Abstract
Deficiency of thymidine kinase 2 (TK2) is a frequent cause of isolated myopathy or encephalomyopathy in children with mitochondrial DNA (mtDNA) depletion. To determine the bases of disease onset, organ specificity and severity of TK2 deficiency, we have carefully characterized Tk2 H126N knockin mice (Tk2−/−). Although normal until postnatal day 8, Tk2−/− mice rapidly develop fatal encephalomyopathy between postnatal days 10 and 13. We have observed that wild-type Tk2 activity is constant in the second week of life, while Tk1 activity decreases significantly between postnatal days 8 and 13. The down-regulation of Tk1 activity unmasks Tk2 deficiency in Tk2−/− mice and correlates with the onset of mtDNA depletion in the brain and the heart. Resistance to pathology in Tk2 mutant organs depends on compensatory mechanisms to the reduced mtDNA level. Our analyses at postnatal day 13 have revealed that Tk2−/− heart significantly increases mitochondrial transcript levels relative to the mtDNA content. This transcriptional compensation allows the heart to maintain normal levels of mtDNA-encoded proteins. The up-regulation in mitochondrial transcripts is not due to increased expression of the master mitochondrial biogenesis regulators peroxisome proliferator-activated receptor-gamma coactivator 1 alpha and nuclear respiratory factors 1 and 2, or to enhanced expression of the mitochondrial transcription factors A, B1 or B2. Instead, Tk2−/− heart compensates for mtDNA depletion by down-regulating the expression of the mitochondrial transcriptional terminator transcription factor 3 (MTERF3). Understanding the molecular mechanisms that allow Tk2 mutant organs to be spared may help design therapies for Tk2 deficiency.
INTRODUCTION
Thymidine kinase 2 (TK2) is a constitutively expressed mitochondrial enzyme that phosphorylates deoxythymidine (dT), deoxycytidine (dC) and deoxyuridine (dU) to their corresponding monophosphates (1–3). This is the first and rate-limiting step for the salvage pathway synthesis of deoxypyrimidine nucleoside triphosphates required for mitochondrial DNA (mtDNA) replication and maintenance in post-mitotic cells. In contrast, TK2 activity has minor functional significance in proliferating cells due to the overwhelming activity of cell-cycle-regulated cytosolic enzymes involved in the deoxynucleoside triphosphate (dNTP) synthesis, including thymidylate synthase (TS) and ribonuclotide reductase (R1–R2) in de novo synthesis, or thymidine kinase 1 (TK1) and deoxycytidine kinase (dCK) in the salvage pathways (4–6). Strict homeostatic maintenance of the mitochondrial deoxythymidine monophosphate (dTMP) and deoxycytidine monophosphate (dCMP) levels is achieved by counteracting TK2 activity with the catabolic actions of cytosolic thymidine phosphorylase (TP) that decreases the levels of TK2 substrates and of mitochondrial deoxynucleotidase 2 that dephosphorylates dTMP to dT (7,8).
TK2 activity became clinically relevant when autosomal recessive TK2 gene mutations were identified as a frequent cause of severe mtDNA depletion syndrome (MDS) in humans (9). Although there is variability in age at onset and survival, MDS due to TK2 mutations (TK2 MDS) typically begins in the first 2 years of life and is fatal in childhood. Cases reported in the past few years have expanded the phenotype of Tk2 MDS from pure myopathy to myopathy plus rigid spine or central nervous system involvement, including hearing loss, spinal-muscular atrophy-like cases and severe encephalomyopathy (10–14). It is not known why some organs are spared from the pathogenic effects of the ubiquitously mutated TK2 enzyme. Transcriptional compensatory mechanisms have been proposed in a TK2-deficient patient with high levels of MT-CO2 mRNA and MT-RNR1 rRNA in severely mtDNA-depleted muscle fibers (15). Alternative compensatory mechanisms to preserve normal levels of mtDNA in selected myofibers were hypothesized in one patient with severe TK2 myopathy with abnormally slow progression and long survival (16,17), and in TK2 deficient fibroblasts without mtDNA depletion (18).
We have described a homozygous H126N Tk2 mutant knockin mouse (Tk2−/−) model of human TK2 MDS (19). Tk2−/− mice are normal at birth and show normal early growth. Around postnatal day 10, the mutant animals stop gaining weight, after which the disease progresses rapidly with reduced spontaneous and locomotor activity, tremor, ataxic gait, muscle weakness and severe encephalomyopathy, so that 75% of the Tk2−/− mice die by postnatal day 14. Tk2−/−mice show mtDNA depletion in multiple tissues, including the brain, heart and muscle. However, only the brain showed significant deficiency of respiratory chain complexes (OXPHOS), protein levels and activities (the activity of complex I was 40% and that of complex IV 50% in brain of Tk2−/− versus Tk2+/+ mice). We postulated that the heart and muscle were spared by compensatory mechanisms.
In this work, we have used the homozygous H126N Tk2 mutant knockin mouse (Tk2−/−), to characterize the activities of the Tk1 and Tk2 enzymes in relation to disease onset, phenotypic severity and organ involvement. For this purpose, we have compared the biochemical and molecular genetic features of two organs with high energy-demands, the brain, which is affected, and the heart, which is spared in our Tk2−/− mouse model. We have focused our study in two time points, postnatal days 8 and 13, to span the transition from pre-symptomatic to symptomatic states. Our results demonstrate biochemical factors that contribute to the brain pathology and in vivo transcriptional compensatory mechanisms in Tk2−/− heart.
RESULTS
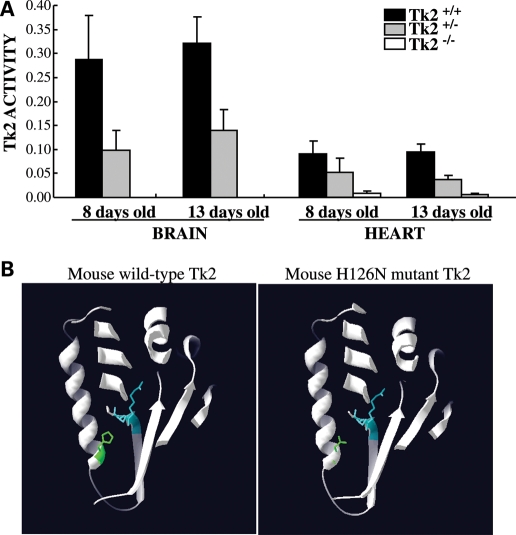
Using the Tk2-specific radiolabeled substrate 3H-bromovinyl deoxyuridine (3H-BVDU) (20,21), we measured Tk2 activity in the heart and the brain from healthy (TK2+/+and Tk2+/−) mice and from Tk2−/− mice at the onset of disease manifestations. The results (Fig. 1A) showed no significant increase in Tk2 activity between postnatal days 8 and 13. As expected, Tk2 activities were higher in organs from Tk2+/+than Tk2+/− mice; and 2- to 3-fold higher in the brain than in the heart, in agreement with published data (19,22). Remarkably, Tk2−/− brain showed no detectable Tk2 activity and Tk2−/− heart showed less than 10% of the wild-type metabolism rate (probably due to crossover activity of abundant Tk1 in this organ). These findings indicated that our Tk2−/− knockin mouse is comparable with a Tk2 knockout mouse (23). Our structural model of mouse Tk2 (Fig. 1B) localizes histidine 126 at the C-terminal end of the α helix (α4) that faces the “phosphate transferring” ERS (E138-R139-S140) domain. Therefore, the substitution of a charged histidine by a neutral asparagine (H126N) could alter the positioning of the critical residue R139 and hinder the transfer of γ-phosphate from the ATP-binding pocket to the nucleoside-binding pocket, thus impairing Tk2 function.
Figure 1.
(A) Tk2 activity does not change in the brain or heart of healthy mice (Tk2+/+, Tk2+/−) between postnatal days 8 and 13. Tk2 activity is expressed in pmol/min/mg protein as mean ± SD of n ≥ 5 at each day for each group of mice. (B) Computer-generated model of mouse Tk2 structure. In green, wild-type histidine 126 (left panel) and mutant asparagine 126 (right panel). In blue, the phosphate-transferring domain ERS (E138-R139-S140).
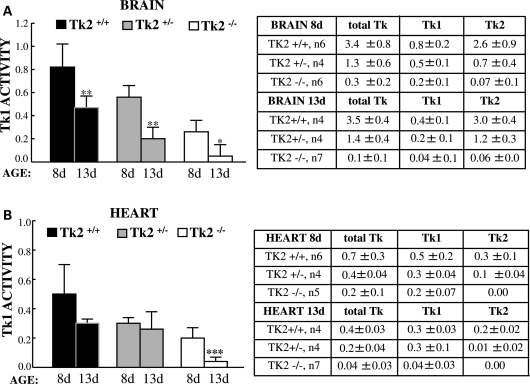
As Tk2 activity did not increase between days 8 and 13, we investigated whether the onset of disease in Tk2−/− mice was due to a down-regulation of Tk1 activity during this time period. We performed 3H-thymidine assays to measure total Tk (Tk1 + Tk2) and Tk1 activity (determined using excess cold dCyt as a specific competitor of Tk2). The results showed significant reductions of Tk1 activity in the brain at postnatal day 13 compared with postnatal day 8 in all three mouse lines: Tk2+/+(58 ± 7%), Tk2+/− (41 ± 14%) and Tk2−/− (23 ± 29%) (Fig. 2A). Similarly, in the heart, Tk2−/− mice showed a significant reduction in Tk1 activity between postnatal days 8 and 13 (29 ± 22%), whereas Tk2+/+and Tk2+/− mice showed trends toward lower Tk1 activity over this time period (Fig. 2B). We confirmed that Tk2 was the main Tk in the brain as the total 3H-thymidine phosphorylation was inhibited by more than 75% with cold dCyt (Table in Fig. 2A). Conversely, in the heart, Tk1 was the main Tk component, as total Tk activity was reduced by less than 50% with cold dCyt (Table in Fig. 2B). On the other hand, there was evidence of cross-regulation between Tk1 and Tk2 enzymes in the brain and the heart, because decreases in Tk2 activity were associated with secondary reductions of Tk1 activity (i.e. Tk1 activities were highest in Tk2+/+mice, intermediate in Tk2+/− animals and lowest in Tk2−/− mice) (Fig. 2).
Figure 2.
Tk1 activity is down-regulated in mice brain and heart between postnatal days 8 and 13. Tk1 activity is expressed in pmol/min/mg as mean ± SD of n (indicated in the tables) *P < 0.05, **P < 0.01 and ***P < 0.001 Tk2−/− versus Tk2+/+. Representative of the experiments performed.
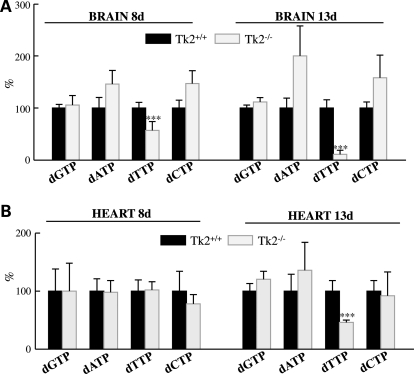
To determine whether down-regulation of Tk1 activity between days 8 and 13 correlated with the onset of imbalanced dNTPs’ pool in Tk2−/− mice heart and brain, we measured total dNTPs by an optimized method that excluded interference from ribonucleotides (24). Already at postnatal day 8, the dTTP levels were markedly decreased in the brain of Tk2−/− and decreased even further at Day 13 (dTTP/dNTP levels in Tk2−/− relative to Tk2+/+brains 57 ± 17% at Day 8 and 11 ± 8% at Day 13) (Fig. 3). The heart of Tk2−/− mice did not show dNTPs imbalance at Day 8 but revealed clearly reduced dTTP/dNTP levels at Day 13 (46 ± 4% relative to dTTP/dNTP in Tk2+/+). Therefore, the down-regulation of Tk1 activity levels unmasks Tk2 deficiency in Tk2−/− organs and triggers reductions in dTTP levels.
Figure 3.
The brain and heart of Tk2−/− mice have reduced dTTP levels at postnatal day 13 but only Tk2−/− brain has dNTPs imbalance at Day 8. Measurement of total dNTPs expressed in % of each nucleotide/dNTPs relative to Tk2+/+ (100%) as mean ± SD of n = 5 for each group of mice. ***P < 0.001 Tk2−/− versus Tk2+/+.
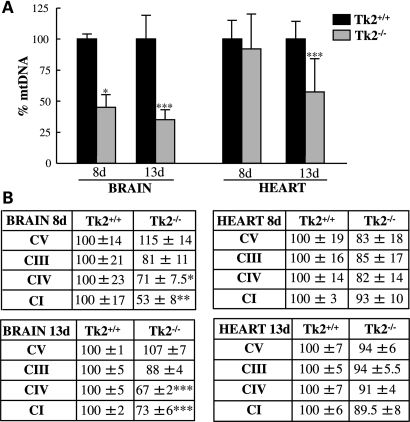
The temporal relationship between Tk1 down-regulation and development of dTTP pool imbalance prompted us to assess if there were also parallel decreases in mtDNA levels in the Tk2−/− mice. Quantitative PCR (Fig. 4A) of the brain showed that the significant mtDNA depletion was already present at Day 8 (mtDNA level 45 ± 10% in Tk2−/− versus Tk2+/+ litter-mates) and became more severe at Day 13 (35 ± 8% residual mtDNA in Tk2−/− mice). In contrast, mtDNA levels in Tk2−/− heart were normal at Day 8, but depleted at postnatal Day 13 (mtDNA 58 ± 27% in Tk2−/− versus Tk2+/+ heart).
Figure 4.
The brain and heart of Tk2−/− mice have mtDNA depletion but only the brain has mt-encoded proteins deficiency. (A) Quantitative real-time PCR using COXI as a probe for mtDNA and GAPDH as nuclear control. (B) Quantification of western blot performed with OXPHOS cocktail of antibodies against the following subunits of the respiratory chain enzyme complexes: subunit α (complex V), core2 (complex III), COXI (complex IV) and ND6 (complex I). In (A) and (B), results are the mean percent relative to Tk2+/+ (100%) ± SD of n = 6 per group per day (n = 15 for mtDNA in 13-day heart). *P < 0.05, **P < 0.01 and ***P < 0.001 Tk2−/− versus Tk2+/+.
Next, we analyzed whether the reduction in the amount of mtDNA affected levels of mtDNA-encoded proteins (mtDNA proteins). We performed western blot with a cocktail of antibodies against subunits of the respiratory chain enzyme complexes. The quantification of bands intensity relative to the nuclear-encoded complex II as a control is shown in Fig. 4B. Compared with Tk2+/+ brain at postnatal day 8, age-matched Tk2−/− brain showed significant reductions of the mtDNA-encoded proteins cytochrome c oxidase I (COXI, complex IV subunit 1) (71 ± 7.5%) and subunit 6 of NADH dehydrogenase (ND6, complex I subunit 6) (53 ± 8%). Consistently, Tk2−/− brain showed reduction of both mtDNA-encoded subunits of the respiratory chain at postnatal day 13; COXI was 67 ± 2% and ND6 was 73 ± 6% relative to Tk2+/+ litter-mates (we had previously reported OXPHOS subunit levels in brain at Day 13) (19). Nevertheless, at this time point, the ratio of the mtDNA protein/mtDNA level in the brain was ∼1.5, indicating that the reduction in mtDNA-encoded protein expression did not correlate exactly with the reduction in the mtDNA amount. In contrast with the brain, in Tk2−/− heart, the levels of respiratory chain proteins were normal at postnatal day 8 (consistent with the normal amount of mtDNA in this organ at this age) but strikingly, they were also normal at postnatal day 13, despite the marked mtDNA depletion. In fact, Tk2−/− heart, like brain, showed ∼1.5-fold more protein relative to mtDNA at age 13 days. These results indicate that, in Tk2−/− mice, both the brain and the heart manifest compensatory mechanisms for mtDNA depletion.
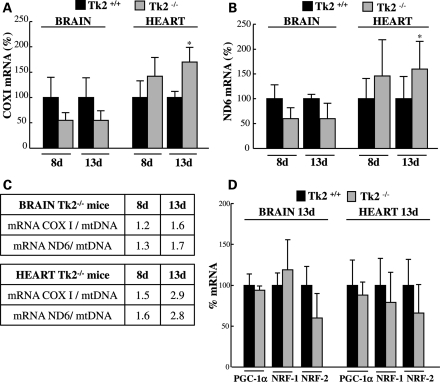
To investigate whether compensation for mtDNA depletion may occur through transcriptional up-regulation, we used real-time quantitative reverse transcriptase–PCR (qRT–PCR) to measure transcripts for COXI and ND6 (transcribed from the heavy and the light strand of mtDNA, respectively) at presymptomatic Day 8 and at symptomatic Day 13. The amounts of COXI mRNA in Tk2−/− brain (Fig. 5A) was clearly reduced both at Day 8 (55 ± 15%) and at Day 13 (55 ± 19%) compared with the levels in corresponding Tk2+/+ litter-mates. Similarly, ND6 transcripts (Fig. 5B) were reduced to 60 ± 22% at Day 8 and 60 ± 31% at Day 13 in brain of Tk2−/− mice. The reductions of mRNA in Tk2−/− brain coincided temporally with the depletion of mtDNA and accounted for the deficiency in COXI and ND6 protein levels. However, amounts of mtDNA-encoded transcripts were disproportionately less severely reduced than the levels of mtDNA in Tk2−/− brain, indicating up-regulation of mtDNA-encoded mRNA. This increase in mtDNA transcripts was most evident at postnatal Day 13 for ND6, which showed 1.7-fold more mRNA in Tk2−/− mice compared with Tk2+/+ litter-mates when normalized to mtDNA (Fig. 5C). At age 8 days, Tk2−/− heart showed normal levels of mtDNA and transcripts for COXI and ND6; however, at postnatal day 13, Tk2−/− heart had increased levels of transcripts for COXI (170 ± 29%) and ND6 (160 ± 50%) relative to Tk2+/+ litter-mates. Furthermore, when normalized to mtDNA levels (Fig. 5C), COXI and ND6 transcripts were ∼3-fold higher in Tk2−/− hearts than in Tk2+/+ hearts at age 13 days. Thus, mtDNA transcriptional up-regulation in the heart of 13-day-old Tk2−/− mice compensates for mtDNA depletion and prevents deficits of the COXI and ND6 proteins.
Figure 5.
Up-regulation of mt-encoded COXI and ND6 transcripts in Tk2−/− heart at symptomatic Day 13. This up-regulation is not associated with increased mRNA expression of PGC-1α or NRF-1/2. Quantification of mRNA levels by quantitative RT–PCR for the target genes COXI (A), ND6 (B) or PGC-1α, NRF-1 and NRF-2 (D), normalized to values of GAPDH gene as control. Results are expressed as mean percents of arbitrary units relative to Tk2+/+ (100%) ± SD of n ≥ 5 at each day for each group of mice. In (A) and (B), *P < 0.05 Tk2−/− versus Tk2+/+. In (C), mRNA/mtDNA in Tk2−/− mice tissues indicates the ratio of the percent of transcript levels [relative to Tk2+/+ (100%)] and the percent of mtDNA [relative to Tk2+/+ (100%)] in each tissue at each time point.
To determine which factor(s) may be responsible for the increased mitochondrial mRNA levels, we studied the expression of master-regulators of mitochondrial gene expression, including the following: peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1α), nuclear respiratory factor 1 (NRF-1) and nuclear respiratory factor 2 (NRF-2) (25). Quantitative RT–PCR (Fig. 5C) indicated that, at age 13 days, Tk2−/− and Tk2+/+ mice did not manifest significant differences in the expression of these genes in the brain and the heart.
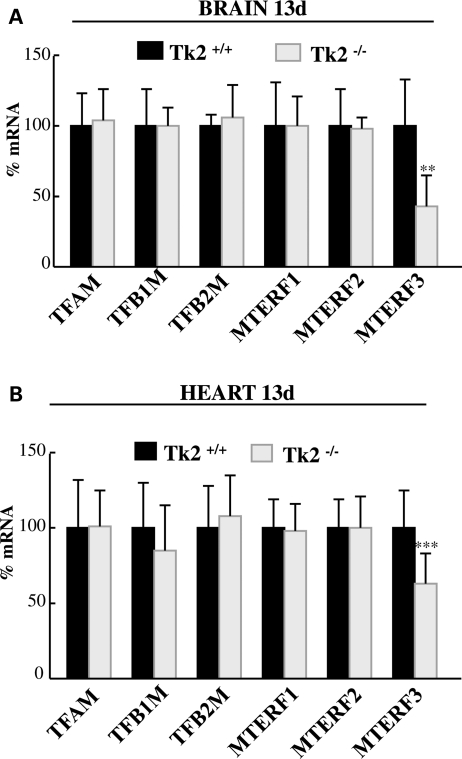
We then analyzed downstream factors known to be directly involved in mitochondrial transcription, including mitochondrial transcriptional activator A (TFAM), mitochondrial transcription factor B1 (TFB1M) and mitochondrial transcription factor B2 (TFB2M), as well as the mitochondrial transcriptional terminator factors 1, 2 and 3 (MTERF 1, 2 and 3) (26–29). Quantitative RT–PCR (Fig. 6) demonstrated that mRNA levels of the transcriptional repressor MTERF3 were significantly reduced in the brain (43 ± 22%, Fig. 6A) and the heart (63 ± 21%, Fig. 6B) of Tk2−/− mice at Day 13 when compared with MTERF3 transcript expression in Tk2+/+ litter-mates. This result explained how Tk2−/− heart and brain were able to produce increased amounts of mtDNA-encoded transcripts relative to the amount of mtDNA at postnatal day 13.
Figure 6.
The expression of the transcriptional repressor MTERF3 is down-regulated in the brain and heart of Tk2−/− mice. Quantification of mRNA levels of the target genes by quantitative RT–PCR, normalized to values of GAPDH gene as control. Results are mean percents relative to Tk2+/+(100%) ± SD of n = 6 for each factor in each group of mice (for MTERF3 in heart n = 19 per mice group). **P < 0.01 and ***P < 0.001 Tk2−/− versus Tk2+/+.
DISCUSSION
In our previous study of Tk2 H126N knockin mice, we identified mtDNA depletion in most tissues with the brain and heart among the most severely affected (19). Curiously, brain was the only organ that also showed defects of respiratory chain complex proteins and activities. To better understand which factors are responsible for disease onset, survival and for the encephalopathic phenotype of Tk2−/− mice, we have compared biochemical and molecular features of one affected tissue, the brain, and one biochemically unaffected organ, the heart, before and after the appearance of physical signs of disease.
During the first 2 weeks of life, mice undergo critical growth and maturation, which involves the arrest of cell proliferation and differentiation in all organs. During this transition from cell division to quiescence, Tk1 activity declines sharply from high activity in the S-phase to inactivity, via proteolytic degradation, immediately after mitosis (4,6). In contrast, Tk2 expression is not cell-cycle dependent, but its activity becomes functionally relevant after the cell-cycle arrests and the cytosolic machinery that provides dNTPs is down-regulated (with the exception of the de novo synthesis subunit, ribonucleotide reductase R1-p53R2, which is cytosolic and remains active in post-mitotic cells) (30). Thus, the period of pyrimidine salvage transition from the predominantly cytosolic Tk1 to the mitochondrial Tk2 determines when cells in different organs become susceptible to Tk2 deficiency. Using 3H-Thy incorporation into DNA to quantify cell division, developmental studies from the 1970s had demonstrated the timing of arrest of cell proliferation in rodents’ brain and heart. The brain revealed intense cell proliferation until postnatal day 6, followed by a rapid decline to almost undetectable levels at postnatal day 10 (31,32). Paralleling changes in Thy uptake, Tk activity was maximal in the brain at postnatal day 6, before declining to undetectable values during the second week of life (33). In contrast to the brain, murine cardiomyocytes proliferate mainly in the fetal period, although cell proliferation is maintained in newborn hearts until postnatal day 10, coinciding with the plateau of cardiomyocyte binucleation (34).
Our data extend the reported 3H-Thy incorporation findings by documenting that Tk2 has a stable activity in the brain and heart through the second week of murine life (Fig. 1). Tk2 activity reaches critical importance at this time due to the progressive down-regulation of Tk1 activity (Fig. 2). According to rodent brain developmental studies, overall Tk activity starts to decline at postnatal day 6. This explains why Tk2−/− brains from 8-day-old mice already show imbalanced dNTPs’ pool and mtDNA depletion (Figs 3 and 4). In fact, some parts of the brain could be post-mitotic at birth, accounting for the moderate mtDNA depletion shown in Tk2−/− newborn mice [our unpublished data and (23)]. In contrast, arrest of cell proliferation occurs later in the heart (between postnatal days 10 and 13), explaining the absence of defects in the hearts of 8-day-old Tk2−/− mice (Figs 3 and 4), and the appearance of mild cardiac mtDNA depletion at postnatal day 13. Therefore, the onset of disease in Tk2−/− mice coincides with the time of cell proliferation arrest and Tk1 activity down-regulation in vital organs. By postnatal day 13, the lowered Tk1 activity is no longer able to mask the Tk2 deficiency in the brain and the heart of Tk2−/− mice. In support of this notion, peripheral blood cells, which include a high proportion of proliferating cells, do not develop mtDNA depletion in Tk2 knockout mice (35).
Human patients with TK2 deficiency manifest myopathy or encephalomyopathy of varying severity (10,12–14). It has been proposed that the basal low activity of TK2 in muscle accounts for the vulnerability of this tissue (36). We further hypothesize that the residual activity of the mutant Tk2 protein also determines the phenotype. With the exception of low TK2 activity in muscle (9), little is known about the activity of TK2 mutant proteins in affected tissues from patients, because most of the measurements has been performed in patients’ fibroblasts, or in recombinant proteins harboring mutations identified in patients (13,37,38). However, based on those limited data, the phenotype in patients appears to correlate with the severity of TK2 deficiency; partial reductions of TK2 activity (14–45% of normal) produce myopathy, whereas severe reductions (less than 10% of normal) cause encephalomyopathy. In mice, basal Tk2 activity is low in muscle and high in brain (19,22). In humans, TK2 activity is also low in muscle and presumably higher in brain. Thus, brain can tolerate mild TK2 deficiency (40% residual activity) but muscle cannot. In contrast, severe TK2 deficiency (90% loss of TK2 activity) affects brain and muscle.
Accordingly, mice with partial Tk2 activity would be expected to manifest myopathy while severe or complete Tk2 deficiency would be predicted to develop encephalomyopathy. In fact, our Tk2 knockin mouse, like the Tk2 knockout mouse, is an excellent model of the human TK2 deficient infantile fatal encephalomyopathy (19,39). The murine homozygous H126N Tk2 mutation has negligible Tk2 activity in Tk2−/− brain (<10%, Fig. 1) and produces clear central nervous system pathology, and death within the first 3 weeks of life (19), similar to Tk2 knockout mice (39). The lack of activity of the H126N Tk2 mutant enzyme could be due to impaired protein dimerization, defective nucleoside binding or both, as proposed for the homologous human H121N mutation (10,13). An alternative explanation offered by our structure model for mouse Tk2 (Fig. 1B) is that the H126N substitution could alter the positioning of the critical residue R139 and hinder the transfer of γ-phosphate from the ATP-binding pocket to the nucleoside-binding pocket (Fig. 1B). Still, we cannot exclude that the H126N mutation may also cause instability and premature degradation of Tk2 protein, because the anti-murine Tk2 antibodies that we have tested have failed to recognize wild-type Tk2 in tissue extracts.
Clearly both human disease and the mouse model indicate that TK2 deficiency causes dNTP pool imbalances in mitochondria leading to mtDNA depletion. Although cytosolic and mitochondrial dNTP pools are physically separated by mitochondrial membranes, exchange of deoxynucleotides between the two cellular compartments is rapid and constant (40,41). Therefore, measurements of total cellular dNTP pools reflect mitochondrial dNTP levels (7,40–42). Taking these observations into account, we circumvented the technical challenge of measuring dNTPs in isolated mitochondria from 13-day-old mice heart (≤30 mg tissue weight) by determining dNTP pools in total cellular extracts. The correctness of this assumption was fully supported by our observation that dTTP levels in total cellular extracts were reduced in Tk2-deficient tissues (Fig. 3). Although Tk2 phosphorylates deoxycytidine in addition to thymidine, in contrast to the expected decrease of dTTP, we found normal levels of dCTP in the brain and heart of Tk2−/− mice. Why dTTP and dCTP levels should diverge in Tk2−/− mice is not clear. Cytosolic dCK, which is active throughout the cell cycle, could theoretically compensate for the loss of Tk2 phosphorylation of deoxycytidine, but dCK activity is undetectable in murine brain and heart (43). More probably, de novo cytosolic synthesis via ribonucleotide reductase R1-p53R2 produces sufficient dCMP to maintain dCTP levels in the brain and heart of Tk2−/− mice, but cannot generate enough dTMP to compensate for Tk2 deficiency.
While deficiencies of dTTP trigger widespread mtDNA depletion in Tk2−/− mice, analyses of activities and levels of mitochondrial respiratory chain complexes showed significant defects only in the brain (19). In the heart of 12–13-day-old mutant mice, the discrepancy between the substantial mtDNA depletion (20–58% of normal) and the lack of OXPHOS deficiency (Fig. 4B) was especially striking (19) and suggested the existence of transcriptional or translational compensatory mechanisms for mtDNA depletion. In fact, evidence of transcriptional compensation was noted in two TK2-deficient patients, who had increased mt-transcripts contrasting with the severe mtDNA depletion in muscle (15–17) and in cultured skin TK2 deficient fibroblasts (18). In addition, transcriptional compensation has been observed in TFAM and Mpv17 knockout mouse models of MDS (44,45). In our Tk2 H126N knockin mouse, the heart is spared at Day 13 by virtue of the increased amount of transcripts of mtDNA-encoded proteins (ND6 of complex I and COXI of complex IV) relative to its residual mtDNA (Fig. 5). We observed the same trend in the brain; however, mtDNA was so severely reduced in this tissue (12–30% of normal) (Fig. 4B) (19) that the increase in transcripts (1.6–1.7-fold elevation) was insufficiently compensatory. Based on our data, we can define the mtDNA depletion threshold as the reduction in mtDNA that overwhelms transcriptional compensation.
We hypothesize that mitochondria depleted of mtDNA signal the nucleus to up-regulate the expression of genes involved in mitochondrial transcription. Mitochondrial retrograde signaling has been described in yeast and mammalian cells under normal and pathological conditions (46,47), including mtDNA depletion, which has been associated with increased mRNA levels of respiratory chain subunits (48), enhanced expression of TFAM and NRF-1 (transcription factors of mitochondrial genes) (49), increased intracellular Ca2+ levels and activated cAMP response-element-binding protein (50). We postulated that PGC-1α could be the molecule responsible for enhancing mitochondrial gene expression in Tk2−/− mice, because PGC-1α induces mitochondrial biogenesis by increasing the expression and the transactivational capacity of NRF-1 and NRF-2 (transcriptional activators of nuclear mitochondrial genes), which, in turn, up-regulate the expression of their downstream targets TFAM, TFB1M and TFB2M (29,51–54) directly involved in up-regulating mitochondrial transcription. However, we did not find any difference in the transcript levels of PGC-1α, NRF-1 or NRF-2 in the heart and brain between 13-day-old Tk2−/− and Tk2+/+ mice (Fig. 5). Furthermore, we did not observe signs of PGC-1α-induced generalized mitochondrial proliferation by histology (e.g. increased SDH histochemical staining) or biochemical assays (e.g. increased citrate synthase activity) in 13-day-old Tk2−/− brain or heart (data not shown). Accordingly, neither did we observe increased mRNA levels of TFAM, TFB1M and TFB2M, downstream targets of PGC-1α, in 13-day-old Tk2−/− mice versus their Tk2+/+ litter-mates (Fig. 6).
Mitochondrial transcription is regulated, not only by transcriptional initiators, but also by mitochondrial termination factors (MTERFs). MTERF1 terminates transcription by binding to a specific site downstream the 16S rRNA gene, whereas MTERF 2 and MTERF3 are thought to regulate transcription by binding to the mtDNA heavy-strand promoter (55,56). While MTERF2 knockout mouse showed decreased mitochondrial mRNAs levels (57), these transcripts were increased in the MTERF3 knockout mouse (58). MTERF1, but not MTERF3, is regulated by the PGC-1 NRF1/2 axis (59) while MTERF2 appears to be insignificant in heart (57). Taken together, these reports indicate that a fine-tuned interaction among these three MTERF factors regulates mitochondrial transcription in a cell state-dependent and tissue-specific manner. The down-regulation of MTERF3 expression that we observed in the brain and the heart of 13-day-old Tk2−/− mice (Fig. 6) enhances the initiation of mitochondrial transcription, which may account for the increased levels of mitochondrial transcripts in the Tk2−/− mice (Fig. 5A and B); analogous to the increased transcripts from both mtDNA strands observed in MTERF3 knockout mice (58). Thus, in Tk2−/− mice, down-regulation of the transcriptional repressor MTERF3 appears to be a compensatory mechanism for the partial mtDNA depletion in the heart, but is probably insufficient to compensate for the more severe mtDNA depletion in the brain.
In summary, our homozygous Tk2 knockin mice show ubiquitous Tk2 deficiency, which becomes functionally significant in tissues after Tk1 activity decreases. The onset of mtDNA depletion in each tissue correlates with the down-regulation of Tk1 activity. The severity and tissue specificity of human MDS due to TK2 deficiency appear to correlate with the residual TK2 activity of Tk2 mutant proteins. In addition, Tk2-deficient tissues that trigger MTERF3-mediated transcriptional compensatory mechanisms prior to the onset of severe mtDNA depletion can be spared from OXPHOS deficiency. In mitochondrial diseases, understanding how some tissues are spared from the effects of mutations in ubiquitously expressed proteins is important for targeting therapies to affected tissues. The development of compounds that modulate mitochondrial transcription, such as MTERF3 inhibitors, may have therapeutic potential in TK2 deficiency.
MATERIALS AND METHODS
Mice
Mutant Tk2 H126N knockin mice were reported (19). Mating heterozygous Tk2 mutant male and female mice produced litters with Tk2 wild-type (Tk2+/+), heterozygous (Tk2+/−) and homozygous (Tk2−/−) mice with expected Mendelian frequency.
Whole tissue protein extracts
Tissues were homogenized on ice with 5 volumes (w/v) of cold buffer [10 mm Tris–HCl pH 7.5, 0.5% Triton X, 2 mm EDTA, 1 mm DTT and Complete Protease Inhibitor Cocktail Tablets (Roche, Indianapolis, IN, USA)]. 0.2 m NaCl was added prior to centrifugation at 19 000g for 20 min at 4°C. Supernatants were aliquoted and stored at −80°C until needed.
Tk2 activity measurement
We used [5′-3H]5-(2-bromovinyl)-2′-deoxyuridine (BVDU) (Moravek Biochemicals, Brea, CA, USA) as radiolabeled Tk2 substrate and followed a published protocol (20). Two quantities of total protein extracts were used for each determination: 17.5 and 35 μg for the brain, and 30 and 60 μg for the heart. Briefly, 40 μl volume reactions were established by dissolving proteins in 20 μl of dilution buffer (1 mg/ml BSA, 10 mm Tris–HCl, 2 mm DTT and 5 mm ATP) followed by mixing with 20 μl of 2× reaction mix (100 mm Tris–HCl, pH 7.5, 10 mm ATP, 10 mm MgCl2, 1 mg/ml BSA, 10 mm NaF, 4 mm DTT, 100 μm 5-bromouracil (TP inhibitor) and 2500 cpms/pmol 0.4 μm 3H-BVDU). Parallel samples without protein were used as blank. After 60 min at 37°C, 30 μl of reaction was spotted on Whatman DE81 filters and air dried. Subsequently, filters were washed three times for 5 min in 5 mm ammonium formate and placed in vials to elute with 2 ml of 0.1 m HCl, 0.2 m NaCl for 30 min at room temperature. Radioactivity in the elution buffer was counted after adding 9 ml of Ultima Gold scintillation fluid (Perkin Elmer, Waltham, MA, USA) in a Packard Tri-Carb 2900TR counter (Perkin Elmer). Enzyme activity was calculated as pmol product/min/mg protein.
Modeling of mouse thymidine kinase 2 structure
The modeling was performed using SwissModel software (spdv 4.0.1) with the crystal structure of Drosophila melanogaster deoxynucleoside kinase (dmNK) as template (PBD_2vp4). To visualize the structure and predict the effect of the H16N mutation on the conformation, SpdbViewer 4.0.1 software was used. Protein alignments were performed using ClustalW (8).
Tk1 activity measurement
We used [methyl-3H] thymidine (Perkin-Elmer) as the radiolabeled substrate. We followed the same protocol as for Tk2 activity measurement, except the 2× reaction mix did not include 5′-bromouracil and the substrate was 2500 cpms/pmol 20 μm 3H-thymidine in the presence or absence of 10 mm deoxycytidine. Tk1 activity (pmol/min/mg) was estimated as the remaining Tk activity in the presence of a molar excess of cold deoxycytidine as inhibitor of Tk2 (60).
dNTPs pool determination
dNTP extracts were obtained as described (40). Briefly, tissues were homogenized on ice in 10 volumes (w/v) of cold MTSE buffer (210 mm mannitol, 70 mm sucrose, 10 mm Tris–HCl pH 7.5, 2 mm EGTA, 0.2 mg/ml BSA) and centrifuged twice at 1000g for 3 min at 4°C. Supernatants were precipitated with 100% methanol, kept 1 h at −20°C, boiled 3 min, stored at −20°C (from 1 h to overnight) and centrifuged at 20 800g for 20 min at 4°C. Supernatants were evaporated until dry, resuspended in 200 μl of water and stored at −80°C until needed.
To minimize ribonucleotide interference, total dNTP pools were determined following a reported protocol (24). Briefly, 20 μl volume reactions were generated by mixing 10 μl of sample or standard with 10 μl of 2× reaction buffer [0.04U/ml ThermoSequenase DNA polymerase (GE Healthcare, Piscataway, NJ, USA) or Taq DNA polymerase (Roche), 0.5 μm 3H-dTTP or 3H-dATP (Moravek Biochemicals), 0.5 μm specific oligonucleotide, 40 mm Tris–HCl, pH 7.5, 20 mm MgCl2, 10 mm DTT]. After 60 min at 48°C, 15 μl of reaction was spotted on Whatman DE81 filters, air dried and washed three times for 10 min with 5% Na2HPO4, once in distilled water and once in absolute ethanol. The retained radioactivity was determined by scintillation counting.
mtDNA quantification
Real-time PCR was performed with the primers and probes (Applied Biosystems, Invitrogen, Foster City, CA, USA) for murine COX I gene (mtDNA) and mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH, nDNA) as described (61) using standard curve quantification, in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems). mtDNA values were normalized by nDNA values and expressed as percent relative to wild-type (100%).
Western blot analyses
Thirty micrograms of whole tissue extracts, prepared as described in the Tk2 enzyme assay, were electrophoresed in an SDS-12%-PAGE gel, transferred to Immun-BlotTM PVDF membranes (Biorad, Hercules, CA, USA) and probed with Rodent Total OXPHOS Complexes Detection Kit cocktail of antibodies (MitoSciences, Eugene, OR, USA). Protein–antibody interaction was detected with peroxidase-conjugated mouse anti-mouse IgG antibody (Sigma-Aldrich, St Louis, MO, USA), using SuperSignal® chemiluminiscence detection kit (Thermo Fisher, Waltham, MA, USA). Quantification of proteins was carried out using NIH ImageJ 1.37V software. Average gray value was calculated within selected areas as the sum of the gray values of all the pixels in the selection divided by the number of pixels.
Quantification of mRNA levels
Tissue samples ≤30 were used to extract RNA with RNeasy kit (Qiagen, Valencia, CA, USA), treated with RNase-Free DNase (Qiagen), electrophoresed in agarose 1% to check integrity and quantified by optical density at 260 nm.
cDNA was obtained with SuperScriptTM III First-Strand Synthesis for RT–PCR (Invitrogen). Quantification was performed with real-time PCR, by standard curve method, with specific murine primers and Taqman® probes (from Applied Biosystems, Invitrogen) for the targeted genes and the mouse glyceraldehyde-3-phosphate dehydrogenase probe as a standard loading control (AB 4352339E, Invitrogen). Applied-Biosystems Taqman® gene expression assays for specific genes were the following: Mm_01208835_m1 for PGC-1α, Mm_01135609_m1 for NRF-1, Mm_00477784_m1 for NRF-2, Mm_00447485_m1 for TFAM, Mm_00524825-m1 for TFB1M and Mm_00481557_m1 for MTERF3.
Sequence (5′–3′) of primer forward (F), reverse (R) and 6FAM probe of Applied-Biosystems Taqman assays on demand for ND6, COXI, TFB2M and MTERF1 were: ND6-F, AGCTACTGAGGAATATCCAGAGACTTG; ND6-R, TCAACCAATCTCCCAAACCATCAA; ND6 probe, ATGAAGTTGGAGTAATTAATC; COXI-F, TGCTAGCCGCAGGCATTACT; COXI-R, CGGGATCAAAGAAAGTTGTGTTT; COXI probe, TACTACTAACAGACCGCAACC; TFB2M-F, AAACGCAATGCCCCAATAATACG; TFB2M-R, GGGTTTTTTCTTATCTGCCTCAGGAT; TFB2M probe, ACCGTACTCAGTGAACGTA; MTERF1-F, GGCGGAAGTGAAAGGTGTCTA; MTERF1-R, CCATAATCATCAGGTAGCCCAAAGT; MTE RF-1 probe, CCGGGAGCGTTGCATT.
Thermal cycling was performed in ABI PRISM 7000 Sequence Detector System (Applied Biosystems, Invitrogen)
FUNDING
The work was supported in part by the Muscular Dystrophy Association, USA, and the FUNDISMUN Foundation. The authors are supported by grants from the Muscular Dystrophy Association, National Institutes of Health (R01 HD057543, R01 HD056103, RC1 NS070232, P01 HD032062) and the Marriott Mitochondrial Disorder Clinical Research Fund (MMDCRF).
ACKNOWLEDGMENTS
We thank Drs E.A. Schon and S. DiMauro for useful comments.
References
- 1.Johansson M., Karlsson A. Cloning of the cDNA and chromosome localization of the gene for human thymidine kinase 2. J. Biol. Chem. 1997;272:8454–8458. doi: 10.1074/jbc.272.13.8454. [DOI] [PubMed] [Google Scholar]
- 2.Wang L., Munch-Petersen B., Herrstrom Sjoberg A., Hellman U., Bergman T., Jornvall H., Eriksson S. Human thymidine kinase 2: molecular cloning and characterisation of the enzyme activity with antiviral and cytostatic nucleoside substrates. FEBS Lett. 1999;443:170–174. doi: 10.1016/s0014-5793(98)01711-6. [DOI] [PubMed] [Google Scholar]
- 3.Wang L., Eriksson S. Cloning and characterization of full-length mouse thymidine kinase 2: the N-terminal sequence directs import of the precursor protein into mitochondria. Biochem. J. 2000;351:469–476. [PMC free article] [PubMed] [Google Scholar]
- 4.Coppock D.L., Pardee A.B. Control of thymidine kinase mRNA during the cell cycle. Mol. Cell. Biol. 1987;7:2925–2932. doi: 10.1128/mcb.7.8.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 6.Ke P.Y., Chang Z.F. Mitotic degradation of human thymidine kinase 1 is dependent on the anaphase-promoting complex/cyclosome-CDH1-mediated pathway. Mol. Cell. Biol. 2004;24:514–526. doi: 10.1128/MCB.24.2.514-526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rampazzo C., Fabris S., Franzolin E., Crovatto K., Frangini M., Bianchi V. Mitochondrial thymidine kinase and the enzymatic network regulating thymidine triphosphate pools in cultured human cells. J. Biol. Chem. 2007;282:34758–34769. doi: 10.1074/jbc.M705923200. [DOI] [PubMed] [Google Scholar]
- 8.Rampazzo C., Miazzi C., Franzolin E., Pontarin G., Ferraro P., Frangini M., Reichard P., Bianchi V. Regulation by degradation, a cellular defense against deoxyribonucleotide pool imbalances. Mutat. Res. 2010 doi: 10.1016/j.mrgentox.2010.06.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 9.Saada A., Shaag A., Mandel H., Nevo Y., Eriksson S., Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat. Genet. 2001;29:342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- 10.Mancuso M., Salviati L., Sacconi S., Otaegui D., Camano P., Marina A., Bacman S., Moraes C.T., Carlo J.R., Garcia M., et al. Mitochondrial DNA depletion: mutations in thymidine kinase gene with myopathy and SMA. Neurology. 2002;59:1197–1202. doi: 10.1212/01.wnl.0000028689.93049.9a. [DOI] [PubMed] [Google Scholar]
- 11.Oskoui M., Davidzon G., Pascual J., Erazo R., Gurgel-Giannetti J., Krishna S., Bonilla E., De Vivo D.C., Shanske S., DiMauro S. Clinical spectrum of mitochondrial DNA depletion due to mutations in the thymidine kinase 2 gene. Arch. Neurol. 2006;63:1122–1126. doi: 10.1001/archneur.63.8.1122. [DOI] [PubMed] [Google Scholar]
- 12.Gotz A., Isohanni P., Pihko H., Paetau A., Herva R., Saarenpaa-Heikkila O., Valanne L., Marjavaara S., Suomalainen A. Thymidine kinase 2 defects can cause multi-tissue mtDNA depletion syndrome. Brain. 2008;131:2841–2850. doi: 10.1093/brain/awn236. [DOI] [PubMed] [Google Scholar]
- 13.Lesko N., Naess K., Wibom R., Solaroli N., Nennesmo I., von Dobeln U., Karlsson A., Larsson N.G. Two novel mutations in thymidine kinase-2 cause early onset fatal encephalomyopathy and severe mtDNA depletion. Neuromuscul. Disord. 2010;20:198–203. doi: 10.1016/j.nmd.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Marti R., Nascimento A., Colomer J., Lara M.C., Lopez-Gallardo E., Ruiz-Pesini E., Montoya J., Andreu A.L., Briones P., Pineda M. Hearing loss in a patient with the myopathic form of mitochondrial DNA depletion syndrome and a novel mutation in the TK2 gene. Pediatr. Res. 2010;68:151–154. doi: 10.1203/PDR.0b013e3181e33bbe. [DOI] [PubMed] [Google Scholar]
- 15.Barthelemy C., Ogier de Baulny H., Diaz J., Cheval M.A., Frachon P., Romero N., Goutieres F., Fardeau M., Lombes A. Late-onset mitochondrial DNA depletion: DNA copy number, multiple deletions, and compensation. Ann. Neurol. 2001;49:607–617. [PubMed] [Google Scholar]
- 16.Vila M.R., Segovia-Silvestre T., Gamez J., Marina A., Naini A.B., Meseguer A., Lombes A., Bonilla E., DiMauro S., Hirano M., et al. Reversion of mtDNA depletion in a patient with TK2 deficiency. Neurology. 2003;60:1203–1205. doi: 10.1212/01.wnl.0000055928.58122.47. [DOI] [PubMed] [Google Scholar]
- 17.Vila M.R., Villarroya J., Garcia-Arumi E., Castellote A., Meseguer A., Hirano M., Roig M. Selective muscle fiber loss and molecular compensation in mitochondrial myopathy due to TK2 deficiency. J. Neurol. Sci. 2008;267:137–141. doi: 10.1016/j.jns.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villarroya J., de Bolos C., Meseguer A., Hirano M., Vila M.R. Altered gene transcription profiles in fibroblasts harboring either TK2 or DGUOK mutations indicate compensatory mechanisms. Exp. Cell Res. 2009;315:1429–1438. doi: 10.1016/j.yexcr.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akman H.O., Dorado B., Lopez L.C., Garcia-Cazorla A., Vila M.R., Tanabe L.M., Dauer W.T., Bonilla E., Tanji K., Hirano M. Thymidine kinase 2 (H126N) knockin mice show the essential role of balanced deoxynucleotide pools for mitochondrial DNA maintenance. Hum. Mol. Genet. 2008;17:2433–2440. doi: 10.1093/hmg/ddn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzolin E., Rampazzo C., Perez-Perez M.J., Hernandez A.I., Balzarini J., Bianchi V. Bromovinyl-deoxyuridine: a selective substrate for mitochondrial thymidine kinase in cell extracts. Biochem. Biophys. Res. Commun. 2006;344:30–36. doi: 10.1016/j.bbrc.2006.03.147. [DOI] [PubMed] [Google Scholar]
- 21.Wang L., Eriksson S. 5-Bromovinyl 2′-deoxyuridine phosphorylation by mitochondrial and cytosolic thymidine kinase (TK2 and TK1) and its use in selective measurement of TK2 activity in crude extracts. Nucleosides Nucleotides Nucleic Acids. 2008;27:858–862. doi: 10.1080/15257770802146510. [DOI] [PubMed] [Google Scholar]
- 22.Rylova S.N., Mirzaee S., Albertioni F., Eriksson S. Expression of deoxynucleoside kinases and 5′-nucleotidases in mouse tissues: implications for mitochondrial toxicity. Biochem. Pharmacol. 2007;74:169–175. doi: 10.1016/j.bcp.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X., Solaroli N., Bjerke M., Stewart J.B., Rozell B., Johansson M., Karlsson A. Progressive loss of mitochondrial DNA in thymidine kinase 2-deficient mice. Hum. Mol. Genet. 2008;17:2329–2335. doi: 10.1093/hmg/ddn133. [DOI] [PubMed] [Google Scholar]
- 24.Ferraro P., Franzolin E., Pontarin G., Reichard P., Bianchi V. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 2010;38:e85. doi: 10.1093/nar/gkp1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scarpulla R.C. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann. N. Y. Acad. Sci. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asin-Cayuela J., Gustafsson C.M. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007;32:111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N.G., Gustafsson C.M. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 28.Falkenberg M., Larsson N.G., Gustafsson C.M. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 29.Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 30.Pontarin G., Ferraro P., Hakansson P., Thelander L., Reichard P., Bianchi V. p53R2-dependent ribonucleotide reduction provides deoxyribonucleotides in quiescent human fibroblasts in the absence of induced DNA damage. J. Biol. Chem. 2007;282:16820–16828. doi: 10.1074/jbc.M701310200. [DOI] [PubMed] [Google Scholar]
- 31.Haas R.J., Werner J., Fliedner T.M. Cytokinetics of neonatal brain cell development in rats as studied by the ‘complete 3H-thymidine labelling’ method. J. Anat. 1970;107:421–437. [PMC free article] [PubMed] [Google Scholar]
- 32.Mori K., Yamagami S., Kawakita Y. Thymidine metabolism and deoxyribonucleic acid synthesis in the developing rat brain. J. Neurochem. 1970;17:835–843. doi: 10.1111/j.1471-4159.1970.tb02237.x. [DOI] [PubMed] [Google Scholar]
- 33.Yamagami S., Mori K., Kawakita Y. Changes of thymidine kinase in the developing rat brain. J. Neurochem. 1972;19:369–376. doi: 10.1111/j.1471-4159.1972.tb01346.x. [DOI] [PubMed] [Google Scholar]
- 34.Soonpaa M.H., Kim K.K., Pajak L., Franklin M., Field L.J. Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 1996;271:H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X., Johansson M., Solaroli N., Rozell B., Grandien A., Karlsson A. Hematopoiesis in the thymidine kinase 2 deficient mouse model of mitochondrial DNA depletion syndrome. J. Inherit. Metab. Dis. 2010;33:231–236. doi: 10.1007/s10545-010-9102-x. [DOI] [PubMed] [Google Scholar]
- 36.Saada A., Shaag A., Elpeleg O. mtDNA depletion myopathy: elucidation of the tissue specificity in the mitochondrial thymidine kinase (TK2) deficiency. Mol. Genet. Metab. 2003;79:1–5. doi: 10.1016/s1096-7192(03)00063-5. [DOI] [PubMed] [Google Scholar]
- 37.Saada A., Ben-Shalom E., Zyslin R., Miller C., Mandel H., Elpeleg O. Mitochondrial deoxyribonucleoside triphosphate pools in thymidine kinase 2 deficiency. Biochem. Biophys. Res. Commun. 2003;310:963–966. doi: 10.1016/j.bbrc.2003.09.104. [DOI] [PubMed] [Google Scholar]
- 38.Wang L., Limongelli A., Vila M.R., Carrara F., Zeviani M., Eriksson S. Molecular insight into mitochondrial DNA depletion syndrome in two patients with novel mutations in the deoxyguanosine kinase and thymidine kinase 2 genes. Mol. Genet. Metab. 2005;84:75–82. doi: 10.1016/j.ymgme.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Bartesaghi S., Betts-Henderson J., Cain K., Dinsdale D., Zhou X., Karlsson A., Salomoni P., Nicotera P. Loss of thymidine kinase 2 alters neuronal bioenergetics and leads to neurodegeneration. Hum. Mol. Genet. 2010;19:1669–1677. doi: 10.1093/hmg/ddq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferraro P., Nicolosi L., Bernardi P., Reichard P., Bianchi V. Mitochondrial deoxynucleotide pool sizes in mouse liver and evidence for a transport mechanism for thymidine monophosphate. Proc. Natl Acad. Sci. USA. 2006;103:18586–18591. doi: 10.1073/pnas.0609020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pontarin G., Gallinaro L., Ferraro P., Reichard P., Bianchi V. Origins of mitochondrial thymidine triphosphate: dynamic relations to cytosolic pools. Proc. Natl Acad. Sci. USA. 2003;100:12159–12164. doi: 10.1073/pnas.1635259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leanza L., Ferraro P., Reichard P., Bianchi V. Metabolic interrelations within guanine deoxynucleotide pools for mitochondrial and nuclear DNA maintenance. J. Biol. Chem. 2008;283:16437–16445. doi: 10.1074/jbc.M801572200. [DOI] [PubMed] [Google Scholar]
- 43.Wang L., Eriksson S. Tissue specific distribution of pyrimidine deoxynucleoside salvage enzymes shed light on the mechanism of mitochondrial DNA depletion. Nucleosides Nucleotides Nucleic Acids. 2010;29:400–403. doi: 10.1080/15257771003741042. [DOI] [PubMed] [Google Scholar]
- 44.Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 45.Viscomi C., Spinazzola A., Maggioni M., Fernandez-Vizarra E., Massa V., Pagano C., Vettor R., Mora M., Zeviani M. Early-onset liver mtDNA depletion and late-onset proteinuric nephropathy in Mpv17 knockout mice. Hum. Mol. Genet. 2009;18:12–26. doi: 10.1093/hmg/ddn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butow R.A., Avadhani N.G. Mitochondrial signaling: the retrograde response. Mol. Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z., Butow R.A. Mitochondrial retrograde signaling. Annu. Rev. Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 48.Li K., Neufer P.D., Williams R.S. Nuclear responses to depletion of mitochondrial DNA in human cells. Am. J. Physiol. 1995;269:C1265–C1270. doi: 10.1152/ajpcell.1995.269.5.C1265. [DOI] [PubMed] [Google Scholar]
- 49.Miranda S., Foncea R., Guerrero J., Leighton F. Oxidative stress and upregulation of mitochondrial biogenesis genes in mitochondrial DNA-depleted HeLa cells. Biochem. Biophys. Res. Commun. 1999;258:44–49. doi: 10.1006/bbrc.1999.0580. [DOI] [PubMed] [Google Scholar]
- 50.Biswas G., Adebanjo O.A., Freedman B.D., Anandatheerthavarada H.K., Vijayasarathy C., Zaidi M., Kotlikoff M., Avadhani N.G. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 52.Gleyzer N., Vercauteren K., Scarpulla R.C. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelly D.P., Scarpulla R.C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 54.Virbasius J.V., Scarpulla R.C. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl Acad. Sci. USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kruse B., Narasimhan N., Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58:391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- 56.Martin M., Cho J., Cesare A.J., Griffith J.D., Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 57.Wenz T., Luca C., Torraco A., Moraes C.T. mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab. 2009;9:499–511. doi: 10.1016/j.cmet.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Park C.B., Asin-Cayuela J., Camara Y., Shi Y., Pellegrini M., Gaspari M., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 59.Bruni F., Polosa P.L., Gadaleta M.N., Cantatore P., Roberti M. Nuclear respiratory factor 2 induces the expression of many but not all human proteins acting in mitochondrial DNA transcription and replication. J. Biol. Chem. 2010;285:3939–3948. doi: 10.1074/jbc.M109.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arner E.S., Spasokoukotskaja T., Eriksson S. Selective assays for thymidine kinase 1 and 2 and deoxycytidine kinase and their activities in extracts from human cells and tissues. Biochem. Biophys. Res. Commun. 1992;188:712–718. doi: 10.1016/0006-291x(92)91114-6. [DOI] [PubMed] [Google Scholar]
- 61.Spinazzola A., Viscomi C., Fernandez-Vizarra E., Carrara F., D'Adamo P., Calvo S., Marsano R.M., Donnini C., Weiher H., Strisciuglio P., et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]