Abstract
Using multilocus sequence analyses (MLSA), we investigated the phylogenetic relationship of spirochaete strains from North America previously assigned to the genospecies Borrelia bissettii. We amplified internal fragments of 8 housekeeping genes (clpA, clpX, nifS, pepX, pyrG, recG, rplB, and uvrA) located on the main linear chromosome by polymerase chain reaction. Phylogenetic analysis of concatenated sequences of the 8 loci showed that the B. bissettii clade consisted of 4 closely related clusters which included strains from California (including the type strain DN127-Cl9-2/p7) and Colorado that were isolated from Ixodes pacificus, I. spinipalpis, or infected reservoir hosts. Several strains isolated from I. scapularis clustered distantly from B. bissettii. Genetic distance analyses confirmed that these strains are more distant to B. bissettii than they are to B. carolinensis, a recently described Borrelia species, which suggests that they constitute a new Borrelia genospecies. We propose that it be named Borrelia kurtenbachii sp. nov. in honour of the late Klaus Kurtenbach. The data suggest that ecological differences between B. bissettii and the new Borrelia genospecies reflect different transmission cycles. In view of these findings, the distinct vertebrate host-tick vector associations and the distributions of B. bissettii and B. kurtenbachii require further investigation.
Keywords: Borrelia bissettii, Borrelia kurtenbachii sp. nov, Ixodes, Multilocus sequence analysis, Molecular ecology
Introduction
Lyme borreliosis (LB) is caused by several species of bacteria belonging to the LB group of spirochaetes, also referred to as Borrelia burgdorferi sensu lato (s.l.) species complex. These bacteria are maintained in nature by numerous vertebrate hosts and are transmitted by ticks in the Ixodes persulcatus species complex (reviewed by Kurtenbach et al., 2006). Today, LB is the most frequently reported vector-borne disease in the temperate zones of the Northern Hemisphere. In the United States, human cases exceeding 20,000 per year currently are being reported with 90% of the cases occurring in 2 regional foci, the Northeast and Upper Midwest. Human cases are far less frequently reported in the western and southern regions of the country (Bacon et al., 2008). In North America, B. burgdorferi sensu stricto (hereinafter referred to as B. burgdorferi) is the sole species associated with human disease (Mathiesen et al., 1997; Wormser et al., 2008). The reasons for the skewed spatial distribution of LB cases in the United States are not entirely clear but may be due to ecological factors, such as the abundance of, and spirochaete infection prevalences in, tick vectors, the presence or absence of disseminating, disease-causing B. burgdorferi genotypes, differences in tick host-seeking behaviour (in particular the immature stages), or possibly other factors such as land-use changes and regional differences in human behaviour (Burgdorfer et al., 1985; Oliver, 1996; Lane and Quistad, 1998; Wright et al., 2000; Slowik and Lane, 2001; Lane et al., 2004; Diuk-Wasser et al., 2006; Eisen et al., 2006; Killilea et al., 2008; Gatewood et al., 2009; Hamer et al., 2010; Humphrey et al., 2010).
The aetiological agent of LB was identified in 1981 and named Borrelia burgdorferi after its discoverer, Willy Burgdorfer (Burgdorfer et al., 1982; Johnson et al., 1984). Over the following 2 decades, many isolates derived from human patients, ticks, or reservoir host animals were analysed biochemically and genetically, and a high diversity within B. burgdorferi was found which led to the delineation of new genospecies in Europe, Asia, and North America (Baranton et al., 1992; Kawabata et al., 1993; Maupin et al., 1994; Postic et al., 1994; Marconi et al., 1995; Fukunaga et al., 1996; Le Fleche et al., 1997; Mathiesen et al., 1997; Wang et al., 1997, 1999; Postic et al., 1998; Masuzawa et al., 1999, 2001; Lin et al., 2001). The LB group of spirochaetes now forms a species complex consisting of 17 named genospecies (Kurtenbach et al., 2010) reflecting the complex ecology of these bacteria (Kurtenbach et al., 2006). Of the named species, 6 occur in North America including B. andersonii, B. bissettii, B. californiensis, B. carolinensis, B. americana, and B. burgdorferi. The true species richness doubtless is higher as several strains phylogenetically distinct from all other species have not yet been named (Postic et al., 1998, 2007).
B. bissettii enzootic transmission cycles in the United States involving tick species such as I. spinipalpis, I. minor, I. affinis, and various vertebrate host species were found in Colorado, California, and some Southern states (Bissett and Hill, 1987; Brown and Lane, 1992; Maupin et al., 1994; Lane and Quistad, 1998; Schneider et al., 2000; Oliver et al., 2003). In 1990, Anderson and coworkers described a non-pathogenic variant of B. burgdorferi, strain 25015. This strain was isolated from 4 blood-engorged I. scapularis larvae (then termed I. dammini) that had been removed from 3 Peromyscus leucopus mice collected in Dutchess County, New York, in 1987 (Anderson et al., 1988, 1990). In 1998, the species B. bissettii was delineated using restriction patterns of the rrf-rrl intergenic spacer as well as sequences of the intergenic spacer and 16S rDNA. Samples included mainly strains isolated from I. pacificus or I. neotomae (now I. spinipalpis) in California (Postic et al., 1998). Strain 25015 was included in the genospecies B. bissettii, although differences in rrf-rrl restriction patterns between strain 25015 and DN127 were observed (Mathiesen et al., 1997; Postic et al., 1998; Lin et al., 2003). In 1996, 1997, and 1998, Borrelia isolates were obtained from the white-footed mouse (Peromyscus leucopus), the meadow vole (Microtus pennsylvanicus), and the meadow jumping mouse (Zapus hudsonius) in forested, suburban areas of Chicago, Illinois (Picken and Picken, 2000). These isolates were compared to B31, DN127, and 25015 by RFLP analysis. They were designated B. bissettii because most of them exhibited a restriction pattern similar to that of strain 25015, and 2 strains exhibited a macrorestriction pattern like that of the type strain DN127 (Picken and Picken, 2000).
DNA-DNA hybridization and sequence analyses of the 16S rRNA locus were used as standard procedures to delineate bacterial species for many years, but were not without drawbacks (Gevers et al., 2005; Stackebrandt and Ebers, 2006). The invention of multilocus sequence typing (MLST) during the late 1990s (Maiden et al., 1998; Enright and Spratt, 1999; Spratt, 1999; Feil and Enright, 2004) led to the development of multilocus sequence analysis (MLSA), a tool that has been used since to resolve inter-species relationships of bacteria and to delineate bacterial species (Gevers et al., 2005; Bishop et al., 2009). Several different multiple-loci schemes have been used for the LB group of spirochaetes (Richter et al., 2006; Postic et al., 2007; Margos et al., 2008; Rudenko et al., 2009a, 2009b). One of them employs housekeeping genes (Margos et al., 2008) and resembles typical MLSA schemes reported for other microorganisms (see http://www.mlst.net).
Here, we used this MLSA system (Margos et al., 2008, 2009) to investigate the phylogenetic relationships among B. bissettii strains isolated from reservoir hosts or ticks removed from them in different regions of North America. The data confirm that B. bissettii forms a heterogeneous clade (Postic et al., 1998). We also demonstrate that strain 25015 and several other strains collected in Illinois and Nova Scotia that were assigned previously to B. bissettii represent a phylogenetically distinct group, and we propose to name this group Borrelia kurtenbachii sp. nov.
Materials and methods
Tick collection and isolation of LB spirochaetes
The samples used in this study are listed in Table 1. They mainly represent cultured isolates principally obtained from ticks that were collected from rodents trapped in California, Colorado, Illinois, and New York. Isolation of LB spirochaetes followed standard procedures (Maupin et al., 1994). One sample, NS07-121, was not a cultured isolate but DNA-extracted from a nymphal I. scapularis collected in Nova Scotia from a human in 2007.
Table 1.
Borrelia isolates evaluated during this study.
| ST | Strain ID | Country of origin | Region | Species | Biological origin | Year of collection | Collected by |
|---|---|---|---|---|---|---|---|
| 156 | CA128 | USA | Hopland Field Station, Mendocino County, CA | B. bissettii | Ixodes neotomae (now known as I. spinipalpis) ex Neotoma fuscipes | 1991 | R.S. Lane |
| 282 | CA370 | USA | Alameda County, CA | B. bissettii | N. fuscipes ear biopsy | 1992 | R.S. Lane |
| 283 | CA371 | USA | Alameda County, CA | B. bissettii | N. fuscipes ear biopsy | 1992 | R.S. Lane |
| 270 | CA389 | USA | Tilden Regional Park, Berkeley, CA | B. bissettii | I. pacificus | 1993 | R.S. Lane |
| 272 | DN127-Cl9-2/p7 | USA | Del Norte County, CA | B. bissettii | I. pacificus | 1985 | J.R. Clover |
| gom93-267a | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex Neotoma mexicana | 1993 | Gary O. Maupin | |
| 273 | gom93-268 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| gom93-270a | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin | |
| gom93-271a | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin | |
| gom93-272a | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin | |
| 273 | gom93-274 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| 273 | gom93-275 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| gom93-277a | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin | |
| 273 | gom93-278 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| gom93-280a | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin | |
| 160 | gom93-283 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| 273 | gom93-284 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| 274 | gom93-286 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| 273 | gom93-287 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| gom93-289a | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin | |
| 271 | gom93-296 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| 273 | gom93-297 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| gom93-298a | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin | |
| 158 | gom93-299 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| gom93-300a | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex Peromyscus difficilis | 1993 | Gary O. Maupin | |
| gom93-304a | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin | |
| 275 | gom93-305 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex P. difficilis | 1993 | Gary O. Maupin |
| 276 | gom93-310 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| 277 | gom93-501 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| 277 | gom93-543 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| 277 | gom93-544 | USA | Larimer County, CO | B. bissettii | I. spinipalpis ex N. mexicana | 1993 | Gary O. Maupin |
| 278 | IL 96-255 | USA | Poplar Creek, Cook County, IL | B. kurtenbachiib | Microtus pennsylvanicus | 1996 | R. Picken |
| 279 | IL 97-236u | USA | Poplar Creek, Cook County, IL | B. kurtenbachiib | M. pennsylvanicus | 1996 | R. Picken |
| 280 | 25015 | USA | New York State | B. kurtenbachiib | I. scapularis | 1987 | J.F. Anderson |
| 281 | NS07-121 | Canada | Nova Scotia | B. kurtenbachiib | I. scapularis | 2007 | N. Ogden |
These strains showed mixed infections and could not be included into phylogenetic and multilocus sequence analysis.
These strains form a distinct group that is genetically sufficiently distant from B. bissettii to represent a new species. We propose the name Borrelia kurtenbachii spec. nov. in honour of Klaus Kurtenbach.
Molecular methods and MLST analyses
Borrelia DNA was extracted from cultured isolates or frozen stocks using DNA STAT-60 (Tel-Test, Friendswood, TX, USA). Briefly, cells were lysed in 1 ml of DNA isolation reagent and 0.2 ml chloroform. After centrifugation (12,000 × g for 15 min at 4°C), the upper layer was isolated and added to an equal volume of isopropanol, followed by incubation at room temperature for 5 min. The sample was centrifuged at 12,000 × g for 10 min at 4°C, followed by a wash in 75% ethanol. After drying, the sample was resuspended in 50 μl H2O. For strain NS07-121, no isolation was attempted, but tick and Borrelia DNA was purified as described earlier (Ogden et al., 2010).
Amplification of the 8 housekeeping genes (clpA, clpX, nifS, pepX, pyrG, recG, rplB, and uvrA) was conducted by nested PCR using primers and conditions principally as described previously (Margos et al., 2009; Ogden et al., 2010; see also http://borrelia.mlst.net). Template DNA was diluted to 10 pg/μl, and 2 μl were used in a total reaction volume of 15 μl for the first amplification step except for clpA for which 4 μl of template DNA were used instead. The nested step of amplification was slightly modified: HotStarTaq Mastermix (Qiagen, Germany) was used and 1 pmol/μl of primer in a final reaction volume of 30 μl. Three μl from the first amplification step were used as template DNA. A touchdown PCR starting at 58°C with a decrease of 1°C at each cycle until 50°C was used for the second amplification step, followed by 34 cycles of amplification at an annealing temperature of 50°C. All PCR products were sequenced in both forward and reverse directions. Quality control of DNA traces was conducted manually in DNASTAR (Lasergene plc, USA). Sequences with more than one base peak at the same position in forward and reverse sequence were considered mixed isolates and removed from further analysis.
Sequences of individual housekeeping genes were compared to each other and to sequences in the MLST database. Sequences were assigned allelic numbers, and novel sequences received consecutive numbers. New consecutive sequence type numbers were assigned to allelic profiles with novel combinations or containing novel alleles (Table 2) (Margos et al., 2008). Sequences were aligned and concatenated in Mega 4.0 (Kumar et al., 2004), which also was used to determine genetic distances and to generate a neighbour-joining tree using the Maximum Composite Likelihood model. All sequences are available on the MLST website, http://borrelia.mlst.net, hosted at Imperial College London, U.K.
Table 2.
Sequence types and allelic profiles of B. bissettii and B. kurtenbachii isolates.
| Strain | ST | clpA | clpX | nifS | pep | pyrG | recG | rplB | uvrA |
|---|---|---|---|---|---|---|---|---|---|
| DN127T, Cl9-2/p7 | 272 | 117 | 69 | 68 | 102 | 99 | 99 | 83 | 89 |
| CA128 | 156 | 91 | 69 | 68 | 81 | 77 | 73 | 69 | 73 |
| CA389 | 270 | 118 | 70 | 83 | 103 | 100 | 100 | 84 | 90 |
| CA370 | 282 | 119 | 83 | 88 | 81 | 101 | 101 | 89 | 73 |
| CA371 | 283 | 120 | 69 | 68 | 104 | 99 | 74 | 83 | 91 |
| Gom93-268 | 273 | 121 | 84 | 83 | 103 | 102 | 102 | 84 | 92 |
| Gom93-274 | 273 | 121 | 84 | 83 | 103 | 102 | 102 | 84 | 92 |
| Gom93-275 | 273 | 121 | 84 | 83 | 103 | 102 | 102 | 84 | 92 |
| Gom93-278 | 273 | 121 | 84 | 83 | 103 | 102 | 102 | 84 | 92 |
| Gom93-283 | 160 | 92 | 70 | 69 | 82 | 78 | 74 | 70 | 74 |
| Gom93-284 | 273 | 121 | 84 | 83 | 103 | 102 | 102 | 84 | 92 |
| Gom93-286 | 274 | 92 | 70 | 69 | 105 | 78 | 103 | 84 | 93 |
| Gom93-287 | 273 | 121 | 84 | 83 | 103 | 102 | 102 | 84 | 92 |
| Gom93-296 | 271 | 118 | 70 | 83 | 103 | 100 | 100 | 84 | 94 |
| Gom93-297 | 273 | 121 | 84 | 83 | 103 | 102 | 102 | 84 | 92 |
| Gom93-299 | 158 | 93 | 71 | 70 | 83 | 79 | 74 | 71 | 75 |
| Gom93-305 | 275 | 122 | 70 | 84 | 106 | 103 | 104 | 84 | 94 |
| Gom93-310 | 276 | 92 | 70 | 69 | 105 | 78 | 74 | 84 | 93 |
| Gom93-501 | 277 | 123 | 70 | 85 | 103 | 100 | 100 | 84 | 95 |
| Gom93-543 | 277 | 123 | 70 | 85 | 103 | 100 | 100 | 84 | 95 |
| gom93-544 | 277 | 123 | 70 | 85 | 103 | 100 | 100 | 84 | 95 |
| IL 96-255 | 278 | 124 | 85 | 86 | 107 | 104 | 105 | 85 | 96 |
| IL 97-236u | 279 | 125 | 86 | 87 | 108 | 105 | 106 | 86 | 97 |
| 25015 | 280 | 126 | 87 | 87 | 109 | 106 | 107 | 87 | 97 |
| NS07-121 | 281 | 127 | 88 | 87 | 107 | 105 | 108 | 88 | 98 |
Results
A total of 33 strains designated previously as B. bissettii (Postic et al., 1998; Picken and Picken, 2000) was analysed by MLST. Of these isolates, 30% (n=10) contained mixed infections probably reflecting isolation from hosts infected with multiple strains. This included the type strain DN127, but further analysis of a cloned isolate (cl9-2/p7; 2/18/93) yielded unambiguous results. The remaining 23 samples fell into 2 clades. One clade consisted of 4 closely related clusters from California (i.e., CA128, CA389, CA370, CA371, and DN127-Cl9-2/p7) and Colorado (all strains isolated in 1993, designated here gom93-267 to gom93-544). The second clade was genetically distant and included strain 25015 (Fig. 1, Table 3). The pair-wise genetic similarities of strains in the first clade were >98.5% (Table 3), which is above the genetic distance used to designate distinct genospecies in the MLST scheme used here (i.e. 98.3%) (Margos et al., 2009). Some isolates of different geographic origin (i.e. Colorado – isolate gom93-299, California – DN127-Cl9-2/p7) have a high degree of similarity, and therefore occurred in the same cluster (Fig. 1). The number of alleles for the B. bissettii strains found in individual genes varied from 4 to 9 (Table 4). The G+C contents of the gene fragments ranged from 26.6 to 36.5% and are close to the average G+C contents of the B. burgdorferi main chromosome (Fraser et al., 1997). The nucleotide diversity per site was particularly high for the pyrG locus (Table 4) due to high diversity of this gene in DN127-Cl9-2/p7.
Fig. 1.
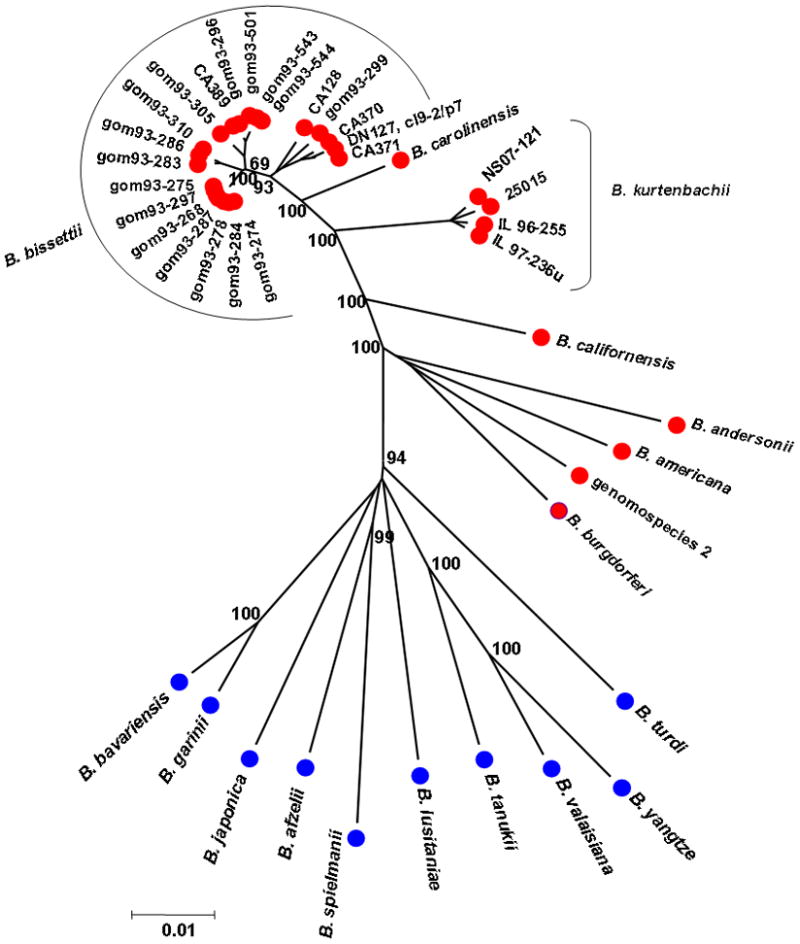
Phylogenetic analysis of Lyme borreliosis (LB) genospecies. The tree was generated using concatenated sequences of 8 chromosomally located housekeeping gene fragments. For the phylogenetic reconstruction, the maximum composite likelihood model available in MEGA 4 was used with 1000 bootstrap replicates. Scale bar indicates 1% divergence.
Table 3.
Genetic distances of LB spirochaetes. The lower left panel shows genetic distances as calculated in MEGA 4.0 using Kimura-2 parameter Model. The upper right panel shows percent genetic similarity, calculated by the fomula: 100 − (genetic distance × 100). B. bissettii strains are highlighted in yellow, B. kurtenbachii strains are highlighted in blue, percent genetic distance between B. bissettii and B. kurtenbachii are highlighted in purple.
| DN127 | CA128 | CA389 | CA370 | CA371 | gom93-268 | gom93-274 | gom93-275 | gom93-278 | gom93-283 | gom93-284 | gom93-286 | gom93-287 | gom93-296 | gom93-297 | gom93-299 | gom93-305 | gom93-310 | gom93-501 | gom93-543 | gom93-544 | IL 96-255 | IL 97_236u | 25015 | NS07-121 | SCJ1 | CA443 | QLM4P1 | B31 | VS461 | 20047 | VS116 | B. andersoni | A14S | PBi | CA2 | CA8 | PoTiBL37 | HK501 | HO14 | Ya501 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DN127,cl9-2/p7- B. bissettii | 98.97 | 98.54 | 99.5 | 99.81 | 98.71 | 98.71 | 98.71 | 98.71 | 98.58 | 98.71 | 98.58 | 98.71 | 98.52 | 98.71 | 99.05 | 98.45 | 98.6 | 98.34 | 98.34 | 98.34 | 96.5 | 96.55 | 96.37 | 96.53 | 97.32 | 95.47 | 90.56 | 94.13 | 91.81 | 92.03 | 91.33 | 93.36 | 90.97 | 91.91 | 94.54 | 94.04 | 91.82 | 91.72 | 91.71 | 91.53 | |
| CA128 | 0.0103 | 98.86 | 99.1 | 99.12 | 98.9 | 98.9 | 98.9 | 98.9 | 98.73 | 98.9 | 98.73 | 98.9 | 98.88 | 98.9 | 99.03 | 98.77 | 98.75 | 98.71 | 98.71 | 98.71 | 96.84 | 96.88 | 96.7 | 96.86 | 97.69 | 95.42 | 90.56 | 94.1 | 94.191.56 | 91.98 | 91.23 | 93.36 | 90.75 | 91.89 | 94.24 | 93.99 | 91.7 | 91.72 | 91.53 | 91.41 | |
| CA389 | 0.0146 | 0.0114 | 98.63 | 98.6 | 99.41 | 99.41 | 99.41 | 99.41 | 99.16 | 99.41 | 99.16 | 99.41 | 99.98 | 99.41 | 98.86 | 99.54 | 99.18 | 99.81 | 99.81 | 99.81 | 96.61 | 96.65 | 96.55 | 96.63 | 97.87 | 95.26 | 90.23 | 93.91 | 91.45 | 91.72 | 90.98 | 93.14 | 90.84 | 91.57 | 93.75 | 93.78 | 91.48 | 91.59 | 91.28 | 91.41 | |
| CA370 | 0.005 | 0.009 | 0.0137 | 99.6 | 98.84 | 98.84 | 98.84 | 98.84 | 98.67 | 98.84 | 98.67 | 98.84 | 98.65 | 98.84 | 99.1 | 98.63 | 98.69 | 98.48 | 98.48 | 98.48 | 96.6 | 96.64 | 96.46 | 96.62 | 97.46 | 95.51 | 90.74 | 94.2 | 91.92 | 92.2 | 91.48 | 93.31 | 91.02 | 92.06 | 94.56 | 94.08 | 91.8 | 91.82 | 91.8 | 91.73 | |
| CA371 | 0.0019 | 0.0088 | 0.014 | 0.0040 | 98.72 | 98.72 | 98.72 | 98.72 | 98.65 | 98.72 | 98.65 | 98.72 | 98.58 | 98.72 | 99.12 | 98.56 | 98.67 | 98.47 | 98.41 | 98.41 | 96.62 | 96.66 | 96.48 | 96.64 | 97.44 | 95.54 | 90.76 | 94.2 | 91.94 | 92.1 | 91.4 | 93.43 | 91.04 | 91.96 | 94.6 | 94.18 | 91.9 | 91.82 | 91.82 | 91.63 | |
| gom93-268 | 0.0129 | 0.011 | 0.0059 | 0.0116 | 0.0123 | 100 | 100 | 100 | 99.37 | 100 | 99.61 | 100 | 99.43 | 100 | 98.9 | 99.28 | 99.39 | 99.26 | 99.26 | 99.26 | 96.72 | 96.74 | 96.64 | 96.72 | 97.82 | 95.44 | 90.43 | 94.01 | 91.52 | 91.81 | 91.13 | 93.29 | 90.89 | 91.72 | 94.01 | 93.92 | 91.63 | 91.69 | 91.43 | 91.39 | |
| gom93-274 | 0.0129 | 0.011 | 0.0059 | 0.0116 | 0.0123 | 0 | 100 | 100 | 99.37 | 100 | 99.41 | 100 | 99.43 | 100 | 98.9 | 99.28 | 99.39 | 99.26 | 99.26 | 99.26 | 96.72 | 96.74 | 96.64 | 96.72 | 97.82 | 95.44 | 90.43 | 94.01 | 91.52 | 91.81 | 91.13 | 93.29 | 90.89 | 91.72 | 94.01 | 93.92 | 91.63 | 91.69 | 91.43 | 91.39 | |
| gom93-275 | 0.0129 | 0.011 | 0.0059 | 0.0116 | 0.0123 | 0 | 0 | 100 | 99.37 | 100 | 99.41 | 100 | 99.43 | 100 | 98.9 | 99.28 | 99.39 | 99.26 | 99.26 | 99.26 | 96.72 | 96.74 | 96.64 | 96.72 | 97.82 | 95.44 | 90.43 | 94.01 | 91.52 | 91.81 | 91.13 | 93.29 | 90.89 | 91.72 | 94.01 | 93.92 | 91.63 | 91.69 | 91.43 | 91.39 | |
| gom93-278 | 0.0129 | 0.011 | 0.0059 | 0.0116 | 0.0123 | 0 | 0 | 0 | 99.37 | 100 | 99.41 | 100 | 99.43 | 100 | 98.9 | 99.28 | 99.39 | 99.26 | 99.26 | 99.26 | 96.72 | 96.74 | 96.64 | 96.72 | 97.82 | 95.44 | 90.43 | 94.01 | 91.52 | 91.81 | 91.13 | 93.29 | 90.89 | 91.72 | 94.01 | 93.92 | 91.63 | 91.69 | 91.43 | 91.39 | |
| gom93-283 | 0.0142 | 0.0127 | 0.0084 | 0.0133 | 0.0136 | 0.0063 | 0.0063 | 0.0063 | 0.0063 | 99.37 | 99.83 | 99.37 | 99.18 | 99.37 | 98.86 | 99.24 | 99.85 | 99.01 | 99.01 | 99.01 | 96.66 | 96.7 | 96.59 | 96.72 | 97.69 | 95.47 | 90.33 | 93.89 | 91.43 | 91.84 | 91.03 | 93.1 | 90.79 | 91.7 | 93.91 | 93.87 | 91.63 | 91.57 | 91.38 | 91.39 | |
| gom93-284 | 0.0129 | 0.011 | 0.0059 | 0.0116 | 0.0123 | 0 | 0 | 0 | 0 | 0.0063 | 99.41 | 100 | 99.43 | 100 | 98.9 | 99.28 | 99.39 | 99.26 | 99.26 | 99.26 | 96.72 | 96.74 | 96.64 | 96.72 | 97.82 | 95.44 | 90.43 | 94.01 | 91.52 | 91.81 | 91.13 | 93.29 | 90.89 | 91.72 | 94.01 | 93.92 | 91.63 | 91.69 | 91.43 | 91.39 | |
| gom93-286 | 0.0142 | 0.0127 | 0.0084 | 0.0133 | 0.0136 | 0.0059 | 0.0059 | 0.0059 | 0.0059 | 0.0017 | 0.0059 | 99.41 | 99.18 | 99.41 | 98.86 | 99.24 | 99.85 | 99.01 | 99.01 | 99.01 | 96.66 | 96.7 | 96.59 | 96.72 | 97.69 | 95.47 | 90.33 | 93.89 | 91.43 | 91.84 | 91.03 | 93.1 | 90.79 | 91.7 | 93.91 | 93.87 | 91.63 | 91.57 | 91.38 | 91.39 | |
| gom93-287 | 0.0129 | 0.011 | 0.0059 | 0.0116 | 0.0123 | 0 | 0 | 0 | 0 | 0.0063 | 0 | 0.0059 | 99.43 | 100 | 98.9 | 99.28 | 99.39 | 99.26 | 99.26 | 99.26 | 96.72 | 96.74 | 96.64 | 96.72 | 97.82 | 95.44 | 90.43 | 94.01 | 91.52 | 91.81 | 91.13 | 93.29 | 90.89 | 91.72 | 94.01 | 93.92 | 91.63 | 91.69 | 91.43 | 91.39 | |
| gom93-296 | 0.0148 | 0.0112 | 0.0002 | 0.0136 | 0.0142 | 0.0057 | 0.0057 | 0.0057 | 0.0057 | 0.0082 | 0.0057 | 0.0082 | 0.0057 | 99.43 | 98.88 | 99.56 | 99.2 | 99.83 | 99.83 | 99.83 | 96.63 | 96.68 | 96.57 | 96.66 | 97.89 | 95.28 | 90.26 | 93.94 | 91.47 | 91.74 | 91.01 | 93.17 | 90.87 | 91.6 | 93.77 | 93.8 | 91.51 | 91.62 | 91.31 | 91.44 | |
| gom93-297 | 0.0129 | 0.011 | 0.0059 | 0.0116 | 0.0123 | 0 | 0 | 0 | 0 | 0.0063 | 0 | 0.0059 | 0 | 0.0057 | 98.9 | 99.28 | 99.39 | 99.26 | 99.26 | 99.26 | 96.72 | 96.74 | 96.64 | 96.72 | 97.82 | 95.44 | 90.43 | 94.01 | 91.52 | 91.81 | 91.13 | 93.29 | 90.89 | 91.72 | 94.01 | 93.92 | 91.63 | 91.69 | 91.43 | 91.39 | |
| gom93-299 | 0.0095 | 0.0097 | 0.0114 | 0.0091 | 0.0089 | 0.011 | 0.011 | 0.011 | 0.011 | 0.0114 | 0.011 | 0.0114 | 0.011 | 0.0112 | 0.011 | 98.77 | 98.88 | 98.71 | 98.71 | 98.71 | 96.48 | 96.52 | 96.35 | 96.5 | 97.39 | 95.4 | 90.33 | 94.1 | 91.52 | 91.86 | 91.11 | 93.21 | 90.77 | 91.69 | 94.44 | 93.94 | 91.6 | 91.52 | 91.28 | 91.29 | |
| gom93-305 | 0.0155 | 0.0123 | 0.0046 | 0.0138 | 0.0144 | 0.0072 | 0.0072 | 0.0072 | 0.0072 | 0.0076 | 0.0072 | 0.0076 | 0.0072 | 0.0044 | 0.0072 | 0.0123 | 99.26 | 99.52 | 99.52 | 99.52 | 96.75 | 96.79 | 96.68 | 96.77 | 97.95 | 95.37 | 90.41 | 94.01 | 91.57 | 91.65 | 91.13 | 93.19 | 90.99 | 91.58 | 93.87 | 93.85 | 91.56 | 91.6 | 91.46 | 91.54 | |
| gom93-310 | 0.014 | 0.0125 | 0.0082 | 0.0131 | 0.0133 | 0.0061 | 0.0061 | 0.0061 | 0.0061 | 0.0015 | 0.0061 | 0.0002 | 0.0061 | 0.008 | 0.0061 | 0.0112 | 0.0074 | 99.03 | 99.03 | 99.03 | 96.68 | 96.72 | 96.61 | 96.7 | 97.72 | 95.44 | 90.3 | 93.91 | 91.4 | 91.77 | 91.06 | 93.07 | 90.77 | 91.72 | 93.89 | 93.85 | 91.61 | 91.59 | 91.4 | 91.41 | |
| gom93-501 | 0.0166 | 0.0129 | 0.0019 | 0.0153 | 0.0159 | 0.0074 | 0.0074 | 0.0074 | 0.0074 | 0.0099 | 0.0074 | 0.0099 | 0.0074 | 0.0017 | 0.0074 | 0.0129 | 0.0048 | 0.0097 | 100 | 100 | 96.5 | 96.54 | 96.43 | 96.52 | 97.85 | 95.33 | 90.31 | 93.89 | 91.4 | 91.65 | 91.11 | 93.14 | 90.91 | 91.53 | 93.75 | 93.8 | 91.39 | 91.57 | 91.28 | 91.36 | |
| gom93-543 | 0.0166 | 0.0129 | 0.0019 | 0.0153 | 0.0159 | 0.0074 | 0.0074 | 0.0074 | 0.0074 | 0.0099 | 0.0074 | 0.0099 | 0.0074 | 0.0017 | 0.0074 | 0.0129 | 0.0048 | 0.0097 | 0 | 100 | 96.5 | 96.54 | 96.43 | 96.52 | 97.85 | 95.33 | 90.31 | 93.89 | 91.4 | 91.65 | 91.11 | 93.14 | 90.91 | 91.53 | 93.75 | 93.8 | 91.39 | 91.57 | 91.28 | 91.36 | |
| gom93-544 | 0.0166 | 0.0129 | 0.0019 | 0.0153 | 0.0159 | 0.0074 | 0.0074 | 0.0074 | 0.0074 | 0.0099 | 0.0074 | 0.0099 | 0.0074 | 0.0017 | 0.0074 | 0.0129 | 0.0048 | 0.0097 | 0 | 0 | 96.5 | 96.54 | 96.43 | 96.52 | 97.85 | 95.33 | 90.31 | 93.89 | 91.4 | 91.65 | 91.11 | 93.14 | 90.91 | 91.53 | 93.75 | 93.8 | 91.39 | 91.57 | 91.28 | 91.36 | |
| IL 96-255 - B. kurtenbachii | 0.035 | 0.0316 | 0.0339 | 0.0341 | 0.0339 | 0.0328 | 0.0328 | 0.0328 | 0.0328 | 0.0334 | 0.0328 | 0.0334 | 0.0328 | 0.0337 | 0.0328 | 0.0352 | 0.0325 | 0.0332 | 0.035 | 0.035 | 0.035 | 99.66 | 99.39 | 99.52 | 96.95 | 95.46 | 90.39 | 94.01 | 91.74 | 91.99 | 91.21 | 93.4 | 90.87 | 91.99 | 93.94 | 93.66 | 91.56 | 91.89 | 91.87 | 91.51 | |
| IL 97-236u | 0.0345 | 0.0312 | 0.0335 | 0.0337 | 0.0334 | 0.0326 | 0.0326 | 0.0326 | 0.0326 | 0.033 | 0.0326 | 0.033 | 0.0326 | 0.0332 | 0.0326 | 0.0348 | 0.0321 | 0.0328 | 0.0346 | 0.0346 | 0.0346 | 0.0034 | 99.56 | 99.56 | 97.01 | 95.46 | 90.36 | 94.12 | 91.74 | 91.91 | 91.14 | 93.43 | 90.82 | 91.84 | 93.98 | 93.66 | 91.56 | 91.77 | 91.89 | 91.39 | |
| 25015 | 0.0363 | 0.033 | 0.0345 | 0.0354 | 0.0352 | 0.0336 | 0.0336 | 0.0336 | 0.0336 | 0.0341 | 0.0336 | 0.0341 | 0.0336 | 0.0343 | 0.0336 | 0.0365 | 0.0332 | 0.0339 | 0.0357 | 0.0357 | 0.0357 | 0.0061 | 0.0044 | 99.41 | 96.9 | 95.28 | 90.24 | 93.92 | 91.68 | 91.77 | 91.02 | 93.31 | 90.8 | 91.73 | 93.85 | 93.5 | 91.59 | 91.7 | 91.83 | 91.35 | |
| NS07-121 | 0.0347 | 0.0314 | 0.0337 | 0.0339 | 0.0336 | 0.0328 | 0.0328 | 0.0328 | 0.0328 | 0.0328 | 0.0328 | 0.0332 | 0.0328 | 0.0334 | 0.0328 | 0.035 | 0.0323 | 0.033 | 0.0348 | 0.0348 | 0.0348 | 0.0048 | 0.0044 | 0.0059 | 96.97 | 95.37 | 90.32 | 93.98 | 91.7 | 91.89 | 91.04 | 93.24 | 90.7 | 91.75 | 93.82 | 93.59 | 91.61 | 91.72 | 91.77 | 91.27 | |
| SCJ1 - B. carolinensis | 0.0268 | 0.0231 | 0.0213 | 0.0255 | 0.0257 | 0.0218 | 0.0218 | 0.0218 | 0.0218 | 0.0231 | 0.0218 | 0.0231 | 0.0218 | 0.0211 | 0.0218 | 0.0261 | 0.0205 | 0.0228 | 0.0215 | 0.0215 | 0.0215 | 0.0305 | 0.0299 | 0.031 | 0.0303 | 95.38 | 90.37 | 93.97 | 91.41 | 91.43 | 91.05 | 93.22 | 90.82 | 91.53 | 94.08 | 93.92 | 91.62 | 91.55 | 91.8 | 91.42 | |
| CA443 - B. californensis | 0.0453 | 0.0458 | 0.0474 | 0.0449 | 0.0447 | 0.0456 | 0.0456 | 0.0456 | 0.0456 | 0.0453 | 0.0456 | 0.0458 | 0.0456 | 0.0472 | 0.0456 | 0.046 | 0.0463 | 0.0456 | 0.0467 | 0.0467 | 0.0467 | 0.0454 | 0.0454 | 0.0472 | 0.0463 | 0.0462 | 91.3 | 94.47 | 92.35 | 92.39 | 91.92 | 93.9 | 91.55 | 92.37 | 94.89 | 94.45 | 92.07 | 92.01 | 92.08 | 91.63 | |
| QLM4P1 - B. yangtze | 0.0944 | 0.0944 | 0.0977 | 0.0927 | 0.0934 | 0.0957 | 0.0957 | 0.0957 | 0.0957 | 0.0967 | 0.0957 | 0.0972 | 0.0957 | 0.0974 | 0.0957 | 0.0967 | 0.0959 | 0.097 | 0.0969 | 0.0969 | 0.0969 | 0.0961 | 0.0964 | 0.0976 | 0.0968 | 0.0963 | 0.087 | 91.43 | 91.83 | 92.05 | 96.45 | 90.31 | 90.94 | 92.22 | 91.3 | 90.8 | 92.18 | 94.18 | 91.71 | 91.39 | |
| B31 - B. burgdorferi | 0.0587 | 0.059 | 0.0609 | 0.0581 | 0.0581 | 0.0599 | 0.0599 | 0.0599 | 0.0599 | 0.0611 | 0.0599 | 0.0611 | 0.0599 | 0.0606 | 0.0599 | 0.059 | 0.0599 | 0.0609 | 0.0611 | 0.0611 | 0.0611 | 0.0599 | 0.0588 | 0.0608 | 0.0602 | 0.0603 | 0.0553 | 0.0857 | 92.18 | 92.21 | 91.9 | 93.97 | 91.44 | 92.12 | 95.43 | 94.79 | 92.12 | 92.07 | 91.6 | 91.54 | |
| VS461 - B. afzelii | 0.0819 | 0.0835 | 0.0855 | 0.0809 | 0.0806 | 0.0848 | 0.0848 | 0.0848 | 0.0848 | 0.0857 | 0.0848 | 0.0863 | 0.0848 | 0.0853 | 0.0848 | 0.0848 | 0.0843 | 0.086 | 0.086 | 0.086 | 0.086 | 0.0826 | 0.0826 | 0.0832 | 0.083 | 0.0859 | 0.0765 | 0.0817 | 0.0782 | 93.31 | 92.47 | 91.9 | 93.32 | 93.55 | 92.61 | 91.52 | 93.04 | 92.61 | 93.22 | 92.51 | |
| 20047 - B. garinii | 0.0797 | 0.0802 | 0.0828 | 0.0780 | 0.0790 | 0.0819 | 0.0819 | 0.0819 | 0.0819 | 0.0816 | 0.0819 | 0.0826 | 0.0819 | 0.0826 | 0.0819 | 0.0814 | 0.0835 | 0.0823 | 0.0835 | 0.0835 | 0.0835 | 0.0801 | 0.0809 | 0.0823 | 0.0811 | 0.0857 | 0.0761 | 0.0795 | 0.0779 | 0.0669 | 92.94 | 91.71 | 92.22 | 98 | 92.78 | 91.76 | 92.99 | 93.09 | 93.22 | 92.7 | |
| VS116 - B. valaisiana | 0.0867 | 0.0877 | 0.0902 | 0.0852 | 0.0860 | 0.0887 | 0.0887 | 0.0887 | 0.0887 | 0.0897 | 0.0887 | 0.0897 | 0.0887 | 0.0899 | 0.0887 | 0.0889 | 0.0887 | 0.0894 | 0.0889 | 0.0889 | 0.0889 | 0.0879 | 0.0886 | 0.0898 | 0.0896 | 0.0895 | 0.0808 | 0.0355 | 0.081 | 0.0753 | 0.0706 | 90.69 | 91.77 | 93.11 | 91.97 | 91.45 | 92.47 | 95.12 | 92.46 | 91.88 | |
| B. andersoni | 0.0664 | 0.0664 | 0.0686 | 0.0669 | 0.0657 | 0.0671 | 0.0671 | 0.0671 | 0.0671 | 0.069 | 0.0671 | 0.0695 | 0.0671 | 0.0683 | 0.0671 | 0.0679 | 0.0681 | 0.0693 | 0.0686 | 0.0686 | 0.0686 | 0.066 | 0.0657 | 0.0669 | 0.0676 | 0.0678 | 0.061 | 0.0969 | 0.0603 | 0.081 | 0.0829 | 0.0931 | 90.49 | 91.52 | 94.2 | 94.02 | 91.41 | 91.27 | 91.37 | 90.51 | |
| A14S - B. spielmanii | 0.0903 | 0.0925 | 0.0916 | 0.0898 | 0.0896 | 0.0911 | 0.0911 | 0.0911 | 0.0911 | 0.0921 | 0.0911 | 0.0926 | 0.0911 | 0.0913 | 0.0911 | 0.0923 | 0.0901 | 0.0923 | 0.0909 | 0.0909 | 0.0909 | 0.0913 | 0.0918 | 0.092 | 0.093 | 0.0918 | 0.0845 | 0.0906 | 0.0856 | 0.0668 | 0.0778 | 0.0823 | 0.0951 | 92.41 | 91.7 | 90.98 | 92.08 | 91.72 | 91.95 | 91.52 | |
| PBi - B. bavariensis | 0.0809 | 0.0811 | 0.0843 | 0.0794 | 0.0804 | 0.0828 | 0.0828 | 0.0828 | 0.0828 | 0.083 | 0.0828 | 0.083 | 0.0828 | 0.084 | 0.0828 | 0.0831 | 0.0842 | 0.0828 | 0.0847 | 0.0847 | 0.0847 | 0.0801 | 0.0816 | 0.0827 | 0.0825 | 0.0847 | 0.0763 | 0.0778 | 0.0788 | 0.0645 | 0.02 | 0.0689 | 0.0848 | 0.0759 | 92.71 | 91.81 | 93.28 | 93.21 | 93.39 | 92.83 | |
| CA2 - genomospec 2 | 0.0546 | 0.0576 | 0.0625 | 0.0544 | 0.0539 | 0.0599 | 0.0599 | 0.0599 | 0.0599 | 0.0609 | 0.0599 | 0.0613 | 0.0599 | 0.0623 | 0.0599 | 0.0556 | 0.0613 | 0.0611 | 0.0625 | 0.0625 | 0.0625 | 0.0606 | 0.0602 | 0.0615 | 0.0618 | 0.0592 | 0.0511 | 0.087 | 0.0457 | 0.0739 | 0.0722 | 0.0803 | 0.058 | 0.083 | 0.0729 | 95.01 | 92.62 | 91.97 | 92.08 | 91.7 | |
| CA8 - B. americana | 0.0596 | 0.0601 | 0.0622 | 0.0592 | 0.0587 | 0.0608 | 0.0608 | 0.0608 | 0.0608 | 0.0613 | 0.0608 | 0.0613 | 0.0608 | 0.062 | 0.0608 | 0.0606 | 0.0615 | 0.0615 | 0.062 | 0.062 | 0.062 | 0.0634 | 0.0634 | 0.065 | 0.0641 | 0.0608 | 0.0555 | 0.092 | 0.0521 | 0.0848 | 0.0824 | 0.0855 | 0.0598 | 0.0902 | 0.0819 | 0.0499 | 91.67 | 91.78 | 91.27 | 91.06 | |
| PoTiBL37 - B. lusitaniae | 0.0818 | 0.083 | 0.0852 | 0.0820 | 0.0811 | 0.0837 | 0.0837 | 0.0837 | 0.0837 | 0.0837 | 0.0837 | 0.0842 | 0.0837 | 0.0849 | 0.0837 | 0.084 | 0.0844 | 0.0839 | 0.0861 | 0.0861 | 0.0861 | 0.0844 | 0.0844 | 0.0841 | 0.0839 | 0.0838 | 0.0793 | 0.0782 | 0.0788 | 0.0696 | 0.0701 | 0.0753 | 0.0859 | 0.0792 | 0.0672 | 0.0738 | 0.0833 | 92.78 | 92.73 | 92.18 | |
| HK501 - B. tanukii | 0.0828 | 0.0828 | 0.0841 | 0.0819 | 0.0819 | 0.0831 | 0.0831 | 0.0831 | 0.0831 | 0.0843 | 0.0831 | 0.0843 | 0.0831 | 0.0838 | 0.0831 | 0.0848 | 0.084 | 0.0841 | 0.0843 | 0.0843 | 0.0843 | 0.0811 | 0.0823 | 0.083 | 0.0828 | 0.0845 | 0.0799 | 0.0582 | 0.0793 | 0.0739 | 0.0691 | 0.0488 | 0.0873 | 0.0828 | 0.0679 | 0.0803 | 0.0824 | 0.0722 | 92.75 | 92.3 | |
| HO14 - B. japonica | 0.0828 | 0.0847 | 0.0872 | 0.0821 | 0.0818 | 0.0857 | 0.0857 | 0.0857 | 0.0857 | 0.0862 | 0.0857 | 0.0857 | 0.0857 | 0.0869 | 0.0857 | 0.0872 | 0.0854 | 0.086 | 0.0872 | 0.0872 | 0.0872 | 0.0813 | 0.0811 | 0.0817 | 0.0823 | 0.082 | 0.0792 | 0.0829 | 0.0804 | 0.0678 | 0.0678 | 0.0754 | 0.0863 | 0.0805 | 0.0661 | 0.0792 | 0.0873 | 0.0727 | 0.0725 | 92.03 | |
| Ya501 - B. turdi | 0.0847 | 0.0859 | 0.0859 | 0.0827 | 0.0837 | 0.0861 | 0.0861 | 0.0861 | 0.0861 | 0.0861 | 0.0861 | 0.0861 | 0.0861 | 0.0856 | 0.0861 | 0.0871 | 0.0846 | 0.0859 | 0.0864 | 0.0864 | 0.0864 | 0.0849 | 0.0861 | 0.0865 | 0.0873 | 0.0858 | 0.0837 | 0.0861 | 0.0846 | 0.0749 | 0.073 | 0.0814 | 0.0949 | 0.0848 | 0.0717 | 0.083 | 0.0894 | 0.0782 | 0.077 | 0.0797 |
Table 4.
Genetic diversity within B. bissettiia.
| Locus | Gene product/Description | Fragment length analysed (bp) | % G/C | π value | No. of alleles |
|---|---|---|---|---|---|
| clpA | Clp protease subunit A | 579 | 26.6 | 0.00929 | 8 |
| clpX | Clp protease subunit X | 624 | 32.2 | 0.00349 | 4 |
| nifS | aminotransferase | 564 | 28.6 | 0.00427 | 6 |
| pepX | dipeptidyl aminopeptidase | 570 | 30.6 | 0.00349 | 7 |
| pyrG | CTP synthase | 603 | 32.0 | 0.02009 | 7 |
| recG | DNA recombinase | 651 | 31.0 | 0.00631 | 6 |
| rplB | 50S ribosomal protein L2 | 624 | 36.5 | 0.00865 | 5 |
| uvrA | exonuclease ABC, SU A | 570 | 35.7 | 0.00882 | 9 |
| concatenated | 4785 | 31.7 | 0.00767 |
Only samples identified as B. bissettii (Table 1) are included.
The genetic similarity of the strains that formed the second clade (Fig. 1) (i.e. 25015, 96-255, 97-236u, NS07-121) ranged from 98.7 to 99.9%. The genetic distance of these strains from those of B. bissettii and B. carolinensis was >3%, and it was even higher for all the other Borrelia species included (Table 3). Therefore, we conclude that the strains belonging to the second clade represent a novel genospecies. In fact, the genetic distances between those strains and the remaining B. bissettii strains was higher than it was between B. bissettii and B. carolinensis (Fig. 1, Table 2), a recently described species from South Carolina (Rudenko et al., 2009a). The second clade included 2 samples from Illinois (96-255, 97-236u), one from New York (25015), and one from Nova Scotia, (NS07-121) (Table 1).
Overall, the phylogenetic tree produced 2 major clades – one that clustered the Eurasian species, the other clustering the North American species. One genospecies, B. sinica, was not included in this study, but we consider it highly unlikely that the strains we propose as a new genospecies are similar to B. sinica. Firstly, B. sinica is only known from I. ovatus and murid rodents in southern China (Masuzawa et al. 2001). Secondly, B. bissettii strain DN127, a phylogenetic neighbour of 25015, was quite distant from B. sinica when analyzed by DNA-DNA hybridization (Masuzawa et al. 2001). Thirdly, published DraI and MseI restriction fragment length patterns of the 5S-23S rrf-rrl intergenic spacer of strain 25015 and B. sinica differ considerably (Masuzawa et al., 2001; Chu et al., 2008; Lin et al., 2001).
Discussion
In this study, we have reinvestigated the phylogenetic relatedness of spirochaete strains previously assigned to the genospecies B. bissettii (Postic et al., 1998; Picken and Picken, 2000). We found that while strains from California and Colorado clustered with the B. bissettii type strain DN127-Cl9-2/p7, other strains from Illinois and New York fell into a different cluster. A genetic distance analysis indicated that these strains are more distant to B. bissettii than they are to B. carolinensis, a newly described Borrelia species (Rudenko et al., 2009a). Thus, we propose to name this group Borrelia kurtenbachii sp. nov., in honour of the late Klaus Kurtenbach who contributed significantly to our understanding of the ecology and evolution of the LB spirochaetes (Kurtenbach et al., 2006).
All isolates from Colorado were obtained either from I. spinipalpis removed from the Mexican woodrat Neotoma mexicana or from the woodrats themselves, whereas strains from California were either isolated from I. pacificus or I. spinipalpis that had been removed from the dusky-footed woodrat Neotoma fuscipes. These strains clustered together phylogenetically, and the genetic distance analysis indicated that they are true members of the species B. bissettii. Natural transmission cycles of B. bissettii involve either I. spinipalpis or I. pacificus; in contrast, strain 25015 and the Canadian strain were both detected in the eastern North American tick vector, I. scapularis. Although it has been demonstrated experimentally that B. bissettii can be transmitted between hosts by I. scapularis (Oliver et al., 2003), our data support the notion that B. kurtenbachii strains may be maintained in enzootic transmission cycles that differ ecologically from those of B. bissettii. This needs, however, to be further explored since isolation of strain 25015 from blood-fed larvae does not comprise proof of vector competence. Both strains from Illinois were isolated from the rodent Microtus pennsylvanicus (Picken and Picken, 2000). In the southeastern United States, strains assigned to B. bissettii have been isolated from the eastern woodrat (Neotoma floridana), the cotton rat (Sigmodon hispidus), the cotton mouse (Peromyscus gossypinus), and the vectors I. affinis and I. minor (Lin et al., 2001, 2005; Oliver et al., 2003). However, the data presented here render the association of B. bissettii with I. minor uncertain because the strain isolated from this tick showed a DraI restriction pattern similar to that of strain 25015, and it also differed in its MseI restriction pattern from all other B. bissettii isolates (Lin et al., 2001). Taken together, our data suggest that the transmission cycles of B. bissettii and B. kurtenbachii differ, but the vector(s) for B. kurtenbachii needs to be determined throughout its geographic distribution, particularly in Illinois.
In Europe, B. bissettii DNA has been detected in, and was isolated from, human patients (Picken et al., 1996a, 1996b; Strle et al., 1997; Fingerle et al., 2008; Rudenko et al., 2008, 2009c), but it rarely has been found in questing I. ricinus ticks (Hulinska et al., 2007). It is, therefore, tempting to speculate that the transmission of B. bissettii is maintained by an unknown vector in an enzootic cycle with occasional spill over into the human population. Alternatively, if B. bissettii is transmitted by I. ricinus, the spirochaete distribution may be highly focal and restricted by its host associations since human cases have been recorded quite regionally. However, any association of B. bissettii with particular tick vectors and hosts in Europe is currently at best speculative and requires further investigation. This is also true for B. kurtenbachii(-related) strains that may occur in Europe (Picken et al., 1996a, 1996b). Of the tick species known to transmit B. bissettii in the United States, i.e., I. pacificus and I. spinipalpis in the Far West and Southwest and I. affinis in the Southeast (Bissett and Hill, 1987; Maupin et al., 1994; Lin et al., 2001, 2003), only I. pacificus attaches to humans with any frequency. This may partly explain why B. bissettii has not been incriminated as a human pathogen in the United States so far (Maupin et al., 1994). In addition, using a murine model for LB, it was demonstrated that B. bissettii strains in the United States differed considerably in their potential to induce arthritis in mice (Schneider et al., 2008). Early investigations into the pathogenic potential of strain 25015 revealed that this strain was non-arthritogenic in mice (Anderson et al., 1990), although subsequent studies demonstrated that it might be mildly pathogenic when passaged repeatedly through laboratory mice (Fikrig et al., 1992). In North America, strain 25015 and related strains have been detected in I. scapularis, a tick that frequently attaches to humans. However, the rarity of finding B. bissettii-like strains in I. scapularis in recent years (Hoen et al., 2009, Hamer et al., 2010) or in patients underpins the need for further studies to clarify whether I. scapularis is the primary vector for B. kurtenbachii or whether the rare findings of this Borrelia species in I. scapularis can be attributed to spill-over from another enzootic transmission cycle. Although the number of B. kurtenbachii strains evaluated during the present study is restricted, most of the strains found in the previous study in Illinois revealed a macrorestriction pattern similar to that of strain 25015 (Picken and Picken, 2000). Such strains also have been described from the southeastern United States and perhaps Wisconsin indicating that B. kurtenbachii may be more widespread in its distribution and common in certain habitats than realized so far (Picken and Picken, 2000; Lin et al., 2001, 2005; Oliver et al., 2003; Caporale et al., 2005). At the time when B. bissettii was described, strain 25015 was the only example found in I. scapularis in upstate New York (Postic et al., 1998) suggesting that these strains are more common in Illinois and the southern United States. As mentioned above, the discovery of these strains in New York and eastern Canada are unusual because such strains have not been detected in host-seeking I. scapularis during recent studies in the northeastern or upper midwestern United States (Hoen et al., 2009; Hamer et al., 2010). In Illinois, the bacteria were isolated from voles and mice in consecutive years, and a rodent-associated transmission cycle might limit the distributional range. More research is needed to clarify the relationships among strains in these locations.
In the United States, the species with the widest distribution is B. burgdorferi (s.s.), which occurs in the southern, northeastern, Midwestern, and western states (Fig. 2). B. andersonii and B. kurtenbachii seem to be restricted to the east of the Rocky Mountains, whereas B. carolinensis and B. californiensis appear to be limited to South Carolina and California, respectively. Two species, B. bissettii and B. americana, are found in the southeastern and far-western regions, with B. bissettii also being present in Colorado. The occurrence of several genospecies across the continent suggests a dynamic evolutionary history of LB spirochaetes in the United States and is in agreement with recent suggestions that B. burgdorferi has been present in the country for several thousand or even millions of years (Hoen et al., 2009). They also imply that demographic events related to glacial and interglacial periods may have shaped the current population structure. Collectively, our own and previous findings on the diversity of LB spirochaetes in the United States underscore the need for additional research to fully comprehend the ecology and evolutionary history of these spirochaetes and the impact of this spirochaetal diversity, if any, on human health.
Fig. 2.

Present day distribution of LB species in the United States based on published records.
Description of Borrelia kurtenbachii sp. nov
Borrelia kurtenbachii (kur.ten.ba’chi. L. gen in honour of the late Klaus Kurtenbach and his numerous contributions to research on the Lyme borreliosis group of spirochaetes).
The type strain 25015T was isolated from an I. scapularis larva in upstate New York (Anderson et al., 1990). The morphology matches that of previously described Borrelia species in that it displays the typical helical morphology of spirochaetal bacteria (Barbour and Hayes, 1986). Its in vitro culture properties are as described for the genus (Johnson et al., 1984). B. kurtenbachii can be distinguished from all other LB species by means of its restriction pattern of the rrf-rrl intergenic spacer (Postic et al., 1998) and its housekeeping gene sequences using multilocus sequence analysis.
Acknowledgments
This work was funded by NIH-NIAID under grant number 5R21AI065848.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JF, Magnarelli LA, McAninch JB. New Borrelia burgdorferi antigenic variant isolated from Ixodes dammini from upstate New York. J Clin Microbiol. 1988;26:2209–2212. doi: 10.1128/jcm.26.10.2209-2212.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JF, Barthold SW, Magnarelli LA. Infectious but nonpathogenic isolate of Borrelia burgdorferi. J Clin Microbiol. 1990;28:2693–2699. doi: 10.1128/jcm.28.12.2693-2699.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon RM, Kugeler KJ, Mead PS. Surveillance for Lyme disease--United States, 1992–2006. MMWR Surveill Summ. 2008;57:1–9. [PubMed] [Google Scholar]
- Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti JC, Assous M, Grimont PA. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Hayes SF. Biology of Borrelia species. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop CJ, Aanensen DM, Jordan GE, Kilian M, Hanage WP, Spratt BG. Assigning strains to bacterial species via the internet. BMC Biol. 2009;7 doi: 10.1186/1741-7007-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissett ML, Hill W. Characterization of Borrelia burgdorferi strains isolated from Ixodes pacificus ticks in California. J Clin Microbiol. 1987;25:2296–2301. doi: 10.1128/jcm.25.12.2296-2301.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RN, Lane RS. Lyme disease in California: a novel enzootic transmission cycle of Borrelia burgdorferi. Science. 1992;256:1439–1442. doi: 10.1126/science.1604318. [DOI] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease – a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- Burgdorfer W, Lane RS, Barbour AG, Gresbrink RA, Anderson JR. The western black-legged tick, Ixodes pacificus: a vector of Borrelia burgdorferi. Am J Trop Med Hyg. 1985;34:925–930. doi: 10.4269/ajtmh.1985.34.925. [DOI] [PubMed] [Google Scholar]
- Caporale DA, Johnson CM, Millard BJ. Presence of Borrelia burgdorferi (Spirochaetales: Spirochaetaceae) in southern Kettle Moraine State Forest, Wisconsin, and characterization of strain W97F51. J Med Entomol. 2005;42:457–472. doi: 10.1093/jmedent/42.3.457. [DOI] [PubMed] [Google Scholar]
- Chu CY, Liu W, Jiang BG, Wang DM, Jiang WJ, Zhao QM, Zhang PH, Wang ZX, Tang GP, Yang H, Cao WC. Novel genospecies of Borrelia burgdorferi sensu lato from rodents and ticks in Southwestern China. J Clin Microbiol. 2008;46(9):3130–3133. doi: 10.1128/JCM.01195-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diuk-Wasser MA, Gatewood AG, Cortinas MR, Yaremych-Hamer S, Tsao J, Kitron U, Hickling G, Brownstein JS, Walker E, Piesman J, Fish D. Spatiotemporal patterns of host-seeking Ixodes scapularis nymphs (Acari: Ixodidae) in the United States. J Med Entomol. 2006;43:166–176. doi: 10.1603/0022-2585(2006)043[0166:spohis]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Eisen RJ, Lane RS, Fritz CL, Eisen L. Spatial patterns of Lyme disease risk in California based on disease incidence data and modeling of vector-tick exposure. Am J Trop Med Hyg. 2006;75:669–676. [PubMed] [Google Scholar]
- Enright MC, Spratt BG. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Enright MC. Analyses of clonality and the evolution of bacterial pathogens. Curr Opin Microbiol. 2004;7:308–313. doi: 10.1016/j.mib.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Fikrig E, Barthold SW, Persing DH, Sun X, Kantor FS, Flavell RA. Borrelia burgdorferi strain 25015: characterization of outer surface protein A and vaccination against infection. J Immunol. 1992;148:2256–2260. [PubMed] [Google Scholar]
- Fingerle V, Schulte-Spechtel UC, Ružić-Sabljić E, Leonhard S, Hofmann H, Weber K, Pfister K, Strle F, Wilske B. Epidemiological aspects and molecular characterization of Borrelia burgdorferi s.l. from southern Germany with special respect to the new species Borrelia spielmanii sp. nov. Int J Med Microbiol. 2008;298:279–290. doi: 10.1016/j.ijmm.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Fukunaga M, Hamase A, Okada K, Inoue H, Tsuruta Y, Miyamoto K, Nakao M. Characterization of spirochetes isolated from ticks (Ixodes tanuki, Ixodes turdus, and Ixodes columnae) and comparison of the sequences with those of Borrelia burgdorferi sensu lato strains. Appl Environ Microbiol. 1996;62:2338–2344. doi: 10.1128/aem.62.7.2338-2344.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood AG, Liebman KA, Vourc’h G, Bunikis J, Hamer SA, Cortinas MR, Melton F, Cislo P, Kitron U, Tsao J, Barbour AG, Fish D, Diuk-Wasser MA. Climate and tick seasonality predict Borrelia burgdorferi genotype distribution. Appl Environ Microbiol. 2009;75:2476–2483. doi: 10.1128/AEM.02633-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de Peer Y, Vandamme P, Thompson FL, Swings J. Opinion: Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Hamer SA, Tsao JI, Walker ED, Hickling GJ. Invasion of the Lyme disease vector Ixodes scapularis: Implications for Borrelia burgdorferi endemicity. Ecohealth. 2010 doi: 10.1007/s10393-010-0287-0. [DOI] [PubMed] [Google Scholar]
- Hoen AG, Margos G, Bent SJ, Kurtenbach K, Fish D. Phylogeography of Borrelia burgdorferi in the eastern United States reveals multiple independent Lyme disease emergence events. Proc Natl Acad Sci USA. 2009;106:15013–15018. doi: 10.1073/pnas.0903810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulinska D, Votypka J, Kriz B, Holinkova N, Novakova J, Hulinsky V. Phenotypic and genotypic analysis of Borrelia spp. isolated from Ixodes ricinus ticks by using electrophoretic chips and real-time polymerase chain reaction. Folia Microbiol (Praha) 2007;52:315–324. doi: 10.1007/BF02932085. [DOI] [PubMed] [Google Scholar]
- Humphrey PT, Caporale DA, Brisson D. Uncoordinated phylogeography of Borrelia burgdorferi and its tick vector, Ixodes scapularis. Evolution. 2010;64:2653–2663. doi: 10.1111/j.1558-5646.2010.01001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC, Schmidt GP, Hyde FW, Steigerwalt AG, Brenner DJ. Borrelia burgdorferi sp nov.: etiological agent of Lyme disease. Int J Syst Bacteriol. 1984;34:496–497. [Google Scholar]
- Kawabata H, Masuzawa T, Yanagihara Y. Genomic analysis of Borrelia japonica sp. nov. isolated from Ixodes ovatus in Japan. Microbiol Immunol. 1993;37:843–848. doi: 10.1111/j.1348-0421.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- Killilea ME, Swei A, Lane RS, Briggs CJ, Ostfeld RS. Spatial dynamics of Lyme disease: a review. Ecohealth. 2008;5:167–195. doi: 10.1007/s10393-008-0171-3. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. Integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Kurtenbach K, Hanincova K, Tsao JI, Margos G, Fish D, Ogden NH. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat Rev Microbiol. 2006;4:660–669. doi: 10.1038/nrmicro1475. [DOI] [PubMed] [Google Scholar]
- Kurtenbach K, Hoen AG, Bent SJ, Vollmer SA, Ogden NH, Margos G. Population biology of Lyme borreliosis spirochetes. In: Robinson DA, Falush D, Feil EJ, editors. Bacterial Population Genetics in Infectious Disease. John Wiley & Sons, Inc; 2010. [Google Scholar]
- Lane RS, Quistad GB. Borreliacidal factor in the blood of the western fence lizard (Sceloporus occidentalis) J Parasitol. 1998;84:29–34. [PubMed] [Google Scholar]
- Lane RS, Steinlein DB, Mun J. Human behaviors elevating exposure to Ixodes pacificus (Acari: Ixodidae) nymphs and their associated bacterial zoonotic agents in a hardwood forest. J Med Entomol. 2004;41:239–248. doi: 10.1603/0022-2585-41.2.239. [DOI] [PubMed] [Google Scholar]
- Le Fleche A, Postic D, Girardet K, Péter O, Baranton G. Characterization of Borrelia lusitaniae sp. nov. by 16S ribosomal DNA sequence analysis. Int J Syst Bacteriol. 1997;47:921–925. doi: 10.1099/00207713-47-4-921. [DOI] [PubMed] [Google Scholar]
- Lin T, Oliver JH, Jr, Gao L, Kollars TM, Jr, Clark KL. Genetic heterogeneity of Borrelia burgdorferi sensu lato in the southern United States based on restriction fragment length polymorphism and sequence analysis. J Clin Microbiol. 2001;39:2500–2507. doi: 10.1128/JCM.39.7.2500-2507.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Oliver JH, Jr, Gao L. Comparative analysis of Borrelia isolates from southeastern USA based on randomly amplified polymorphic DNA fingerprint and 16S ribosomal gene sequence analyses. FEMS Microbiol Lett. 2003;228:249–257. doi: 10.1016/S0378-1097(03)00763-8. [DOI] [PubMed] [Google Scholar]
- Lin T, Gao L, Seyfang A, Oliver JH., Jr ‘Candidatus Borrelia texasensis’, from the American dog tick Dermacentor variabilis. Int J Syst Evol Microbiol. 2005;55:685–693. doi: 10.1099/ijs.0.02864-0. [DOI] [PubMed] [Google Scholar]
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi RT, Liveris D, Schwartz I. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J Clin Microbiol. 1995;33:2427–2434. doi: 10.1128/jcm.33.9.2427-2434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margos G, Gatewood AG, Aanensen DM, Hanincova K, Terekhova D, Vollmer SA, Cornet M, Piesman J, Donaghy M, Bormane A, Hurn MA, Feil EJ, Fish D, Casjens S, Wormser GP, Schwartz I, Kurtenbach K. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc Natl Acad Sci USA. 2008;105:8730–8735. doi: 10.1073/pnas.0800323105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margos G, Vollmer SA, Cornet M, Garnier M, Fingerle V, Wilske B, Bormane A, Vitorino L, Collares-Pereira M, Drancourt M, Kurtenbach K. A new Borrelia species defined by multilocus sequence analysis of housekeeping genes. Appl Environ Microbiol. 2009;75:5410–5416. doi: 10.1128/AEM.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuzawa T, Fukui T, Miyake M, Oh HB, Cho MK, Chang WH, Imai Y, Yanagihara Y. Determination of members of a Borrelia afzelii-related group isolated from Ixodes nipponensis in Korea as Borrelia valaisiana. Int J Syst Bacteriol. 1999;49(Pt 4):1409–1415. doi: 10.1099/00207713-49-4-1409. [DOI] [PubMed] [Google Scholar]
- Masuzawa T, Takada N, Kudeken M, Fukui T, Yano Y, Ishiguro F, Kawamura Y, Imai Y, Ezaki T. Borrelia sinica sp. nov., a Lyme disease-related Borrelia species isolated in China. Int J Syst Evol Microbiol. 2001;51:1817–1824. doi: 10.1099/00207713-51-5-1817. [DOI] [PubMed] [Google Scholar]
- Mathiesen DA, Oliver JH, Jr, Kolbert CP, Tullson ED, Johnson BJ, Campbell GL, Mitchell PD, Reed KD, Telford SR, 3rd, Anderson JF, Lane RS, Persing DH. Genetic heterogeneity of Borrelia burgdorferi in the United States. J Infect Dis. 1997;175:98–107. doi: 10.1093/infdis/175.1.98. [DOI] [PubMed] [Google Scholar]
- Maupin GO, Gage KL, Piesman J, Montenieri J, Sviat SL, VanderZanden L, Happ CM, Dolan M, Johnson BJ. Discovery of an enzootic cycle of Borrelia burgdorferi in Neotoma mexicana and Ixodes spinipalpis from northern Colorado, an area where Lyme disease is nonendemic. J Infect Dis. 1994;170:636–643. doi: 10.1093/infdis/170.3.636. [DOI] [PubMed] [Google Scholar]
- Ogden NH, Bouchard C, Kurtenbach K, Margos G, Lindsay LR, Trudel L, Nguon S, Milord F. Active and passive surveillance, and phylogenetic analysis of Borrelia burgdorferi elucidate the process of Lyme disease risk emergence in Canada. Environ Health Perspect. 2010;118:909–914. doi: 10.1289/ehp.0901766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JH., Jr Lyme borreliosis in the southern United States: a review. J Parasitol. 1996;82:926–935. [PubMed] [Google Scholar]
- Oliver JH, Jr, Lin T, Gao L, Clark KL, Banks CW, Durden LA, James AM, Chandler FW., Jr An enzootic transmission cycle of Lyme borreliosis spirochetes in the southeastern United States. Proc Natl Acad Sci USA. 2003;100:11642–11645. doi: 10.1073/pnas.1434553100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picken RN, Cheng Y, Strle F, Cimperman J, Maraspin V, Lotric-Furlan S, Ružić-Sabljić E, Han D, Nelson JA, Picken MM, Trenholme GM. Molecular characterization of Borrelia burgdorferi sensu lato from Slovenia revealing significant differences between tick and human isolates. Eur J Clin Microbiol Infect Dis. 1996;15:313–323. doi: 10.1007/BF01695664. [DOI] [PubMed] [Google Scholar]
- Picken RN, Cheng Y, Strle F, Picken MM. Patient isolates of Borrelia burgdorferi sensu lato with genotypic and phenotypic similarities of strain 25015. J Infect Dis. 1996;174:1112–1115. doi: 10.1093/infdis/174.5.1112. [DOI] [PubMed] [Google Scholar]
- Picken RN, Picken MM. Molecular characterization of Borrelia spp. isolates from greater metropolitan Chicago reveals the presence of Borrelia bissettii Preliminary report. J Mol Microbiol Biotechnol. 2000;2:505–507. [PubMed] [Google Scholar]
- Postic D, Assous MV, Grimont PA, Baranton G. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int J Syst Bacteriol. 1994;44:743–752. doi: 10.1099/00207713-44-4-743. [DOI] [PubMed] [Google Scholar]
- Postic D, Ras NM, Lane RS, Hendson M, Baranton G. Expanded diversity among Californian borrelia isolates and description of Borrelia bissettii sp. nov. (formerly Borrelia group DN127) J Clin Microbiol. 1998;36:3497–3504. doi: 10.1128/jcm.36.12.3497-3504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic D, Garnier M, Baranton G. Multilocus sequence analysis of atypical Borrelia burgdorferi sensu lato isolates – description of Borrelia californiensis sp. nov., and genomospecies 1 and 2. Int J Med Microbiol. 2007;297:263–271. doi: 10.1016/j.ijmm.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Richter D, Postic D, Sertour N, Livey I, Matuschka FR, Baranton G. Delineation of Borrelia burgdorferi sensu lato species by multilocus sequence analysis and confirmation of the delineation of Borrelia spielmanii sp. nov. Int J Syst Evol Microbiol. 2006;56:873–881. doi: 10.1099/ijs.0.64050-0. [DOI] [PubMed] [Google Scholar]
- Rudenko N, Golovchenko M, Mokracek A, Piskunova N, Ruzek D, Mallatova N, Grubhoffer L. Detection of Borrelia bissettii in cardiac valve tissue of a patient with endocarditis and aortic valve stenosis in the Czech Republic. J Clin Microbiol. 2008;46:3540–3543. doi: 10.1128/JCM.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko N, Golovchenko M, Grubhoffer L, Oliver JH., Jr Borrelia carolinensis sp.nov – a new (14th) member of Borrelia burgdorferi sensu lato complex from the southeastern United States. J Clin Microbiol. 2009a;47:134–141. doi: 10.1128/JCM.01183-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko N, Golovchenko M, Lin T, Gao L, Grubhoffer L, Oliver JH., Jr Delineation of a new species of the Borrelia burgdorferi sensu lato complex, Borrelia americana sp.nov. J Clin Microbiol. 2009b;47:3875–3880. doi: 10.1128/JCM.01050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko N, Golovchenko M, Ruzek D, Piskunova N, Mallatova N, Grubhoffer L. Molecular detection of Borrelia bissettii DNA in serum samples from patients in the Czech Republic with suspected borreliosis. FEMS Microbiol Lett. 2009c;292:274–281. doi: 10.1111/j.1574-6968.2009.01498.x. [DOI] [PubMed] [Google Scholar]
- Schneider BS, Zeidner NS, Burkot TR, Maupin GO, Piesman J. Borrelia isolates in Northern Colorado identified as Borrelia bissettii. J Clin Microbiol. 2000;38:3103–3105. doi: 10.1128/jcm.38.8.3103-3105.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BS, Schriefer ME, Dietrich G, Dolan MC, Morshed MG, Zeidner NS. Borrelia bissettii isolates induce pathology in a murine model of disease. Vector Borne Zoonotic Dis. 2008;8:623–633. doi: 10.1089/vbz.2007.0251. [DOI] [PubMed] [Google Scholar]
- Slowik TJ, Lane RS. Birds and their ticks in northwestern California: minimal contribution to Borrelia burgdorferi enzootiology. J Parasitol. 2001;87:755–761. doi: 10.1645/0022-3395(2001)087[0755:BATTIN]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Spratt BG. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the Internet. Curr Opin Microbiol. 1999;2:312–316. doi: 10.1016/S1369-5274(99)80054-X. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- Strle F, Picken RN, Cheng Y, Cimperman J, Maraspin V, Lotric-Furlan S, Ruzic-Sabljic E, Picken MM. Clinical findings for patients with Lyme borreliosis caused by Borrelia burgdorferi sensu lato with genotypic and phenotypic similarities to strain 25015. Clin Infect Dis. 1997;25:273–280. doi: 10.1086/514551. [DOI] [PubMed] [Google Scholar]
- Wang G, van Dam AP, Le Fleche A, Postic D, Péter O, Baranton G, de Boer R, Spanjaard L, Dankert J. Genetic and phenotypic analysis of Borrelia valaisiana sp. nov. (Borrelia genomic groups VS116 and M19) Int J Syst Bacteriol. 1997;47:926–932. doi: 10.1099/00207713-47-4-926. [DOI] [PubMed] [Google Scholar]
- Wang G, van Dam AP, Dankert J. Phenotypic and genetic characterization of a novel Borrelia burgdorferi sensu lato isolate from a patient with Lyme borreliosis. J Clin Microbiol. 1999;37:3025–3028. doi: 10.1128/jcm.37.9.3025-3028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormser GP, Brisson D, Liveris D, Hanincova K, Sandigursky S, Nowakowski J, Nadelman RB, Ludin S, Schwartz I. Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J Infect Dis. 2008;198:1358–1364. doi: 10.1086/592279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SA, Thompson MA, Miller MJ, Knerl KM, Elms SL, Karpowicz JC, Young JF, Kramer VL. Ecology of Borrelia burgdorferi in ticks (Acari: Ixodidae), rodents, and birds in the Sierra Nevada foothills, Placer County, California. J Med Entomol. 2000;37:909–918. doi: 10.1603/0022-2585-37.6.909. [DOI] [PubMed] [Google Scholar]


