Abstract
Somatic and germline mutations in PTEN (phosphatase and tensin homolog deleted on chromosome 10) are found in sporadic cancers and Cowden syndrome patients, respectively. Recent identification of naturally occurring cancer and germline mutations within the ATP-binding motifs of PTEN (heretofore referred to as PTEN ATP-binding mutations) has revealed that these mutations disrupted the subcellular localization and tumor-suppressor activity of PTEN. However, very little is known about the underlying mechanisms of PTEN ATP-binding mutations in tumorigenesis. Here we show that these mutations impair PTEN's function both qualitatively and quantitatively. On the one hand, PTEN ATP-binding mutants lose their phosphatase activity and the effect of downregulation of cyclin D1. On the other, the mislocalized mutant PTEN results in a significantly decreased nuclear p53 protein level and transcriptional activity, enhanced production of reactive oxygen species, induction of Cu/Zn superoxide dismutase as well as dramatically increased DNA double-strand breaks (DSBs). When compared with wild-type PTEN, the ATP-binding mutant PTEN has reduced half-life in vitro and decreased protein expression levels in vivo. Our data, thus, reveal a novel mechanism of tumorigenesis in patients with germline or somatic mutations affecting PTEN ATP-binding motifs, i.e. qualitative and quantitative impairment of PTEN due to the loss of its phosphatase activity, and nuclear mislocalization, resulting in rapid PTEN protein degradation, suppression of p53-mediated transcriptional activity, loss of protection against oxidative stress as well as accumulation of spontaneous DNA DSBs.
INTRODUCTION
Breast cancer is the most common malignancy and the second most common cause of cancer-related deaths in women of the western world with an estimated 192 370 new cases, and 40 170 deaths among US women during 2009 (1). The tumor-suppressor PTEN (phosphatase and tensin homolog deleted on chromosome 10) plays an important role in both hereditary and sporadic breast cancer. Our laboratory first reported that germline mutations in PTEN are associated with Cowden syndrome (CS) (2) and Bannayan–Ruvalcaba–Riley syndrome (3), which confers a high risk of breast and other cancers. For CS females, the lifetime risk of developing breast cancer is 25–50%, compared with 13% in the general US population, and at an average age of diagnosis between 38 and 46 years, compared with 55–65 years in the general population (4). Furthermore, somatic loss of PTEN expression and/or function is often detected in a significant fraction of sporadic breast cancers.
Accumulating evidence suggests that the subcellular localization of PTEN may play an important role in cell growth and tumorigenesis. It is clear that the role of nuclear PTEN is not identical to that of cytoplasmic PTEN. Nuclear PTEN plays a powerful role in regulating chromosome stability by binding centrosomes (5), DNA repair (6) and cell cycle arrest (7). There appears to be several mechanisms of PTEN cytoplasmic-nuclear trafficking. The cytoplasm-predominant CS mutation (PTENK289E) has been reported to disrupt PTEN ubiquitination and thus abolish its nuclear import (8). In contrast, we identified that both ATP depletion and germline or somatic mutations in PTEN's ATP-binding motifs (which we will heretofore refer to as PTEN ATP-binding mutations) result in PTEN subcellular mislocalization which we hypothesized results in defects in its nuclear export (9,10).
PTEN contains two ATP-binding motifs and both are localized within its phosphatase domain, with the 3′ one lying within the phosphatase core motif (residues 60–73; residues 122–136) (9). In contrast to PTENWT, both cancer-associated somatic (PTENK62R, PTENY65C and PTENK125E) and germline PTEN missense mutations (PTENR130G and PTENY68H), which lie within the ATP-binding motifs, result in nuclear mislocalization as a cellular phenotypic hallmark (9).
Beyond these observations, little is known about the precise mechanism of breast carcinogenesis induced by the PTEN ATP-binding mutations. Therefore, in the present study, we sought to analyze the functional consequences of nuclear-cytoplasmic mislocalization of these PTEN ATP-binding mutants in breast carcinogenesis.
RESULTS
ATP-binding motif mutants abrogate PTEN's tumor-suppressive capabilities on cell signaling pathways
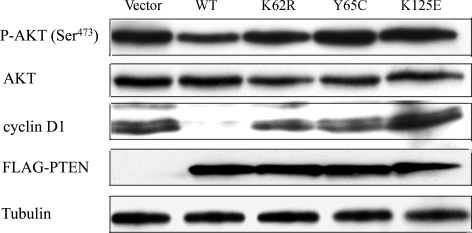
To determine the relative contribution of the PTEN ATP-binding mutations in breast carcinogenesis, we established clones with stable PTEN expression controlled by a Tet-off system to examine the consequences of increased levels of wild-type (WT) and mutant PTEN expression in a well-characterized breast cancer line, MCF-7, as we described before (9,11). These naturally occurring mutations derive from germline and somatic origins. When cultured in the absence of tetracycline (Tet), transfected PTEN constructs in all the stable cell lines (PTENWT, PTENK62R, PTENY65C and PTENK125E) were equally expressed when detected with an antibody against the FLAG tag in the C-terminal of PTEN (Fig. 1 and Supplementary Material, Fig. S1). We then determined the effect of PTEN overexpression on phospho-AKT (P-AKT) and cyclin D1 levels in unstimulated cells. P-AKT was selected as a downstream read-out of the PTEN lipid phosphatase activity; we measured cyclin D1 because it is an important regulator of G1 to S-phase transition downstream of PTEN, and especially nuclear PTEN. We found that the induction of PTENWT expression resulted in a significant decrease in the levels of both P-AKT and cyclin D1; in contrast, the expression of each of the mutant PTENs was unable to alter either P-AKT or cyclin D1 levels (Fig. 1). Together, our results suggest that mutations within ATP-binding motifs impaired PTEN's phosphatase activity and further abolished PTEN's appropriate regulation of G1/S progression, as reflected by unopposed P-AKT and cyclin D1 signaling pathways. This qualitative impairment of PTEN function represents at least one of the mechanisms by which these tumor-derived ATP-binding mutations lead to carcinogenesis.
Figure 1.
The effects of PTENWT and PTEN ATP binding mutants on AKT phosphorylation and cyclin D1 expression. MCF-7 Tet-off cells were stably transfected with plasmids encoding pTre2hyg vector only (Vector), FLAG-PTENWT, FLAG-PTENK62R, FLAG-PTENY65C and FLAG-PTENK125E. After a 24 h induction in the absence of Tet, whole-cell extracts were prepared and probed by immunoblotting for P-AKT, AKT, cyclin D1, FLAG-PTEN and tubulin levels.
Mislocalized PTEN ATP-binding mutants reduce p53 levels in the nucleus
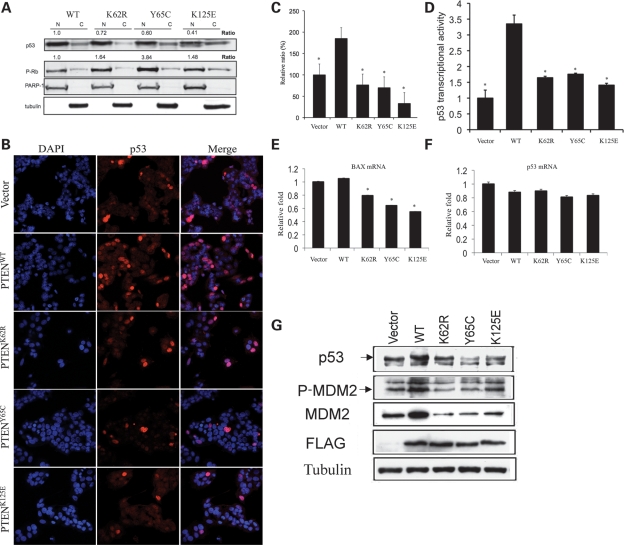
One of the most important intracellular phenotypic changes of the MCF-7 stable cell lines and lymphoblastoid cells harboring patient PTEN ATP-binding mutations is the predominant nuclear localization, which is accompanied by decreased G1/S ratio and apoptotic rate when compared with the PTENWT cells (9). In order to investigate the role of the PTEN ATP-binding mutants in the nucleus, we determined the nuclear phospho-RB (P-RB) and p53 protein expression levels in MCF-7 stable cells by subcellular fractionation after 24 h culture in Tet-free medium. As shown in Figure 2A, a marked reduction in nuclear p53 levels, accompanied by increased P-RB levels, was observed in MCF-7 cells overexpressing K62R, Y65C or K125E mutant PTEN.
Figure 2.
PTEN ATP-binding mutations downregulate p53 in the nucleus. (A) Nuclear (N) and cytoplasmic (C) fractions in the absence of Tet were prepared and examined for protein levels of p53 and P-RB. The data represent the fold change of the normalized nuclear p53 (p53/PARP-1) and nuclear P-RB (P-PB/PARP-1) expressed in MCF-7 cells overexpressing the PTEN mutants, compared with those in MCF-7 cells overexpressing PTENWT, respectively. (B) p53 protein localization and intensity in MCF-7 cells determined by confocal microscope imaging and immunofluorescence. MCF-7 cells stably transfected with PTENWT or PTEN ATP-binding mutations were fixed and analyzed by immunofluorescence for endogenous p53 protein staining. p53 was detected with a polyclonal antibody followed by a goat anti-rabbit secondary antibody conjugated to Alexa Fluor 568 (red). Nuclei were counterstained with DAPI (blue). (C) Quantification of immunofluorescence intensity for p53 in MCF-7 cells overexpressing PTENWT and mutants. The data represent the average of three independent measurements. (D) Luciferase reporter assay. PTENWT but not ATP-binding mutants enhances p53 transcriptional activity. MCF-7 cells stably expressing PTENWT or PTEN ATP-binding mutants, as well as the empty vector, were transfected with PG13-luc plasmid. Forty-eight hours later, luciferase activity was measured in cell lysates using equal amounts of protein. *P < 0.005 versus PTENWT. (E) Transient transfection of PTEN ATP-binding mutants into HEK293 cells decreases Bax mRNA transcription. Following transfection with the PTENs or control vector DNA, total RNA was extracted to measure Bax mRNA levels by qRT–PCR. *P < 0.05 versus PTENWT. (F) Transient transfection of PTEN ATP-binding mutants into HEK293 cells does not change p53 mRNA transcription. Total RNA from the above experiment was also used to measure p53 mRNA levels by qRT–PCR (P > 0.05). (G) Total protein lysates were extracted from MCF-7 cells stably expressing PTENWT, K62R, Y65C, K125E or the empty vector. Immunoblots were performed for p53, MDM2, P-MDM2, FLAG and tubulin.
To further validate these biochemistry results, we carried out immunoflourescence confocal analyses for nuclear p53 immunostaining intensity in MCF-7 stable cell lines. Whereas the overexpression of PTENWT resulted in an 85% increase in p53 immunostaining intensity, when compared with vector-only transfected cells, exogenously expressed K62R, Y65C and K125E PTEN mutants showed significantly decreased p53 immunostaining intensity when compared with PTENWT (Fig. 2B) (P < 0.001, one-way ANOVA). When treated with the proteasome inhibitor, MG132, all MCF-7 lines stably transfected with WT or mutant PTEN had similar levels of p53 protein by immunostaining, suggesting a post-translational degradation mechanism of p53 in the presence of the PTEN ATP-binding mutants (Supplementary Material, Fig. S2) (P > 0.05, one-way ANOVA).
p53 is a transcriptional activator that binds to sequence-specific binding sites at the promoters of several cellular genes and activates their transcription. Recent studies have indicated that PTENWT enhances p53-mediated transactivation of the promoter by stabilization of p53 in a phosphatase-independent manner (12). The decreased p53 abundance in the context of PTEN ATP-binding mutants led us to investigate the p53-mediated transcriptional activation. A luciferase reporter that contains p53-specific binding sites was transfected into MCF-7 stable cells overexpressing PTEN contructs (WT, K62R, Y65C and K125E) or the empty vector, and the p53 transcriptional activities were compared through the luciferase assay. As shown in Figure 2D, p53-mediated transcription was stimulated the greatest when MCF-7 cells were overexpressing PTENWT (a 3.4-fold increase, when compared with the empty vector, P < 0.005). All three PTEN ATP-binding mutants, however, only showed minor increases in the transcriptional activities (1.4–1.7-fold increase, when compared with the empty vector, and 1/3–1/2 that of the WT PTEN context).
To further confirm the decreased transcriptional activity of p53 in PTEN mutants, we next did a quantitative analysis of p53-targeted gene expression by real-time quantitative RT–PCR (qRT–PCR). The mRNA abundance of a p53-inducible gene, BAX, was compared in HEK293 cells after transient transfection with each PTEN construct (WT, K62R, Y65C or K125E) or the empty vector. Consistent with the reduced p53 levels (Fig. 2A and B) and p53 transcriptional activity, decreased BAX mRNA levels were observed in HEK293 cells overexpressing PTEN ATP-binding mutations, when compared with the same cells overexpressing PTENWT or the empty vector (Fig. 2E). p53 mRNA levels, however, did not show significant changes when HEK293 cells were transfected with either PTEN-WT or ATP-binding mutants, which supports our hypothesis that the diminished p53 protein levels in these PTEN mutants are due to accelerated post-translational degradation (Fig. 2F).
We then investigated the post-translational mechanism of p53 degradation by western blot. We first compared the induction of PTEN and p53 levels in MCF-7 cells overexpressing PTEN constructs (WT, K62R, Y65C and K125E) in the absence of Tet (Supplementary Material, Fig. S1). After removal of Tet for 24 h, expression of the FLAG-tagged PTEN was similar for all four PTEN constructs. Interestingly, p53 protein levels increased after removal of Tet only in cells overexpressing PTENWT, suggesting that overexpression of PTENWT, but not PTEN ATP-binding mutants, can increase p53 protein levels. We then compared MDM2 and phospho-MDM2 (P-MDM2) levels in the MCF-7 cell lines stably transfected with each construct, with the rationale that PTEN can stabilize p53 through the degradation of MDM2 by the suppression of the PI3K pathway. Surprisingly, we observed increased MDM2/P-MDM2 in cells overexpressing PTENWT and decreased MDM2/P-MDM2 in cells overexpressing each PTEN ATP-binding mutant (Fig. 2G). This may reflect a possible feedback mechanism in which the mutant cells produce MDM2/P-MDM2 in an attempt to rescue p53 from further degradation. It also suggests that the degradation of p53 in the PTEN ATP-binding mutants is MDM2-independent. Taken together, our data suggest that the PTEN ATP-binding mutants are unable to stabilize p53 in the nucleus and may even induce p53 inactivation through enhanced MDM2-independent degradation and decreased transcriptional activity.
PTEN ATP-binding mutations cause accumulated DNA double-strand breaks
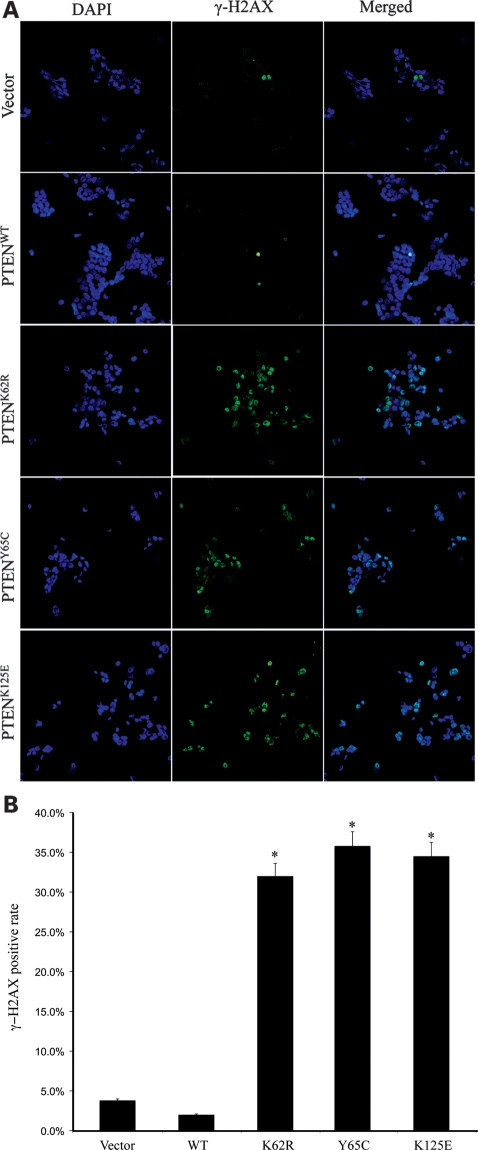
The predominant nuclear localization of the PTEN ATP-binding mutations prompted us to hypothesize that these mutations may disrupt chromosomal integrity by inducing double-strand breaks (DSBs) via a mechanism beyond disrupting binding to centrosomes. To test this, we detected γ-H2AX by immunofluorescence microscopy in senile MCF-7 cells (at passage 40 for all cell lines). After DNA DSBs, histone H2AX is rapidly phosphorylated on serine 139, referred to as γ-H2AX which is emerging as an important marker of DSBs (13).
As expected, overexpression of PTENWT in MCF-7 cells did not change the intensity of γ-H2AX staining, when compared with cells transfected with vector control (Fig. 3A and B). In contrast, MCF-7 cells overexpressing the cancer-associated PTEN mutants, K62R, Y65C or K125E had γ-H2AX intensity much higher than that of the vector only control. More importantly, the increased intensity for γ-H2AX was not found in the earlier passage (<10) cells (data not shown). This suggests that DSBs accumulate in the presence of PTEN ATP-binding mutations when cells develop toward senescence, at least in vitro.
Figure 3.
PTEN ATP-binding mutations lead to increased DSBs. (A) MCF-7 cells stably expressing PTENWT or PTEN ATP-binding mutants were fixed and immunostained for γ-H2AX (green). The cells were counterstained with DAPI (blue). (B) Statistical analysis of the γ-H2AX fluorescence data. *P < 0.001 versus PTENWT.
Breast cancer cells overexpressing PTEN ATP-binding mutations are under oxidative stress
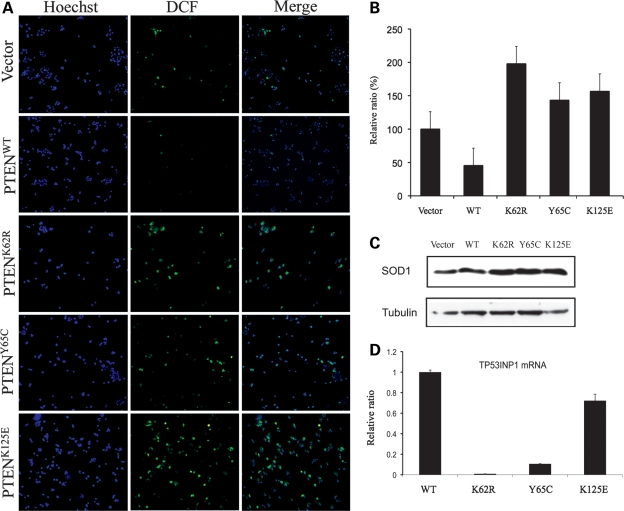
It is well documented that reactive oxygen species (ROS) represents one mechanism whereby stress-induced senescence results from ROS-associated DSBs. Based on the observation of the increased DSBs in senescent (operationally defined as >40 passage cells) MCF-7 cells overexpressing the PTEN ATP-binding mutations, we next examined the basal levels of ROS production in the five MCF-7 stable cell lines. Cells overexpressing PTENWT had a lower level of ROS production compared with control cells expressing empty vector, when not stimulated. In contrast, significantly activated ROS was observed in cells overexpressing PTENK62R, PTENY65C and PTENK125E (Fig. 4A and B). We then detected the quantity of Cu/Zn-SOD (SOD1) by western blot in the same cell lines. Elevated SOD1 levels were detected in cells overexpressing PTENK62R, PTENY65C and PTENK125E (Fig. 4C). Thus, the PTEN ATP-binding mutations lead to increased oxidative stress, characterized by enhanced ROS production and aberrant expression of the antioxidative enzyme, SOD1.
Figure 4.
PTEN ATP-binding mutations increase ROS production and SOD1 expression. (A) MCF-7 cells stably expressing empty vector, PTENWT or PTEN ATP binding mutants were incubated with 25 μm Carboxy-H2DCFDA for 30 min and immunofluorescent images were taken for ROS production (green). DNA was counterstained with Hoechst 33342 (blue). (B) Quantitation of immunofluorescence intensity of DCF [from (A)] in MCF-7 cells overexpressing PTENWT and mutants. The data represent the average of three independent measurements. P < 0.01, one-way ANOVA. (C) SOD1 protein was detected in cell lines expressing WT or mutant PTEN by western blot. (D) Total RNA from MCF-7 cells overexpressing WT or mutant PTEN was extracted and qRT–PCR was performed to measure TP53INP1 mRNA levels. P < 0.01, one-way ANOVA.
Because tumor protein 53-induced nuclear protein 1 (TP53INP1) is a major mediator of p53 antioxidant function, we further compared TP53INP1 mRNA levels in MCF-7 cell lines expressing the FLAG-tagged PTEN (WT and mutants) by qRT–PCR (14). MCF7 cells overexpressing any of the mutant PTEN constructs exhibited a decreased level of TP53INP1 mRNA when compared with those expressing PTENWT (Fig. 4D). This observation is consistent with the decreased p53 protein levels, reduced p53 transcriptional activity and increased ROS in cells overexpressing PTEN ATP-binding mutants.
PTEN ATP-binding mutations reduce protein stability
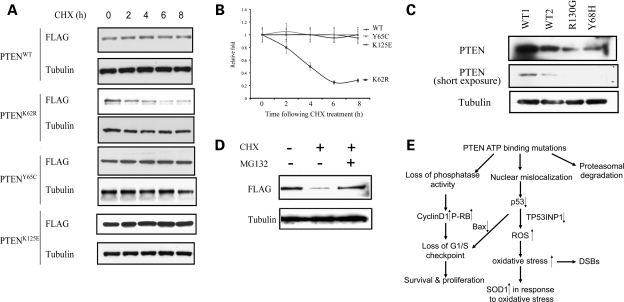
We then examined PTEN stability through pulse-chase experiments using cycloheximide (CHX). We found a significant reduction in the half-lives of PTENK62R but not PTENY65C or PTENK125E proteins, when compared with PTENWT (Fig. 5A and B). We then compared the endogenous PTEN levels in lymphoblastoid cells isolated from two CS patients harboring germline heterozygous PTEN ATP-binding mutations (Y68H and R130G) and from two normal WT controls. Significantly reduced PTEN levels were detected in both CS patients when compared with those in the WT controls (Fig. 5C). These results suggest that some PTEN ATP-binding mutant proteins are unstable proteins which may be degraded.
Figure 5.
PTEN ATP-binding mutations reduce protein stability. (A) CHX-chase analyses of PTEN degradation in MCF-7 cells overexpressing WT or mutant PTEN. Cells were incubated with CHX (50 µg/ml) and were harvested at the indicated time points. Whole protein lysates were harvested and ran for western blots using anti-FLAG antibody for transfectant PTEN and anti-tubulin antibody as a loading control. (B) Protein half-lives of PTENWT, PTENK62R, PTENY65C and PTENK125E. Linear plots represent the averages range of two independent experiments. (C) Western blots of whole cell lysates derived from lyphoblastoid cells isolated from two CS patients with indicated genotypes (PTENY68H/WT and PTENR130G/WT) as well as two normal WT controls. (D) Proteasomal degradation of PTENK62R. PTEN null BT-549 cells were transfected with FLAG-tagged PTENK62R and were treated with either DMSO, CHX (50 µg/ml) or a combination of CHX and MG132 (10 µm) for 16 h. Exogenously expressed PTEN protein levels were determined by western blot using an anti-FLAG antibody. (E) Model for tumorigenesis induced by PTEN ATP-binding mutations. Mutations within PTEN ATP-binding motifs lead to PTEN nuclear mislocalization and decreased p53 level through an MDM2-independent way. Impaired activities of PTEN and p53will increase the vulnerability of cells to oxidative stress, as presented by enhanced ROS, upregulated SOD1 and DNA damage. Loss of phosphatase activities in the PTEN ATP-binding mutants, together with p53 deactivation, can lead to the loss of G1/S checkpoint, as presented with abnormal cell proliferation and survival. For the mutant such as K62R, it undergoes proteasomal degradation. All these quantitative and qualitative impairments compromise PTEN's tumor-suppressor activity.
To determine whether the instability of the K62R mutant protein is through proteasomal degradation, we transfected a PTEN null breast cancer cell line, BT-549, with FLAG-tagged PTENK62R and treated with CHX for 16 h in the presence or absence of MG132. Treatment of BT-549 cells alone with 100 µg/ml CHX reduced PTENK62R protein levels to less than 20% of the untreated levels (Fig. 5D, lane 2). In the presence of the proteasome inhibitor MG132, however, PTENK62R was substantially stabilized (Fig. 5D, lane 3). We therefore conclude that the instability of PTENK62R is mediated by enhanced proteasomal degradation.
DISCUSSION
We have previously shown that PTEN ATP-binding motif mutations alter cellular proliferation, apoptosis and anchorage-dependent growth (9). Our results here demonstrate that PTEN ATP-binding mutations can lead to both qualitative and quantitative impairment of the tumor-suppressive function of PTEN. Mutations within the ATP-binding motifs not only change PTEN subcellular localization, but also lead to deregulated PTEN expression and activity.
The phosphatase activity of PTEN is important to its role in cell cycle arrest. Our current experiments show that all three ATP-binding motif mutations have abrogated phosphatase activity accompanied by unchanged cyclin D1 levels. Considering both ATP-binding motifs are located within the phosphatase domain of PTEN (residues 1–185), especially for PTENK125E, which is located within the catalytic core motif of PTEN (HCXXGXXR; residues 123–130), it is reasonable to postulate that these mutations impair PTEN's phosphatase activity and thus diminish PTEN's tumor-suppressive function (15).
The human breast carcinoma MCF-7 cell line was the main cell line used in our in vitro studies. We chose the MCF-7 cell line (which contains both WT PTEN and WT p53) for two major reasons: (i) we wanted to further investigate the interplay between nuclear PTEN and p53 in breast cancer cells and (ii) more importantly, we wanted to eventually examine mutant PTEN in a heterozygous state that mimics the in vivo situation in CS breast cancer and sporadic primary breast carcinomas (16). PTEN has been reported to upregulate p53 protein levels and transcriptional activity. In the nucleus, PTEN physically interacts with p53 through its C2 domain and forms a PTEN/p53 complex to stabilize the latter (17). Because the ATP-binding mutations are within PTEN's phosphatase domain instead of its C2 domain, it is unlikely that this effect is through the disruption of direct binding between PTEN and p53 in the nucleus.
One of the main pathways for p53 degradation is through MDM2-induced ubiquitination. In our study, however, a significant decrease in the P-MDM2 signal was observed in MCF-7 cells with ectopic expression of the PTEN ATP-binding mutants, suggesting that the downregulation of p53 in the ATP-binding mutants may be mediated via a phosphatase-independent and MDM2-independent manner. In the nucleus, PTEN can form a complex with p300 and plays a role in maintenance of high p53 acetylation, which is important in the control of p53 protein stability and DNA binding (18,19). The accumulated PTEN ATP-binding mutants in the nucleus may interfere with PTENWT forming a complex with p300 for p53 acetylation, and therefore reducing both the stability and the transcriptional activity of p53. Further detailed work will be required in the future to elucidate the precise mechanism of how PTEN ATP-binding mutations/mutants participate in the downregulation of p53 in the nucleus.
As a transcription factor, p53 regulates genes involving apoptosis, cell cycle and oxidative stress. In agreement with the reduced p53 protein levels, repressed p53 transcriptional activity was demonstrated by luciferase assay in cells overexpressing PTEN mutants. Also consistent, we found that transcript levels of a key downstream effector of p53, BAX, were significantly suppressed in the context of PTEN ATP-binding mutants. Loss of BAX transcription likely boosts proliferative and survival signals consistent with our previously published data (9). All in all, this may suggest a fine balance of nuclear PTEN and p53 and that the regulation of this balance is important for normal cell function (Fig. 5E).
The balance of nuclear to cytoplasmic PTEN is important for its tumor-suppressor function. PTEN must associate with the plasma membrane to maintain appropriate steady-state levels of phosphatidylinositol 3,4,5-triphosphate (20,21). At the same time, nuclear PTEN is a key factor for proper cell cycling and chromosomal integrity. Shen et al. (5) reported increased spontaneous DNA DSBs in Pten-deleted mouse embryonic fibroblasts. They also found that a protein from a CS patient with a PTEN R189X mutation does not bind to CENP-C, indicating that PTEN associates with CENP-C in a phosphatase-independent manner (through its C-terminal). These observations together suggest that PTEN's C-terminal domain associates with centrosome proteins to maintain chromosome integrity.
Similar to their results, in our study, we found that MCF-7 cells overexpressing PTEN ATP-binding mutations exhibited spontaneous DNA DSBs. Because all three analyzed mutations contain an intact C-terminal of PTEN, it is unlikely that the spontaneous DNA DSBs caused by these mutations are due to the loss of binding between the mutant PTEN and CENP-C.
Unlike the previous observations, in our study, the DSBs only accumulate with aging of the cells. Oxidative stress plays an important role in breast tumorigenesis through promoting the survival of mammary tumor cells and inducing DNA damages (22,23). ROS is involved in the pathogenesis of aging and cancer, and the escalated ROS generation provides an endogenous source of DNA-damaging agents that promote genetic instability (24). PTEN deletion has been reported to be a key player in ROS-induced oxidative damage of DNA (25). Notably, unlike PTENWT, PTEN-containing ATP-binding mutations are unable to inhibit ROS production, as presented by significantly higher ROS activation (Fig. 4) and consequent elevation of SOD1, suggesting that their anti-oxidative abilities are impaired.
p53 protects the genome from oxidation by ROS through the upregulation of several genes with antioxidant products, associated with a decrease in intracellular ROS (26). The downregulation of p53 results in an increase in intracellular ROS and excessive oxidation of DNA, increased mutation rate and genomic instability (27). TP53INP1 is a major player in p53-driven oxidative stress response (14). Our observation that cells overexpressing PTEN with ATP-binding mutations have decreased p53 protein levels, impaired p53 transcriptional activity and reduced TP53INP1 transcription is consistent with the hypothesis that such mutations result in loss of PTEN's protective effect against ROS-induced oxidative damage, at least as cells age, and therefore may contribute to the oxidative damage of DNA in that setting.
PTEN stability is regulated by ubiquitination, oxidation and phosphorylation (28). In our study we found PTENK62R is unstable, and we also found patients harboring germline mutation of PTENY68H and PTENR130G have decreased PTEN protein levels than the WT controls. Our experiment confirmed that PTENK62R is degraded mainly through the proteasome pathway which underlines the quantitative impairment of the ATP-binding mutations of PTEN.
Our findings therefore beg one question: do these three mutations (K62R, Y65C and K125E) and their consequences promote breast tumorigenesis? K125E, lying within ATP-binding motif B (residues 122–136), is within the core catalytic domain of PTEN (residues 124–129), and it is only one amino acid away from the important C124 residue which confers PTEN's lipid and protein phosphatase activities. We found that K125E mutant cells have the most robust proliferation (9), fastest clonogenic formation ability (9) and the lowest p53 transcriptional activity (as presented by the luciferase assay and BAX mRNA levels; Fig. 2). In contrast, K62R and Y65C, both located within the ATP-binding motif A (residues 60–73), appear to behave more similar to each other than to K125E. K125E is almost certainly exerting its effect in at least a dual manner: as a phosphatase dead (most likely) trapping mutant (dominant negative) and by nuclear mislocalization. We therefore believe that the effect of these ATP motif mutants is an end result of a balance between quantitative (dosage effect) and qualititative (e.g. mislocalization, phosphatase affected, etc.) problems.
In conclusion, we provide evidence that a mechanism for tumorigenesis of cells harboring germline or somatic mutations within PTEN ATP-binding motifs is through the quantitative and qualitative impairment of PTEN tumor-suppressor function. Moreover, our study provides novel insights on oxidative stress which may contribute to breast carcinogenesis in patients with germline or somatic PTEN mutations that result in PTEN nuclear predominance and may help explain why cells become more susceptible to cancer by accumulating DSBs due to loss of PTEN protection against ROS with age. In addition, targeting oxidative stress may help in the development of a therapeutic strategy for patients harboring such mutations or mutations with similar functional effects.
MATERIALS AND METHODS
Cell culture
Tet-off stable MCF-7 cell lines overexpressing FLAG-tagged PTEN (FLAG-PTENWT, FLAG-PTENK62R, FLAG-PTENY65C and FLAG-PTENK125E) were obtained as previously described (9). Tet-off stable MCF-7 cells were grown in DMEM supplemented with 10% fetal bovine serum and 1 μg/ml Tet. Before harvest, MCF-7 stable cells were treated with media in the absence of Tet for 24 h in order to induce overexpression of the transfected PTEN. Immortalized lymphoblastoid cells obtained from CS patients or normal controls were grown in RPMI 1640 supplemented with 10% fetal bovine serum.
Patients
Diagnosis of CS was according to the International Cowden Syndrome Consortium (29). We utilized peripheral blood samples from two CS patients with germline mutations in the ATP-binding domain of PTEN (Y68H and R130G). We also utilized peripheral blood samples from two normal WT controls. Informed consent was obtained from all subjects (CS individuals and controls) in accordance with procedures and protocols approved by the respective Human Subjects Protection Committee of each participating institution. All subjects, whether CS or controls, participated on a voluntary (unpaid) basis.
Protein isolation, SDS–PAGE and western blot analyses
For whole cell protein lysates, cells were washed twice with ice-cold PBS and were harvested in M-PER buffer (Pierce, Rockford, IL, USA) with protease inhibitors and phosphatase inhibitors. After quantitation, proteins were run on 4–15% SDS–PAGE gels and transferred to nitrocellulose. Blots were probed with primary antibodies and followed by incubation with secondary antibody and then visualized using enhanced chemiluminescence detection. The antibodies for cyclin D1, SOD1, P-AKT (Ser473), AKT, P-MDM2 (Ser166) and p53 were obtained from Cell Signaling Technology (Danvers, MA, USA). PTEN antibody was from Cascade Biosciences (Winchester, MA, USA).
Subcellular fractionation
For the nuclear and cytoplasmic fractionation, cells were harvested by trypsinization and then were incubated with buffer A (10 mm MOPS, 1.5 mm MgCl2, 10 mm KCl and 1% Triton X-100) for 15 min on ice with vortex for every 5 min. Cells were then centrifuged at 12 000g for 5 min at 4°C and the supernatant was carefully collected as the cytoplasmic fraction. The pellets were then incubated with buffer A for another 15 min and separated by centrifugation at 12 000g for 5 min at 4°C. The pellets were dissolved in RIPA buffer. The purity of each fraction was analyzed by immunoblotting using antibodies against α-tubulin (Sigma Aldrich, St Louis, MO, USA) for the cytoplasm and PARP-1 (Santa Cruz, CA, USA) for the nucleus.
Indirect immunofluorescence and confocal microscopy
Cells were seeded in six-well plates with cover slips. Cells were fixed in a freshly prepared 100% methanol for 1 min at −20°C. After 5 min incubation with 0.3% Triton X-100 in PBS, the cover slips were washed three times in PBS, blocked in PBS with 10% goat serum for 30 min, incubated with primary antibodies for 1 h, washed three times in PBS and finally incubated with Alexa Fluor® dye-labeled secondary antibodies for 1 h at the concentration of 1:1000 (Invitrogen, Carlsbad, CA, USA). The immunofluorescence antibodies were purchased and used as follows: FLAG M2 monoclonal antibody from Sigma Aldrich (Cat# F1804, 1:300 dilution), p53 rabbit antibody from Cell Signaling (Cat#9282, 1:50 dilution), γ-H2AX (Cat# 05–636; Upstate Biotechnology, Charlottesville, VA, USA, 1:800 dilution). Cells were mounted on glass slides with ProLong® Gold antifade reagent with DAPI (Invitrogen) and visualized on a Leica TCS-SP spectral laser scanning confocal microscope as described previously (9). For quantitation of the immunostaining intensity of p53 in MCF-7 cells, regions of interest were defined so that they would fit in all nuclei, and the intensity was measured for all the individual regions of interest, using the Leica confocal image analysis software. At least 100 cells, from at least two different slides, were used for quantitation.
p53 luciferase assay
The PG13-luc plasmid, which contains a p53 reporter with a firefly luciferase gene under the control of 13 p53 response elements, was a kind gift from Dr Bert Vogelstein (30) (Addgene plasmid 16442). MCF-7 stable cells (Vector, WT, K62R, Y65C and K125E) were plated at 0.5 × 106 cells/60 mm dish and grew overnight before transfection. PG13-luc plasmid was transfected for 48 h followed by the Renilla luciferase assays according to the manufacturer's instruction (Promega, Cat#E2810). Luciferase activities were normalized to Renilla control activities.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from HEK293 transiently transfected or MCF7 stably transfected cells using RNeasy® Mini Kit (QIAGEN, Inc., Valencia, CA, USA) according to the manufacturer's protocol, and subsequently treated with DNase I (Invitrogen). DNase-treated total RNA was reverse-transcribed into cDNA using random primers and SuperTranscript™ III Reverse Transcriptase (Invitrogen), as specified by the manufacturer. Polymerase chain reaction was performed using SYBR Green PCR Master Mix and run on 7500 Real Time PCR machine (Applied Biosystem, Carlsbad, CA). Primer sequences are listed in Table S1, Supplementary Material. The relative mRNA amounts were determined by normalizing each set of samples using the difference in threshold cycles (ΔCT) between interested genes and GAPDH as reference gene (ΔCTSamples = ΔCTinterested − ΔCTGAPDH).
Fluorescence analysis of ROS
The measurement of ROS in live cells was performed using the image-iTTM LIVE Green ROS Detection Kit according to the manufacturer's instructions (Cat# I36007; Molecular Probes, Invitrogen). This assay is based on the finding that oxidation of the non-fluorescent carboxy-dichlorodihydrofluorescein diacetate (Carboxy-H2DCFDA) by ROS can convert it to the fluorescent dichlorofluorescein (DCF) (31). Briefly, MCF-7 stable cells were incubated with 25 μm Carboxy-H2DCFDA for 30 min at 37°C, and the fluorescence intensity of DCF is proportional to the ROS activity. Nuclei were counterstained with Hoechst 33342 and the percentage of ROS-positive cells/total cells of each cell line was compared.
Statistical analysis
The immunostaining intensity of p53 was compared by one-way ANOVA. P < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported, in part, by R01CA118980 from the National Cancer Institute, Bethesda, MD (to C.E.). C.E. was a recipient of the Doris Duke Distinguished Clinical Scientist Award, is an American Cancer Society Clinical Research Professor, generously funded, in part, by the F.M. Kirby Foundation and is the Sondra J. and Stephen R. Hardis Endowed Chair of Cancer Genomic Medicine at the Clinical Clinic. Funding to pay the Open Access publication charges for this article was provided by the National Cancer Institute (R01CA118980).
Supplementary Material
ACKNOWLEDGEMENTS
X.H. would like to thank Drs Judith A. Drazba (Confocal and Imaging Core, Lerner Research Institute, Cleveland Clinic), Avi Jacob and Glenn Lobo (both of the Eng Lab) for helpful technical discussions during the very early stages of these experiments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Mahoney M.C., Bevers T., Linos E., Willett W.C. Opportunities and strategies for breast cancer prevention through risk reduction. CA Cancer J. Clin. 2008;58:347–371. doi: 10.3322/CA.2008.0016. [DOI] [PubMed] [Google Scholar]
- 2.Nelen M.R., Padberg G.W., Peeters E.A., Lin A.Y., van den Helm B., Frants R.R., Coulon V., Goldstein A.M., van Reen M.M., Easton D.F., et al. Localization of the gene for Cowden disease to chromosome 10q22–23. Nat. Genet. 1996;13:114–116. doi: 10.1038/ng0596-114. doi:10.1016/S0168-583X(01)00748-0. [DOI] [PubMed] [Google Scholar]
- 3.Marsh D.J., Dahia P.L., Zheng Z., Liaw D., Parsons R., Gorlin R.J., Eng C. Germline mutations in PTEN are present in Bannayan–Zonana syndrome. Nat. Genet. 1997;16:333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 4.Zbuk K.M., Eng C. Cancer phenomics: RET and PTEN as illustrative models. Nat. Rev. Cancer. 2007;7:35–45. doi: 10.1038/nrc2037. [DOI] [PubMed] [Google Scholar]
- 5.Shen W.H., Balajee A.S., Wang J., Wu H., Eng C., Pandolfi P.P., Yin Y. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. doi:10.1016/j.radmeas.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Saal L.H., Gruvberger-Saal S.K., Persson C., Lovgren K., Jumppanen M., Staaf J., Jonsson G., Pires M.M., Maurer M., Holm K., et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat. Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. doi:10.1016/S1350-4487(96)00085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung J.-H., Ostrowski M.C., Romigh T., Minaguchi T., Waite K.A., Eng C. The ERK1/2 pathway modulates nuclear PTEN-mediated cell cycle arrest by cyclin D1 transcriptional regulation Hum. Mol. Genet. 2006;15:2553–2559. doi: 10.1093/hmg/ddl177. doi:10.1016/1350-4487(94)00084-E. [DOI] [PubMed] [Google Scholar]
- 8.Trotman L.C., Wang X., Alimonti A., Chen Z., Teruya-Feldstein J., Yang H., Pavletich N.P., Carver B.S., Cordon-Cardo C., Erdjument-Bromage H., et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lobo G.P., Waite K.A., Planchon S.M., Romigh T., Nassif N.T., Eng C. Germline and somatic cancer-associated mutations in the ATP-binding motifs of PTEN influence its subcellular localization and tumor suppressive function. Hum. Mol. Genet. 2009;18:2851–2862. doi: 10.1093/hmg/ddp220. doi:10.1097/00004032-197108000-00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lobo G.P., Waite K.A., Planchon S.M., Romigh T., Houghton J.A., Eng C. ATP modulates PTEN subcellular localization in multiple cancer cell lines. Hum. Mol. Genet. 2008;17:2877–2885. doi: 10.1093/hmg/ddn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng L.-P., Smith W.M., Dahia P.L.M., Ziebold U., Gil E., Lees J.A., Eng C. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer Res. 1999;59:5808–5814. [PubMed] [Google Scholar]
- 12.Tang Y., Eng C. PTEN autoregulates its expression by stabilization of p53 in a phosphatase-independent manner. Cancer Res. 2006;66:736–742. doi: 10.1158/0008-5472.CAN-05-1557. doi:10.1016/j.radmeas.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Foster E.R., Downs J.A. Histone H2A phosphorylation in DNA double-strand break repair. FEBS J. 2005;272:3231–3240. doi: 10.1111/j.1742-4658.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- 14.Cano C.E., Gommeaux J., Pietri S., Culcasi M., Garcia S., Seux M., Barelier S., Vasseur S., Spoto R.P., Pebusque M.J., et al. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res. 2009;69:219–226. doi: 10.1158/0008-5472.CAN-08-2320. doi:10.1016/j.radmeas.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 15.Lee J.O., Yang H., Georgescu M.M., Di Cristofano A., Maehama T., Shi Y., Dixon J.E., Pandolfi P., Pavletich N.P. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. doi:10.1016/j.radmeas.2009.10.089. [DOI] [PubMed] [Google Scholar]
- 16.Marsh D.J., Dahia P.L., Coulon V., Zheng Z., Dorion-Bonnet F., Call K.M., Little R., Lin A.Y., Eeles R.A., Goldstein A.M., et al. Allelic imbalance, including deletion of PTEN/MMACI, at the Cowden disease locus on 10q22–23, in hamartomas from patients with Cowden syndrome and germline PTEN mutation. Gene Chromosomes Cancer. 1998;21:61–69. doi: 10.1002/(sici)1098-2264(199801)21:1<61::aid-gcc8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 17.Freeman D.J., Li A.G., Wei G., Li H.H., Kertesz N., Lesche R., Whale A.D., Martinez-Diaz H., Rozengurt N., Cardiff R.D., et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell. 2003;3:117–130. doi: 10.1016/s1535-6108(03)00021-7. doi:10.1016/j.radmeas.2009.10.075. [DOI] [PubMed] [Google Scholar]
- 18.Li A.G., Piluso L.G., Cai X., Wei G., Sellers W.R., Liu X. Mechanistic insights into maintenance of high p53 acetylation by PTEN. Mol. Cell. 2006;23:575–587. doi: 10.1016/j.molcel.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Li M., Luo J., Brooks C.L., Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. doi:10.1016/j.radmeas.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Rahdar M., Inoue T., Meyer T., Zhang J., Vazquez F., Devreotes P.N. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc. Natl Acad. Sci. USA. 2009;106:480–485. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker S.M., Leslie N.R., Perera N.M., Batty I.H., Downes C.P. The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. Biochem. J. 2004;379:301–307. doi: 10.1042/BJ20031839. doi:10.1016/j.asr.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtis C.D., Thorngren D.L., Nardulli A.M. Immunohistochemical analysis of oxidative stress and DNA repair proteins in normal mammary and breast cancer tissues. BMC Cancer. 2010;10:9. doi: 10.1186/1471-2407-10-9. doi:10.1093/rpd/ncp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang D.H. Oxidative stress, DNA damage, and breast cancer. AACN Clin. Issues. 2002;13:540–549. doi: 10.1097/00044067-200211000-00007. [DOI] [PubMed] [Google Scholar]
- 24.Schumacker P.T. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Cancer Cell. 2006;10:175–176. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 25.Huo Y.Y., Li G., Duan R.F., Gou Q., Fu C.L., Hu Y.C., Song B.Q., Yang Z.H., Wu D.C., Zhou P.K. PTEN deletion leads to deregulation of antioxidants and increased oxidative damage in mouse embryonic fibroblasts. Free Radic. Biol. Med. 2008;44:1578–1591. doi: 10.1016/j.freeradbiomed.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Sablina A.A., Budanov A.V., Ilyinskaya G.V., Agapova L.S., Kravchenko J.E., Chumakov P.M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pouyet L., Carrier A. Mutant mouse models of oxidative stress. Transgenic Res. 2010;19:155–164. doi: 10.1007/s11248-009-9308-6. [DOI] [PubMed] [Google Scholar]
- 28.Leslie N.R., Batty I.H., Maccario H., Davidson L., Downes C.P. Understanding PTEN regulation: PIP2, polarity and protein stability. Oncogene. 2008;27:5464–5476. doi: 10.1038/onc.2008.243. [DOI] [PubMed] [Google Scholar]
- 29.Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M., Eng C., Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 30.el-Deiry W.S., Tokino T., Velculescu V.E., Levy D.B., Parsons R., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 31.Wardman P. Use of the dichlorofluorescein assay to measure ‘reactive oxygen species. Radiat. Res. 2008;170:406–407. doi: 10.1667/RR1439a.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.