Abstract
In bacterial chromosomes, the position of a gene relative to the single origin of replication generally reflects its replication timing, how often it is expressed, and consequently, its rate of evolution. However, because some archaeal genomes contain multiple origins of replication, bias in gene dosage caused by delayed replication should be minimized and hence the substitution rate of genes should associate less with chromosome position. To test this hypothesis, six archaeal genomes from the genus Sulfolobus containing three origins of replication were selected, conserved orthologs were identified, and the evolutionary rates (dN and dS) of these orthologs were quantified. Ortholog families were grouped by their consensus position and designated by their proximity to one of the three origins (O1, O2, O3). Conserved orthologs were concentrated near the origins and most variation in genome content occurred distant from the origins. Linear regressions of both synonymous and nonsynonymous substitution rates on distance from replication origins were significantly positive, the rates being greatest in the region furthest from any of the origins and slowest among genes near the origins. Genes near O1 also evolved faster than those near O2 and O3, which suggest that this origin may fire later in the cell cycle. Increased evolutionary rates and gene dispensability are strongly associated with reduced gene expression caused in part by reduced gene dosage during the cell cycle. Therefore, in this genus of Archaea as well as in many Bacteria, evolutionary rates and variation in genome content associate with replication timing.
Keywords: Archaea, origin of replication, ortholog, substitution rate, expression
Introduction
Many archaeal proteins involved in DNA replication, transcription, translation, and recombination are more closely related to those found in eukaryotes than bacteria (Olsen and Woese 1997). As a result, archaeal replication mechanisms make good models for studying eukaryotic DNA machinery and may help us understand more complex evolutionary forces acting on the eukaryotic cell cycle. One notable feature distinguishing the genomes and cell cycles of some Archaea from those of Bacteria is the presence of multiple replication origins per chromosome. The single replication origin in Bacteria has been shown to generate gradients both in the rates of transcription and evolutionary change (Sharp et al. 1989; Henry and Sharp 2007), but additional replication origins should theoretically reduce this genome-wide bias in gene dosage. We therefore sought to test the prediction that the evolutionary rates and biases in codon usage that reflect different expression should be less associated with chromosome position in Archaea. In essence, the evolution of multiple replication origins within a relatively small archaeal genome may be equivalent to the evolution of an isochore that generates regional uniformity in mutational and evolutionary dynamics (Eyre-Walker and Hurst 2001).
However, as in eukaryotes, identifying origins of replication in archaeal genomes has been challenging. Much of our knowledge of the foci of archaeal DNA replication comes from bioinformatic studies with limited experimental analysis. Analysis of the skew in base composition, and particularly the skew in (G − C)/(G + C), is one method that has been effective in identifying replication origins in Bacteria (Boulikas 1996; Lobry 1996; Lobry and Sueoka 2002) and as a result many different algorithms have been developed to use this same approach (Grigoriev 1998; McLean et al. 1998; Mrazek and Karlin 1998; Salzberg et al. 1998; Rocha et al. 1999). However, conventional analyses of GC skew have been relatively ineffective in identifying replication origins in fully sequenced archaeal genomes, including Methanococcus jannaschii, Methanobacterium thermoautotrophicum, and Archaeoglobus fulgidus (Mrazek and Karlin 1998), although it proved accurate for the Pyrococcus abyssi genome (Myllykallio et al. 2000). More recently, marker frequency analysis (MFA), which employs whole-genome DNA microarrays to quantify gene dosage during the cell cycle, have enabled the experimental identification of three replication origins in each of two, well-studied Sulfolobus spp., S. solfataricus, and S. acidocaldarius (Lundgren et al. 2004; Robinson et al. 2004; Duggin et al. 2008) and as many as four in Halobacterium NRC-1 (Coker et al. 2009). Some of these additional replication origins had not been previously detected with computational methods (Berquist and DasSarma 2003; Zhang R and Zhang CT 2005).
Many of these studies also noted that replication origins contained a Orc1/Cdc6 homolog, a gene involved in eukaryotic replication initiation, directly downstream (Myllykallio et al. 2000; Berquist and DasSarma 2003). This finding prompted the hypothesis that cdc6 genes are essential for the function of origins and could predict their locations in Archaea. However, some archaeal replication origins have since been identified that lacked proximal cdc6 homologs (Robinson et al. 2004; Coker et al. 2009). Due to the ineffectiveness of conventional GC skew analysis and cdc6 homolog position to locate archaeal replication origins, this study utilizes the Z-curve method that has been shown to accurately locate the origins of replication in Sulfolobus spp. that had been previously found by MFA (Zhang R and Zhang CT 2005). The Z-curve is a 3D curve that integrates the GC skew of a sequence but also its purine-to-pyrimidine skew and amino-to-keto base skew. Adding these dimensions identified the sites of replication origins that had previously been undetectable using only GC skew (Zhang R and Zhang CT 2004).
It has become increasingly apparent that archaeal DNA replication is more complex than previously thought (Olsen and Woese 1997; Myllykallio et al. 2000; Berquist and DasSarma 2003; Kelman LM and Kelman Z 2004; Lundgren et al. 2004; Coker et al. 2009) and that these dynamics may generate heterogeneity within the genome. A recent comparison of seven S. islandicus genomes from three different locations revealed that genome variation tended to be concentrated in a specific chromosome region in which content was strongly associated with strain biogeography (Reno et al. 2009). This raises the question of why certain genome regions are more prone to vary, or alternatively, why dispensable or environment-specific genes tend to cluster in certain locations. Among related bacterial genomes composed of a single chromosome, more variation is typically found near the replication terminus because this region experiences delayed replication, reduced gene dosage, and hence reduced expression (Sharp et al. 1989). These effects also occur in bacterial genomes with multiple chromosomes, in which smaller secondary chromosomes tend to be replicated later and thus accumulate greater variation (Cooper et al. 2010). However, in relatively small (∼2.7 Mb) genomes with multiple origins of replication such as the Archaea discussed here, variation in gene dosage caused by different replication timing should theoretically be minimized. On the other hand, the slower rate of replication by Sulfolobus in comparison with other prokaryotes (Bernander 2007) could amplify gene dosage effects and the relative strength of selection on these genes, especially if replication initiation is asynchronous.
Here, we examine whether genes found near a replication origin in Archaea tend to be more highly conserved than genes distant from an origin. We identified replication origins in six fully sequenced strains of S. islandicus using the Z-curve method described above and validated these findings against experimental studies of replication in S. solfataricus (Lundgren et al. 2004; Robinson et al. 2004). Next, we identified panorthologs, defined as orthologs present in all genomes, and quantified the evolutionary rates of these genes as a function of their position in the chromosome relative to replication origins. Our analyses show that genes closer to origins of replication are more highly conserved and evolve more slowly than genes distant from an origin of replication, which evolve more quickly and are more dispensable. These patterns occur in spite of the action of multiple replication origins within one circular chromosome. These findings demonstrate that the geographic differentiation among genomes of S. islandicus (generated by recombination) is concentrated in regions prone to greater evolutionary rates likely because of their reduced gene dosage and probable reduced expression. More generally, gene proximity to replication origins may explain variation in evolutionary rates within and among many taxonomic groups.
Materials and Methods
Genomes
Annotated gene predictions of six S. islandicus genomes (L.S.2.15, M.14.25, M.16.27, M.16.4, Y.G.57.14, and Y.N.15.51) were downloaded from the Integrated Microbial Genome database (http://img.jgi.doe.gov) in FASTA nucleotide and amino acid formats. A seventh S. islandicus complete genome (Reno et al. 2009) was not included in these analyses because it was unavailable through IMG at the time of analysis.
Z-Curve Analysis
Complete genomes were downloaded from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) in FASTA nucleotide format. The Z-curves were generated as described previously by Zhang et al. (Zhang R and Zhang CT 2003, 2004, 2005) using the plotting software provided by Tianjin University’s Center of Bioinformatics (TUBIC; http://tubic.tju.edu.cn/Ori-Finder/) and simplified to only plot the MK disparity and RY disparity. MK disparity is the amino (A, C) to keto (G, T) base content where y << 0 is indicative of keto bases are in excess. The RY disparity is the purine (A, G) to pyrimidine (C, T) content where y << 0 is indicative of pyrimidines being in excess.
Identification of Panorthologs
Panortholog families were identified as described previously (Cooper et al. 2010). NCBI BlastP (release 2.2.16) was used to analyze all genes in all genomes for sequence similarity. All Blast hits within an E value threshold of 1 were kept for processing. Homologs were identified as gene pairs that had Blast hits in both directions within a given scaled bit score threshold, which has been used previously to identify conserved homologs in bacteria (Lerat et al. 2003). Homolog families were formed by grouping together genes that had been identified as homologs such that if A and B are homologs and B and C are homologs, then they are all grouped into one family. Putative panorthologs were then identified as the genes from homolog families with exactly one gene from each genome. We kept the largest set of panorthologs found by computing the putative panorthologs while varying the scaled bit score threshold from 0.1 to 0.9 in 0.1 increments; the number of panorthologs, 2,073, was maximized at a scaled bit score of 0.6. We subsequently screened the annotations of these panorthologs for any genes associated with mobile genetic elements (transposons or phages), found two families, and removed them.
Measurement of Evolutionary Rate
A pipeline described previously (Cooper et al. 2010) was used. The amino acid sequences of each putative panortholog family were first aligned using ClustalW2 (Larkin et al. 2007). We then used the codon boundaries to align the nucleotide sequences and trim their leading and trailing edges to a consensus, in-frame sequence. We used the cons utility from the EMBOSS suite to infer a consensus sequence. If any gene in the family differed from the consensus by more than five consecutive amino acid differences, the entire family was discarded from further analysis. We also discarded all families whose estimated dS (see below) exceed 1.0, as these estimates are unreliable. This reduced the number of putative panorthologs from 2,073 families to 1,995 families. Phylogenetic trees were then constructed for each family using DNAML (maximum likelihood) in PHYLIP (Felsenstein 1989); these trees were then used as guides for calculating dN and dS from the trimmed nucleic acid alignment using codeml in the PAML package (Yang 2007). Codeml model 0, which allows for a single dN and dS value throughout the phylogeny, was used. Statistical analysis of variation in evolutionary rates among genome regions was conducted using SPSS 17.0, either by analysis of variance (ANOVA) among coarse regions with post hoc tests or by linear regression of rates on ortholog proximity to the nearest replication origin.
Measurement of Codon Usage Bias
We used the SCUO method, which does not require a reference set of genes known to be highly expressed, to calculate codon usage bias (Angellotti et al. 2007). All genes in the genome, including panorthologs, were analyzed using this method and categorized by genome location relative to replication origins. Codon bias measures for each chromosome region were then compared by ANOVA and by Tukey–Kramer post hoc tests. We validated our general findings of codon usage bias by uploading the complete annotations for each genome location into the INCA software (Supek and Vlahovicek 2005) to calculate CAI (Sharp and Li 1987) and MELP (Supek and Vlahovicek 2005) using genes encoding ribosomal proteins as highly expressed reference genes and found the same general patterns for each genome location.
Results
Replication Origins Have Distinct and Diagnostic Nucleotide Compositions
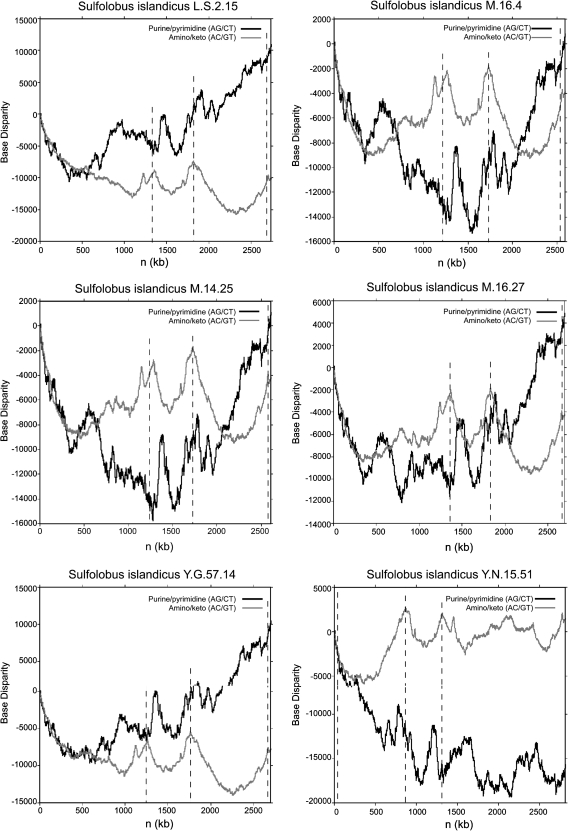
Because replication origins are highly conserved and have functionally important repetitive tracts of DNA, regions of the genome that have distinct differences in nucleotide composition in comparison with the rest of the genome can be indicative of replication origins. The three components of the Z-curve, xn, yn and zn, describe three independent distributions of nucleotide composition of any analyzed DNA sequence (Zhang R and Zhang CT 2003, 2004, 2005). The xn, yn, and zn components represent the distributions of purine to pyrimidine (RY disparity), amino to keto (MK disparity), and strong H-bond to weak H-bond bases (SW disparity) along a sequence, respectively. Lundgren et al. (2004) experimentally identified the locations of the origins of replication of S. acidocaldarius and S. solfataricus to be at base pair positions 1) 101, 2) 578,164, and 3) 1,197,528 and 1) 221,923, 2) 738,069, and 3) 2,010,231 base pairs, respectively (Lundgren et al. 2004). As expected, the Z-curves of these genomes depict two regional maxima in MK disparity and one global maximum in RY disparity precisely where the origins were located experimentally (supplementary fig. S1, Supplementary Material online). This bioinformatic method was therefore applied to the S. islandicus genomes under study here, whose replication origins have not been experimentally found. The origins of replication were determined to be located around 0, 1,250,000 and 1,800,000 base pairs in all genomes except YN.15.51, in which replication origins were predicted at 0, 700,000, and 1,250,000 bp (fig. 1). As further support of these predicted origins, each was located near a cdc6 homolog.
FIG. 1.—
The Z-curve data representing the distribution of purine to pyrimidine (RY disparity) and amino to keto (MK disparity) bases of the (a) Sulfolobus islandicus L.S.2.15 genome, (b) Sulfolobus islandicus M.16.4 genome, (c) Sulfolobus islandicus M.14.25 genome, (d) Sulfolobus islandicus M.16.27 genome, (e) Sulfolobus islandicus Y.G.57.14 genome, and the (f) Sulfolobus islandicus Y.N.15.51 genome. Black lines denote the distribution of purine to pyrimidine bases and gray lines denote the distribution of amino to keto bases throughout the genome. Dashed vertical lines indicate the predicted location of origins of replication.
Panorthologs Are More Numerous and Conserved Near Replication Origins
Six archaeal genomes with multiple origins of replication were selected from the hyperthermophilic, acidophilic species S. islandicus (Reno et al. 2009). To minimize variation in ortholog positions among genomes and to increase resolution of evolutionary rate estimates, genomes from multiple geographically distinct isolates of the same species were chosen rather than genomes from multiple species. “Panorthologs,” or ortholog families with only one ortholog found in each of the genomes under study, were identified using an analysis pipeline based on previous work (Cooper et al. 2010). The stringency of this method eliminated all the genes that were not found in all the genomes, which defined much of the regional specificity of these S. islandicus strains (Reno et al. 2009), including all mobile elements. Panortholog families were then organized into five groups based on their distance from an origin: three consisting of ortholog families within 200 Kbp of an origin designated as O1, O2, and O3, one consisting of orthologs within a 500 Kbp section distant from any origin, N1, and one consisting of ortholog families between O1 and O3, called N2 (fig. 2). Due to variation in genome sizes and ortholog position among the chromosomes, panortholog positions were grouped based on the position of the representative ortholog present in the S. islandicus L.S.2.15 genome. Although these assignments may seem arbitrary, we found nearly perfect synteny between four of the genomes and L.S.2.15 that preserved the relationship between ortholog families and their proximity to replication origins (supplementary fig. S2, Supplementary Material online). This gene order relative to the origins was also preserved in the Y.N.15.51 genome in spite of its large inversion (supplementary fig. S2, Supplementary Material online), thus supporting our use of a single genome to annotate ortholog family position. Interestingly, nearly all the variation in ortholog position among genomes was found in the N1 region distant from replication origins between position 500,000 and 1,000,000 bp (Reno et al. 2009) (supplementary fig. S2, Supplementary Material online). Moreover, the ortholog content of regions distant from replication origins was more weakly conserved: N1 contained the fewest panorthologs (0.28 orthologs/Kbp) followed by N2 (0.74 orthologs/Kbp). In contrast, the regions bounding O1, O2, and O3 contained more panorthologs, with 0.94, 0.89, and 0.92 orthologs/Kbp, respectively.
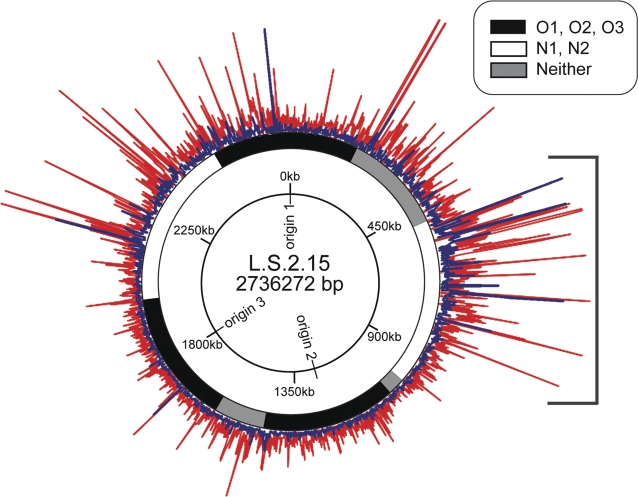
FIG. 2.—
Physical map of the distribution of the dN (blue) and dS (red) values of panortholog families relative to their position in the Sulfolobus islandicus LS.2.15 genome. The outer ring shows the location of panortholog families defined as near an origin (black), distant from an origin (white), or neither (gray). The inner ring shows the location of the origins throughout the genome and relative nucleotide positions. The bracketed region represents the region of greatest variation in genome content, as described previously (Reno et al. 2009).
Rates of Nonsynonymous and Synonymous Substitutions Are Greater in Regions Distant from Replication Origins
We quantified the evolutionary rates (dN and dS) of panorthologs of S. islandicus and analyzed whether genes found in regions distant from origins tend to evolve more quickly. We had predicted that any region-specific variation in evolutionary rates across genomes of S. islandicus would be subtle, so we first compared 400 Kb regions that were either origin-proximal or origin-distal. Both dN and dS varied significantly among genome regions (dN: F = 41.8, P < 10−10; dS: F = 9.62, P = 4 × 109) and were greatest in N1 (fig. 2, table 1). On average, panorthologs in N1 and N2 evolved more rapidly than those near replication origins 1, 2, or 3 (table 1). Interestingly, panorthologs found near O1 tended to evolve more rapidly, with both increased dN and dS, than those near O2 or O3 (table 1). This pattern suggests that either the replication origin located in O1 is used later in the cell cycle and reduces the dosage of nearby genes or these genes tend to be inherently different than those near O2 or O3. We found that the replication origin in O1 differs in its composition as being more pyrimidine (CT)-rich, which could influence its function (fig. 1). We return to this issue below in Discussion. In general, regional variation in rates of synonymous substitutions (dS) was highly correlated with variation in rates of nonsynonymous substitutions (dN), which suggests that a similar process may be affecting both types of substitutions.
Table 1.
Mean Evolutionary Rates of Panorthologs Found in Regions Proximate or Distant to Replication Origins
| Genome Region | N | dN (±95% CI) | Homogeneous Subsets | dS (±95% CI) | Homogeneous Subsets |
| O1 | 373 | 0.00684 (0.000905) | 2, 3 | 0.0303 (0.00444) | 1 |
| O2 | 356 | 0.00432 (0.000926) | 1, 2 | 0.0198 (0.00454) | 1 |
| O3 | 369 | 0.00373 (0.000910) | 1 | 0.0166 (0.00446) | 1 |
| N1 | 135 | 0.0162 (0.0015) | 4 | 0.0685 (0.00738) | 1 |
| N2 | 371 | 0.00782 (0.000907) | 3 | 0.0318 (0.00445) | 2 |
| Neither | 384 | 0.00669 (0.00109) | 2, 3 | 0.0269 (0.00437) | 1 |
Note.—Post hoc comparisons among regions to identify homogeneous groupings were conducted using Tukey’s test. CI, confidence interval.
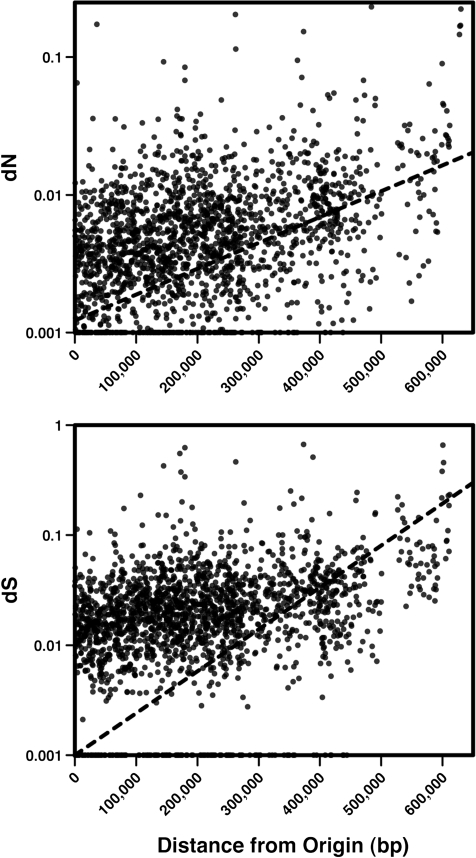
The significant variation among genome blocks either proximate or distant from replication origins prompted us to conduct a regression of substitution rates (dN and dS) on the proximity of each ortholog to its nearest replication origin (fig. 3). Both regressions are statistically significant (dN: F = 179.7, P < 0.001; dS: F = 39.7, P < 0.001), and although their overall explanatory power is low (r2 = 0.083 for dN and 0.02 for dS), they demonstrate that evolutionary rates tend to increase with distance from origins. However, because substitution rates may associate with a range of other factors that could covary with replication timing (such as strand bias, regional clustering of genes of different functions, or variation in nucleotide content), we explored these factors as potential sources of variation.
FIG. 3.—
Linear regressions of (A) the rate of nonsynonymous substitutions (dN) and (B) the rate of synonymous substitutions (dS) among panortholog families on their distance from the nearest replication origin. The dotted line depicts the linear regression function; each is statistically significant (dN: F = 179.7, P < 0.001; dS: F = 39.7, P < 0.001) despite modest explanatory power (dN: r2 = 0.083; dS: r2 = 0.02). Ortholog position was assigned using the Sulfolobus islandicus L.S.2.15 genome, which is justified by the high degree of synteny among the six study genomes (supplementary fig. S2, Supplementary Material online).
Because genes located on different strands may evolve at different rates owing to strand-specific nucleotide biases (Tillier and Collins 2000), we investigated how frequently S. islandicus orthologs switched strands. In 90% of ortholog families, 5/6 or 6/6 genes were found on the same strand, and nearly all the single strand variations occurred in the Y.N.15.51 genome that contains a large inversion. Given that substitution rates were calculated from the phylogeny of ortholog families rather than from averages of pairwise comparisons, it is therefore unlikely that the switching of genes between strands could produce the observed variation in substitution rates. Furthermore, we found no differences in G + C content among ortholog families found in different regions, so substitution patterns specific to higher or lower %G + C could not influence the observed variation in evolutionary rates (supplementary data, Supplementary Material online).
Next, the distributions of orthologs belonging to different functional categories of clusters of orthologous groups (COGs) were compared among origin-proximate and origin-distal regions. The genome-wide distribution of COGs was used to calculate expected numbers of orthologs in each COG category for each genome region; these expected values were then compared with the observed distributions of COGs. Each region (O1, O2, O3, N1, N2) departed significantly from an even distribution (X2 > 45 with 21 degrees of freedom, P < 0.001), but relatively few COG categories explained these differences (supplementary data, Supplementary Material online). Specifically, orthologs contributing to transcription (J) and translation (K) were much more abundant near O2 and O3 and more rare in regions O1, N1, and N2. In contrast, orthologs contributing to energy production and transport, which are more likely to vary among taxa because their functional contribution varies with environment, were more abundant in N1 and N2 and relatively rare in O1, O2, and O3. We interpret these differences as the legacy of selection for more essential genes to be located near origins; conversely, less important or environment-specific genes may more likely be found in late replicated regions.
Codon Usage Bias and Predicted Expression Are Greater among Genes Near Replication Origins
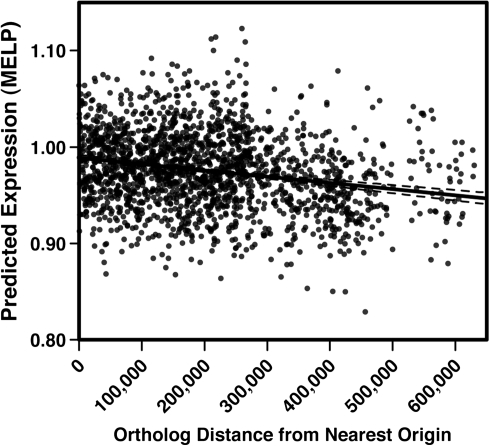
We used two methods for estimating the codon usage bias of panorthologs from different genome regions, CAI (Sharp and Li 1987), which compares codon usage of query genes with genes known to be highly expressed and hence codon-optimized, and SCUO (Angellotti et al. 2007), which uses information theory to quantify bias and is not dependent upon a reference set of genes. Both methods demonstrated that the codon usage bias of genes near O2 and O3 were significantly greater than those near O1, N1, or N2; of these, genes in N2 were the least biased toward preferred codons (supplementary table S1, Supplementary Material online). Although differences among genome regions were quantitatively minor, the slightly stronger average codon bias of genes near replication origins is consistent with their potentially greater expression due to transient increases in gene dosage during the cell cycle. We then used a third algorithm that accurately predicts expression relatively from base composition, MELP (Supek and Vlahovicek 2005), to test for variation in expression as a function of ortholog distance from the nearest replication origin. As expected, predicted expression varied in a manner consistent with the codon usage bias measures: genes near O2 and O3 are predicted to be expressed most and those distant from origins significantly less (supplementary table S2, Supplementary Material online), and the genome-wide regression of MELP on distance to the nearest origin was also highly significant (Fig 4, F = 96.2, P < 0.0001), albeit weakly predictive (r2 = 0.05).
These informatic predictors of gene expression can be useful but they depend strongly on codon usage bias, which can be influenced by factors other than expression frequency, such as GC skew (Wan et al. 2004). A better test of the key prediction of our model, that genes replicated early will be expressed more frequently than genes replicated late, would involve empirical measures of expression from each of these ortholog families. Fortunately, during revisions of this manuscript, a report by Andersson et al. (2010) found exactly this predicted pattern. In the closely related Sulfolobus species S. solfataricus and S. acidocaldarius, highly expressed regions were concentrated near replication origins and expression declined in regions replicated later. Interestingly, the gradient in expression between origin-proximal and origin-distal regions was greater than expected from gene dosage effects alone and greater than the predicted magnitude of differences presented here. Moreover, this pattern was not solely produced by an enrichment of core essential genes near origins but associated with all genes. Thus, Sulfolobus genomes appear to be organized by priority, with more essential conserved genes with higher expression levels replicated early and less conserved and expressed genes replicated late.
Discussion
One of the evolutionary innovations that distinguish some Archaea and Eukarya from Bacteria is the presence of multiple replication origins on each chromosome. When replication occurs from a single site on a bacterial chromosome, genes distant from that site will experience reduced dosage, particularly among fast growing bacteria with multiple active replication forks from the same origin. This gradient in gene dosage influences probability of expression, generating weaker purifying selection on genes nearer the terminus and lesser potential for their repair (Mellon and Hanawalt 1989; Hanawalt and Spivak 2008), and causes them to evolve more rapidly (Sharp et al. 1989; Couturier and Rocha 2006; Cooper et al. 2010). However, with multiple, nonoverlapping replication forks on archaeal chromosomes the transient variation in gene copy number during the cell cycle should be significantly reduced. We therefore hypothesized that archaeal genes should evolve at more uniform rates that are independent of their chromosomal positions. We evaluated this hypothesis using six closely related genomes of S. islandicus; this genus of Crenarchaeota has been frequently studied as a model of archaeal genetics and cell biology (Bernander 2007). However, despite the action of three origins of replication in these small (2.8 Mb) genomes, we found that the substitution rates of genes distant from replication origins were greater at both synonymous and nonsynonymous sites than genes nearby. Moreover, the most variable regions of the S. islandicus genome that define their unique biogeography as thermoacidophiles were precisely the regions most distant from replication origins. Apparently, the genetic signatures of ecological specificity tend to concentrate in regions of the genome that are replicated last.
We propose four explanations for why distance from replication origins in Sulfolobus is still positively associated with evolutionary rates. The first is that replication timing still produces sufficient variation in gene dosage to influence the likelihood of expression and strength of purifying selection, as has been demonstrated in bacterial genomes of various compositions (Sharp and Li 1987; Sharp et al. 1989; Chen et al. 2004; Couturier and Rocha 2006; Rasmussen et al. 2007; Drummond and Wilke 2008; Cooper et al. 2010). In theory, multiple origins of replication should reduce this variation over equivalent chromosomes with only a single origin, and dosage between origin and terminus should never exceed 2-fold in Archaea. However, the root cause of the dosage effect, cell growth rates exceeding replication rates, likely still affects Sulfolobus: MFA studies have recently demonstrated that the Sulfolobus replication rate is an order of magnitude slower than that of Escherichia coli and more similar to that of eukaryotes (Lundgren et al. 2004; Duggin et al. 2008). Slower replication could thus increase the extent to which replication rate lags behind growth rate and strengthen the association between evolutionary rate and replication timing.
Variation in gene copy number correlates with the frequency of expression, so late replicated genes are expected to be expressed less frequently, to experience weaker purifying selection for optimal codon usage, and to display greater dS. Increases in both dN and dS among less expressed genes may reflect a general increase in mutation rate or weaker selection for their robust translation, as the cost of protein misfolding can be quite high for the most highly expressed genes (Drummond and Wilke 2008). It should be noted that highly expressed genes actually experience greater mutation rates than the remainder of the genome owing to their extended exposure as single strands (Ochman 2003; Lind and Andersson 2008), so it is enhanced purifying selection and not reduced mutation that explains their slow evolution. All else being equal, such highly expressed genes should be found close to replication origins. Frequently translated genes should also experience selection for codon usage that is least likely to result in the incorporation of incorrect amino acids and that generally suppresses the substitution rate.
A second explanation invokes weaker, second-order selection on genome architecture (Andersson et al. 2010). If genome location produces systematic biases in expression levels, selection could act on gene position and cause more conserved and expressed genes to become associated with replication origins. The eventual outcome would be that different genome regions would tend to harbor genes optimized for different expression patterns, and those genes expressed least often should become subject to greater effects of drift and potential loss. Consistent with this logic, we found that genes in the Sulfolobus genome distant from replication origins were the least conserved among strains and by definition were most dispensable. Genes distant from origins also tended to represent less essential functional categories (i.e., metabolism and transport). This pattern agrees with the highly variable region in S. islandicus genome content previously reported (Reno et al. 2009) and suggests that the genes in this region are at best only conditionally useful. Whether these dispensable genes tend to be weakly expressed remains a subject for further study, but the predicted patterns of expression (fig. 4) support this possibility.
FIG. 4.—
Linear regression of predicted gene expression (MELP, [Supek and Vlahovicek 2005]) on the distance of orthologs from the nearest origin of replication. The dotted line is the regression function and is statistically significant (F = 96.2, P < 0.0001) but weakly predictive (r2 = 0.05).
Another potential explanation for the observed patterns could be that regions distant from origins of replication are more tolerant and/or more prone to recombination with homologous alleles in Sulfolobus populations. If early replicated genes undergo more efficient repair by gene conversion from the other template, and this accounts in part for the clustering of conserved genes near origins (Andersson et al. 2010), then genes replicated late may become more tolerant to repair by more divergent homologs as they lack a freshly replicated local copy. Reno et al. (2009) also found that much of the variation among these genomes was caused by mobile elements inserted in the region we have termed N1. We emphasize that these mobile elements were not included in the analysis of orthologs presented here, but they could reflect a mechanism of greater rates of recombination in this region. One reason why variation in recombination rates may not explain our findings is that our stringent analysis pipeline is unlikely to retain homolog families in which recombination of diverse alleles have occurred. However, those families subject to recent recombination of very similar alleles may remain in our analysis, thus violating a strict definition of orthology. A general increase in recombination rate with replication timing would also explain the greater substitution patterns in late-replicated regions.
A fourth possible explanation for these patterns that demands further study is that mutation rate increases systematically with the cell cycle, perhaps because of declining efficiency of the replication apparatus (Mira and Ochman 2002), by reduced replication- or transcription-coupled DNA repair (Sweder and Hanawalt 1993; Ochman 2003), or because nucleotide pools become limiting (Wolfe et al. 1989). Such a finding would be unprecedented and did not occur in a detailed study of mutation in Salmonella populations evolved in the absence of selection (Lind and Andersson 2008); however, it would explain the simultaneous increase in both dN and dS with distance from replication origins. Early replication of essential genes would thus be favored to minimize their exposure to damage and to guarantee availability of their gene products. More mutations in late-replicated regions could also increase the frequency of recombination as a means of repair and thus explain why these regions have fewer conserved orthologs.
Each of these dosage-related mechanisms could explain the observed variation in mean substitution rates between origin-proximal and origin-distal regions, which in most pairwise comparisons differ by roughly 2-fold (table 1). However, gene dosage alone is unlikely to explain the recent report that genes in closely related Sulfolobus species found near origins are expressed more than 4-fold more than those near termini (Andersson et al. 2010). Rather, a combination of forces affecting genome architecture seems likely to be at work. These mechanisms include strong selection against translation errors, causing slower evolution of early replicated genes because of the greater likelihood of expression, and a benefit of early replication of essential genes to avoid irreparable damage from the mutagenic environment and maintain their function. Together, these forces lead to a rough ordering of genes by priority or necessity along with replication timing.
Although proximity to a replication origin explained significant variance in evolutionary rates, conserved orthologs near O1 evolved more rapidly than those near either O2 or O3. This region is also the only replication origin that is CT-rich (fig. 1), which could be associated with unique functionality. We propose that the greater evolutionary rates of this region are caused by the delayed initiation of replication at O1. Although computational modeling suggested that the three Sulfolobus origins fire simultaneously (Lundgren et al. 2004), Duggin et al. (2008) showed experimentally that one of the three origins of replication in S. acidocaldarius does exhibit delayed initiation, and this origin is syntenic with O1 of S. islandicus. It is also possible that this origin was acquired by horizontal gene transfer and maintains its anomalous function, although we found no evidence of foreign sequence in this region. Rather we suggest that the potential variation in replication timing in Sulfolobus may be yet another reason why Archaea resemble Eukarya. Because eukaryotic genomes feature two general types of replication origins of replication with different mechanisms of initiation and timing (Gilbert 2001) and Archaea share some of this machinery (Kelman and White 2005), archaeal replication may indeed also be heterogeneous both in time and space.
We acknowledge the need for a more focused analysis of the orthologs found to evolve at greater or lesser rates, including a study of expression in these S. islandicus genomes throughout the cell cycle. The exact positions of the replication origins in S. islandicus and their timing of initiation during the cell cycle also remain to be determined experimentally. It also remains to be studied how multiple replication origins arose in Archaea in general and Sulfolobus in particular, whether by duplication or acquisition of foreign genes from a different archaeon. Nevertheless, we conclude that in at least two domains of life (Bacteria and now in this genus of Archaea), gene evolutionary rates are positively associated with their distance from the nearest replication origin.
Supplementary Material
Supplementary data, figures S1–S2 and table S1–S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank J. Morrow for helpful discussions and to three anonymous reviewers for constructive feedback. This research was supported in part by a President’s Excellence award from the University of New Hampshire.
References
- Andersson A, et al. Replication-biased genome organisation in the crenarchaeon Sulfolobus. BMC Genomics. 2010;11:454. doi: 10.1186/1471-2164-11-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angellotti MC, Bhuiyan SB, Chen G, Wan XF. CodonO: codon usage bias analysis within and across genomes. Nucleic Acids Res. 2007;35:W132–136. doi: 10.1093/nar/gkm392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernander R. The cell cycle of Sulfolobus. Mol Microbiol. 2007;66:557–562. doi: 10.1111/j.1365-2958.2007.05917.x. [DOI] [PubMed] [Google Scholar]
- Berquist BR, DasSarma S. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J Bacteriol. 2003;185:5959–5966. doi: 10.1128/JB.185.20.5959-5966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. Common structural features of replication origins in all life forms. J Cell Biochem. 1996;60:297–316. doi: 10.1002/(sici)1097-4644(19960301)60:3<297::aid-jcb2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH. Codon usage between genomes is constrained by genome-wide mutational processes. Proc Natl Acad Sci U S A. 2004;101:3480–3485. doi: 10.1073/pnas.0307827100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker JA, et al. Multiple replication origins of Halobacterium sp. strain NRC-1: properties of the conserved orc7-dependent oriC1. J Bacteriol. 2009;191:5253–5261. doi: 10.1128/JB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. Why genes evolve faster on secondary chromosomes in bacteria. PLoS Comput Biol. 2010;6:e1000732. doi: 10.1371/journal.pcbi.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier E, Rocha EP. Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes. Mol Microbiol. 2006;59:1506–1518. doi: 10.1111/j.1365-2958.2006.05046.x. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggin IG, McCallum SA, Bell SD. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci U S A. 2008;105:16737–16742. doi: 10.1073/pnas.0806414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD. The evolution of isochores. Nat Rev Genet. 2001;2:549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Mathematics vs. evolution: mathematical evolutionary theory. Science. 1989;246:941–942. doi: 10.1126/science.246.4932.941. [DOI] [PubMed] [Google Scholar]
- Gilbert DM. Making sense of eukaryotic DNA replication origins. Science. 2001;294:96–100. doi: 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Henry I, Sharp PM. Predicting gene expression level from codon usage bias. Mol Biol Evol. 2007;24:10–12. doi: 10.1093/molbev/msl148. [DOI] [PubMed] [Google Scholar]
- Kelman LM, Kelman Z. Multiple origins of replication in archaea. Trends Microbiol. 2004;12:399–401. doi: 10.1016/j.tim.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Kelman Z, White MF. Archaeal DNA replication and repair. Curr Opin Microbiol. 2005;8:669–676. doi: 10.1016/j.mib.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Moran NA. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-Proteobacteria. PLoS Biol. 2003;1:E19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Andersson DI. Whole-genome mutational biases in bacteria. Proc Natl Acad Sci U S A. 2008;105:17878–17883. doi: 10.1073/pnas.0804445105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Lobry JR, Sueoka N. Asymmetric directional mutation pressures in bacteria. Genome Biol. 2002;3:RESEARCH0058. doi: 10.1186/gb-2002-3-10-research0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc Natl Acad Sci U S A. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean MJ, Wolfe KH, Devine KM. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J Mol Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Mira A, Ochman H. Gene location and bacterial sequence divergence. Mol Biol Evol. 2002;19:1350–1358. doi: 10.1093/oxfordjournals.molbev.a004196. [DOI] [PubMed] [Google Scholar]
- Mrazek J, Karlin S. Strand compositional asymmetry in bacterial and large viral genomes. Proc Natl Acad Sci U S A. 1998;95:3720–3725. doi: 10.1073/pnas.95.7.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllykallio H, et al. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288:2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- Ochman H. Neutral mutations and neutral substitutions in bacterial genomes. Mol Biol Evol. 2003;20:2091–2096. doi: 10.1093/molbev/msg229. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR. Archaeal genomics: an overview. Cell. 1997;89:991–994. doi: 10.1016/s0092-8674(00)80284-6. [DOI] [PubMed] [Google Scholar]
- Rasmussen T, Jensen RB, Skovgaard O. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 2007;26:3124–3131. doi: 10.1038/sj.emboj.7601747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. Biogeography of the Sulfolobus islandicus pan-genome. Proc Natl Acad Sci U S A. 2009;106:8605–8610. doi: 10.1073/pnas.0808945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, et al. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Danchin A, Viari A. Universal replication biases in bacteria. Mol Microbiol. 1999;32:11–16. doi: 10.1046/j.1365-2958.1999.01334.x. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Salzberg AJ, Kerlavage AR, Tomb JF. Skewed oligomers and origins of replication. Gene. 1998;217:57–67. doi: 10.1016/s0378-1119(98)00374-6. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987;4:222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Shields DC, Wolfe KH, Li WH. Chromosomal location and evolutionary rate variation in enterobacterial genes. Science. 1989;246:808–810. doi: 10.1126/science.2683084. [DOI] [PubMed] [Google Scholar]
- Supek F, Vlahovicek K. Comparison of codon usage measures and their applicability in prediction of microbial gene expressivity. BMC Bioinformatics. 2005;6:182. doi: 10.1186/1471-2105-6-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweder KS, Hanawalt PC. Transcription-coupled DNA repair. Science. 1993;262:439–440. doi: 10.1126/science.8211165. [DOI] [PubMed] [Google Scholar]
- Tillier ER, Collins RA. Replication orientation affects the rate and direction of bacterial gene evolution. J Mol Evol. 2000;51:459–463. doi: 10.1007/s002390010108. [DOI] [PubMed] [Google Scholar]
- Wan XF, Xu D, Kleinhofs A, Zhou J. Quantitative relationship between synonymous codon usage bias and GC composition across unicellular genomes. BMC Evol Biol. 2004;4:19. doi: 10.1186/1471-2148-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Sharp PM, Li WH. Mutation rates differ among regions of the mammalian genome. Nature. 1989;337:283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang R, Zhang CT. Multiple replication origins of the archaeon Halobacterium species NRC-1. Biochem Biophys Res Commun. 2003;302:728–734. doi: 10.1016/s0006-291x(03)00252-3. [DOI] [PubMed] [Google Scholar]
- Zhang R, Zhang CT. Identification of replication origins in the genome of the methanogenic archaeon, Methanocaldococcus jannaschii. Extremophiles. 2004;8:253–258. doi: 10.1007/s00792-004-0385-4. [DOI] [PubMed] [Google Scholar]
- Zhang R, Zhang CT. Identification of replication origins in archaeal genomes based on the Z-curve method. Archaea. 2005;1:335–346. doi: 10.1155/2005/509646. [DOI] [PMC free article] [PubMed] [Google Scholar]