closed at the top, limiting the accessibility into the groove and, likely, the in vitro lipid loading efficiency in general. Interestingly, it was observed for the human CD1b molecule that the phenylalanine residue in position 84 could, through a change in the conformation of its side chain, modify the hydrophobic binding capacity of the binding groove by packing against residues lining the bottom of the F’ pocket (Phe88, Met90, Phe144) (18). This mechanism appears unlikely to occur for boCD1b3, due to the stronger nature of the interactions mediated by the sidechains of Glu80, Arg84 and Tyr151. However, based on the conservation of charged residues (e.g. Asp60, Glu62, Glu80) potentially involved in a pH-dependent tethering and consistent with the enhanced loading of ligands at acidic pH observed in vitro, we predict that the boCD1b3 α1-α2 superdomain adopts an open conformation under low pH conditions to enable the binding of large antigenic lipids to the groove. Although data regarding the intra-cellular localization of boCD1b3 are at the moment not available, the cytoplasmic tail of boCD1b3 possesses high sequence homology to the same region of huCD1b (22), including an YXXZ motif (where Y is a tyrosine, X a spacer residue and Z a bulky hydrophobic residue, recognized by adaptor proteins 1–3 (3), strongly suggesting that this isoform can localize in late endosomes and lysosomes as the human form. A novel conformation of the C’ portal unique to CD1b molecules was also observed in boCD1b3, with two potential exit routes toward the protein surface. Consistent with this, we also observed weak and unconnected electron density (0.6–0.8σ, not shown) in this portion of the binding groove that would suggest the presence of low-occupancy ligands in at least some of the crystallized molecules. Perhaps most interestingly, the absence of the T’ tunnel in boCD1b3 limits its ability to bind ligands with long acyl chains with the same modality observed for huCD1b. Despite this, it is possible that medium and long ligands, such as C46 and C76–C80 GMM, bind to bovine CD1b3 in an alternative orientation. The conformations of the modeled endogenous ligands PC and PE, which represent the main species present in the binding groove, as determined by ESI-MS, suggest two potential modalities of binding in which the different alkyl chain orientations in the F’ pocket determine the overall length of the bound lipid chain. In particular, as demonstrated by the ligand conformation within the PE bound molecule (Figure 4C, D), long chains could presumably bind in the pocket in a counter-clockwise orientation and possibly egress to the protein surface through the C’ portal. We observed binding of sphingolipids and intermediate-length (C36–C48) mycolic acids derived from the actinomycete R. ruber to boCD1b3, as detected by IEF (Figure 5B) and modeling of a C46 GMM lipid into boCD1b3 illustrates the potential of boCD1b3 to accommodate lipids of this size in two different orientations (Figure 5C), analogous to the two orientations observed for PC and PE. Sufficiently long chains may be required to exchange with the endogenous ligands already present in the groove, as a shorter (C32) mycolic acid failed to yield detectable loading in our in vitro assay. On the other hand, the failure of binding of an extract of long (average length C80) mycolic acids to boCD1b3 might be due to the extremely poor solubility of these compounds. This also raises the question of the importance of lipid transfer proteins, such as saposins (53), in lipid loading onto boCD1b3. Human CD1e has been shown to be involved in the processing and loading of mycobacterial antigens onto human CD1b in the lysosome (6) and we assume that lipid transfer proteins also exist to facilitate lipid binding to boCD1b molecules in general. While attempts at determining the ability of boCD1b3 to present GMM antigens to the human LDN5 cell line demonstrated a lack of cross-reactivity, most likely the result of mutations in key residues at the CD1b-TCR interface, it has been shown that cattle develop strong T cell-based responses to immunization with long (main species C76) GMM molecules (54). Also, animals naturally infected with M. avium paratuberculosis, which contains long chain GMMs, show CD1b-dependent responses against intermediate and long chain GMMs (55). However, it is not clear at this moment if the ability of cattle to respond in a CD1b-dependent manner to GMM is dependent specifically on boCD1b3 or on one of the other two bovine CD1b molecules.
The presence of multiple functional genes for one CD1 isoform is not unique to cattle. Guinea pigs express 4 CD1b and 3 CD1c proteins and dogs express 4 CD1a proteins. In general, the functional capacities of a CD1 molecule in vivo depend on the expression pattern of the molecule, its intracellular trafficking pattern, and the characteristics of its antigen-binding groove. A possible explanation for the presence of multiple CD1b proteins in cattle is that only one of these molecules is a true functional homolog of human CD1b and that the others make up for the lack of bovine CD1c and CD1d protein. However, as we have shown here, the antigen-binding groove of CD1b1 and CD1b5 is closer, although with some remarkable differences, to the human CD1b molecule than to CD1c and CD1d in terms of size and shape. Alternatively, combinations of antigen binding groove and sub-cellular location that are not found in humans are present in cattle and truly perform additional, novel functions in this species. Mutations in the cytoplasmic tail that modify the intra-cellular localization of the CD1 molecule are thought to occur first and induce variation in the binding groove due to the exposure to different ligands in different compartments of the lysosomal pathway (56). Interestingly, the cytoplasmic portion of boCD1b1 does not show homology to the tails of other known CD1 molecules and does not contain any known trafficking motif although the absence of a YXXZ and a dileucine motif suggests a non lysosomal location (22). It is tempting to speculate that in cattle CD1b gene duplicated and diverged to generate, together with a “canonical” CD1b structure (most likely CD1b5), functionally distinct molecules.
In conclusion, boCD1b3 resemble human CD1b in its overall structure and in the fact that it can bind intermediate size lipids and mycolic acids. The presence or absence of a T’ tunnel is not a crucial factor for the binding of these lipids. Our data support the notion that the human family of CD1 proteins, with one functional gene of each isoform, is not representative of the situation in other species. In particular, the bovine CD1b molecules illustrate that variations of the shape of the binding groove as well as variations in tail motifs exist within one species. In addition, the existence of two allelic forms of boCD1b5 with a marked difference in shape of binding groove, suggests that the CD1 family of proteins is more dynamic than previously appreciated.
Acknowledgments
We would like to acknowledge the staff of the Stanford Synchrotron Radiation Laboratory, especially beamline 7-1 for their support during X-ray data collection and the Netherlands Proteomics Centre for the mass spectrometry data collection.
Footnotes
This work is supported in part by grants from the National Institute of Health (AI 074952, DMZ). DMZ is recipient of an investigator award from the Cancer Research Institute. GSB acknowledges support in the form of a Personal Research Chair from Mr. James Badrick, Royal Society Wolfson Research Merit Award, as a former Lister Institute-Jenner Research Fellow, the Medical Council and The Wellcome Trust (084923/B/08/7). IVR is supported by a Meervoud subsidy of the NWO (Nederlandse Organisatie voor Wetenschappelijk Onderzoek).
Structure factors and coordinates are deposited into the PDB database with code 3L9R.
β2m, beta-2-microglobulin; GMM, glucose monomycolate; bo, bovine; hu, human; MS, mass spectrometry; PDB, Protein Data Bank; ES, electrospray ionization; IEF, isoelectric focusing; PI, phosphatidylinositol; GM2, GM2 ganglioside; PC, phosphatidylcholine; PE, phosphatidylethanolamine.
References
- 1.Dye C. Doomsday postponed? Preventing and reversing epidemics of drug-resistant tuberculosis. Nat Rev Microbiol. 2009;7:81–87. doi: 10.1038/nrmicro2048. [DOI] [PubMed] [Google Scholar]
- 2.Colditz GA, Brewer TF, Berkey CS, Wilson ME, Burdick E, Fineberg HV, Mosteller F. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. JAMA. 1994;271:698–702. [PubMed] [Google Scholar]
- 3.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 4.Zajonc DM, Wilson IA. Architecture of CD1 proteins. Curr Top Microbiol Immunol. 2007;314:27–50. doi: 10.1007/978-3-540-69511-0_2. [DOI] [PubMed] [Google Scholar]
- 5.Moody DB, Porcelli SA. Intracellular pathways of CD1 antigen presentation. Nat Rev Immunol. 2003;3:11–22. doi: 10.1038/nri979. [DOI] [PubMed] [Google Scholar]
- 6.de la Salle H, Mariotti S, Angenieux C, Gilleron M, Garcia-Alles LF, Malm D, Berg T, Paoletti S, Maitre B, Mourey L, Salamero J, Cazenave JP, Hanau D, Mori L, Puzo G, De Libero G. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 7.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 8.Grant EP, Degano M, Rosat JP, Stenger S, Modlin RL, Wilson IA, Porcelli SA, Brenner MB. Molecular recognition of lipid antigens by T cell receptors. J Exp Med. 1999;189:195–205. doi: 10.1084/jem.189.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Libero G, Mori L. Recognition of lipid antigens by T cells. Nat Rev Immunol. 2005;5:485–496. doi: 10.1038/nri1631. [DOI] [PubMed] [Google Scholar]
- 10.Schaible UE, Hagens K, Fischer K, Collins HL, Kaufmann SH. Intersection of group I CD1 molecules and mycobacteria in different intracellular compartments of dendritic cells. J Immunol. 2000;164:4843–4852. doi: 10.4049/jimmunol.164.9.4843. [DOI] [PubMed] [Google Scholar]
- 11.Sugita M, Grant EP, van Donselaar E, Hsu VW, Rogers RA, Peters PJ, Brenner MB. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–752. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- 12.Dascher CC, Hiromatsu K, Naylor JW, Brauer PP, Brown KA, Storey JR, Behar SM, Kawasaki ES, Porcelli SA, Brenner MB, LeClair KP. Conservation of a CD1 multigene family in the guinea pig. J Immunol. 1999;163:5478–5488. [PubMed] [Google Scholar]
- 13.Van Rhijn I, Koets AP, Im JS, Piebes D, Reddington F, Besra GS, Porcelli SA, van Eden W, Rutten VP. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d. J Immunol. 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- 14.Looringh van Beeck FA, Zajonc DM, Moore PF, Schlotter YM, Broere F, Rutten VP, Willemse T, Van Rhijn I. Two canine CD1a proteins are differentially expressed in skin. Immunogenetics. 2008;60:315–324. doi: 10.1007/s00251-008-0297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013–1021. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Bohmer G, Prandi J, Mori L, Puzo G, De Libero G. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med. 2004;199:649–659. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moody DB, Guy MR, Grant E, Cheng TY, Brenner MB, Besra GS, Porcelli SA. CD1b-mediated T cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J Exp Med. 2000;192:965–976. doi: 10.1084/jem.192.7.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gadola SD, Zaccai NR, Harlos K, Shepherd D, Castro-Palomino JC, Ritter G, Schmidt RR, Jones EY, Cerundolo V. Structure of human CD1b with bound ligands at 2.3 Å, a maze for alkyl chains. Nat Immunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 19.Batuwangala T, Shepherd D, Gadola SD, Gibson KJ, Zaccai NR, Fersht AR, Besra GS, Cerundolo V, Jones EY. The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol. 2004;172:2382–2388. doi: 10.4049/jimmunol.172.4.2382. [DOI] [PubMed] [Google Scholar]
- 20.Moody DB, Briken V, Cheng TY, Roura-Mir C, Guy MR, Geho DH, Tykocinski ML, Besra GS, Porcelli SA. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol. 2002;3:435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 21.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, Besra GS, Porcelli SA. Structural requirements for glycolipid antigen recognition by CD1b- restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 22.Van Rhijn I, Koets AP, Im JS, Piebes D, Reddington F, Besra GS, Porcelli SA, van Eden W, Rutten VP. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d. J Immunol. 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- 23.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 24.Risler JL, Delorme MO, Delacroix H, Henaut A. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J Mol Biol. 1988;204:1019–1029. doi: 10.1016/0022-2836(88)90058-7. [DOI] [PubMed] [Google Scholar]
- 25.Zajonc DM, Maricic I, Wu D, Halder R, Roy K, Wong CH, Kumar V, Wilson IA. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Heuvel RH, van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJ, Langridge D, van der Oost J, Hoyes J, Heck AJ. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal Chem. 2006;78:7473–7483. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. HKL: Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Vagin AA, Teplyakov A. MOLREP:an automated programm for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 29.CCP4. Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Alles LF, Versluis K, Maveyraud L, Vallina AT, Sansano S, Bello NF, Gober HJ, Guillet V, de la Salle H, Puzo G, Mori L, Heck AJ, De Libero G, Mourey L. Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. Embo J. 2006;25:3684–3692. doi: 10.1038/sj.emboj.7601244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. 2001;D57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: ϕ,ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 34.Ho BK, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y, Mao J, Gunner MR. MCCE2: improving protein pKa calculations with extensive side chain rotamer sampling. J Comput Chem. 2009;30:2231–2247. doi: 10.1002/jcc.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Relloso M, Cheng TY, Im JS, Parisini E, Roura-Mir C, DeBono C, Zajonc DM, Murga LF, Ondrechen MJ, Wilson IA, Porcelli SA, Moody DB. pH-dependent interdomain tethers of CD1b regulate its antigen capture. Immunity. 2008;28:774–786. doi: 10.1016/j.immuni.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleywegt GJ. Validation of protein crystal structures. Acta Crystallogr D Biol Crystallogr. 2000;56:249–265. doi: 10.1107/s0907444999016364. [DOI] [PubMed] [Google Scholar]
- 39.DeLano W. The PyMOL Molecular Graphics System. 2002 www.pymol.org. [Google Scholar]
- 40.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barry CE, 3rd, Lee RE, Mdluli K, Sampson AE, Schroeder BG, Slayden RA, Yuan Y. Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Res. 1998;37:143–179. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- 42.Silk JD, Salio M, Brown J, Jones EY, Cerundolo V. Structural and functional aspects of lipid binding by CD1 molecules. Annu Rev Cell Dev Biol. 2008;24:369–395. doi: 10.1146/annurev.cellbio.24.110707.175359. [DOI] [PubMed] [Google Scholar]
- 43.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 44.Fabini G, Freilinger A, Altmann F, Wilson IB. Identification of core alpha 1,3-fucosylated glycans and cloning of the requisite fucosyltransferase cDNA from Drosophila melanogaster. Potential basis of the neural anti-horseadish peroxidase epitope. J Biol Chem. 2001;276:28058–28067. doi: 10.1074/jbc.M100573200. [DOI] [PubMed] [Google Scholar]
- 45.Staudacher E, Kubelka V, Marz L. Distinct N-glycan fucosylation potentials of three lepidopteran cell lines. Eur J Biochem. 1992;207:987–993. doi: 10.1111/j.1432-1033.1992.tb17134.x. [DOI] [PubMed] [Google Scholar]
- 46.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–1403. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 47.Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruiz Y, Bachi A, Kronenberg M, Wilson IA, Degano M. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol. 2005;175:977–984. doi: 10.4049/jimmunol.175.2.977. [DOI] [PubMed] [Google Scholar]
- 48.Heck AJ. Native mass spectrometry: a bridge between interactomics and structural biology. Nat Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Li Y, Kinjo Y, Mac TT, Gibson D, Painter GF, Kronenberg M, Zajonc DM. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc Natl Acad Sci U S A. 2010;107:1535–1540. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan BA, Nagarajan NA, Wingender G, Wang J, Scott I, Tsuji M, Franck RW, Porcelli SA, Zajonc DM, Kronenberg M. Mechanisms for Glycolipid Antigen-Driven Cytokine Polarization by Va14i NKT Cells. J Immunol. 2010;184:141–153. doi: 10.4049/jimmunol.0902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cantu C, 3rd, Benlagha K, Savage PB, Bendelac A, Teyton L. The paradox of immune molecular recognition of α-galactosylceramide: low affinity, low specificity for CD1d, high affinity for αβ TCRs. J Immunol. 2003;170:4673–4682. doi: 10.4049/jimmunol.170.9.4673. [DOI] [PubMed] [Google Scholar]
- 52.Melian A, Watts GF, Shamshiev A, De Libero G, Clatworthy A, Vincent M, Brenner MB, Behar S, Niazi K, Modlin RL, Almo S, Ostrov D, Nathenson SG, Porcelli SA. Molecular recognition of human CD1b antigen complexes: evidence for a common pattern of interaction with alpha beta TCRs. J Immunol. 2000;165:4494–4504. doi: 10.4049/jimmunol.165.8.4494. [DOI] [PubMed] [Google Scholar]
- 53.De Libero G, Mori L. Mechanisms of lipid-antigen generation and presentation to T cells. Trends Immunol. 2006;27:485–492. doi: 10.1016/j.it.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen TK, Koets AP, Santema WJ, van Eden W, Rutten VP, Van Rhijn I. The mycobacterial glycolipid glucose monomycolate induces a memory T cell response comparable to a model protein antigen and no B cell response upon experimental vaccination of cattle. Vaccine. 2009;27:4818–4825. doi: 10.1016/j.vaccine.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Rhijn I, Nguyen TK, Michel A, Cooper D, Govaerts M, Cheng TY, van Eden W, Moody DB, Coetzer JA, Rutten V, Koets AP. Low cross-reactivity of T-cell responses against lipids from Mycobacterium bovis and M. avium paratuberculosis during natural infection. Eur J Immunol. 2009;39:3031–3041. doi: 10.1002/eji.200939619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dascher CC, Brenner MB. Evolutionary constraints on CD1 structure: insights from comparative genomic analysis. Trends Immunol. 2003;24:412–418. doi: 10.1016/s1471-4906(03)00179-0. [DOI] [PubMed] [Google Scholar]