The ‘Crop Ontology’ database we describe provides a controlled vocabulary for several economically important crops. It facilitates data integration and discovery from global databases and digital literature. This allows researchers to exploit comparative phenotypic and genotypic information of crops to elucidate functional aspects of traits.
Abstract
Background and aims
Agricultural crop databases maintained in gene banks of the Consultative Group on International Agricultural Research (CGIAR) are valuable sources of information for breeders. These databases provide comparative phenotypic and genotypic information that can help elucidate functional aspects of plant and agricultural biology. To facilitate data sharing within and between these databases and the retrieval of information, the crop ontology (CO) database was designed to provide controlled vocabulary sets for several economically important plant species.
Methodology
Existing public ontologies and equivalent catalogues of concepts covering the range of crop science information and descriptors for crops and crop-related traits were collected from breeders, physiologists, agronomists, and researchers in the CGIAR consortium. For each crop, relationships between terms were identified and crop-specific trait ontologies were constructed following the Open Biomedical Ontologies (OBO) format standard using the OBO-Edit tool. All terms within an ontology were assigned a globally unique CO term identifier.
Principal results
The CO currently comprises crop-specific traits for chickpea (Cicer arietinum), maize (Zea mays), potato (Solanum tuberosum), rice (Oryza sativa), sorghum (Sorghum spp.) and wheat (Triticum spp.). Several plant-structure and anatomy-related terms for banana (Musa spp.), wheat and maize are also included. In addition, multi-crop passport terms are included as controlled vocabularies for sharing information on germplasm. Two web-based online resources were built to make these COs available to the scientific community: the ‘CO Lookup Service’ for browsing the CO; and the ‘Crops Terminizer’, an ontology text mark-up tool.
Conclusions
The controlled vocabularies of the CO are being used to curate several CGIAR centres' agronomic databases. The use of ontology terms to describe agronomic phenotypes and the accurate mapping of these descriptions into databases will be important steps in comparative phenotypic and genotypic studies across species and gene-discovery experiments.
Introduction
The challenge of addressing climate change for food security and adaptation of agricultural systems led, in 2004, to the launch of the 10-year Generation Challenge Programme (GCP). This is an agricultural research consortium hosted by international agricultural research centres of the Consultative Group on International Agricultural Research (CGIAR). The GCP involves 22 research institutes in partnership with external collaborators. The GCP research agenda focuses on producing drought-tolerant varieties through comparative genomics-driven improvement and high-throughput molecular characterization of genetic resources in order to introduce favourable alleles into plant-breeding programmes. For decades, CGIAR centres and their gene banks have accumulated considerable amounts of valuable data on germplasm traits. The GCP is now adding new data sets related to genotype and phenotype, which need to be released and made accessible to breeders online.
Scientists are overwhelmed by data: the amount of biological and genetic information has increased dramatically with the advent of high-throughput data collection in the fields of molecular biology and biotechnology. Researchers need a multidisciplinary approach to understand the biological processes from genes to the expression of traits in crops. This approach requires the extraction of biological data sets from a wide range of sources. The interoperability between these sources enables scientists to exploit comparative genomic information, elucidate functional aspects of plant biology and conduct studies of synteny and homology. However, the GCP has not yet achieved the level of interoperability required for providing access to comprehensive sets of biological data. One obstacle to the seamless combination of genetic trait and experimental data is the variability of the terms and concepts used to describe comparable objects across databases. In agronomy, phenotype information has traditionally been captured in a free-text manner. In addition, many traits are crop specific and some have complex trait names, thus making it difficult to understand their exact meaning without further description. Developing trait ontology for economically important crops is crucial to overcoming the inconsistencies between GCP data sources and sharing this knowledge among researchers.
In bioinformatics, an ontology is a formal representation of a set of concepts within a specific discipline or domain and the relationship between those concepts. It provides a shared and controlled vocabulary that can be used to model the domain in terms of the types of object or concept, and their properties and relationships. Ontology is more complex than systematics used for species classification because it involves multiple parents and the opportunity for different relationships. While the structure of an ontology is a strict hierarchy, it is represented by a directed acyclic graph in which multiple types can have parents, with different relationships between them.
The crop ontology (CO) needed to address the concept of an agronomic trait and define how many other domains and publicly available bio-ontologies were needed to fully reflect the concept (Shrestha et al., 2010). An agronomic trait is measured using different plant characters or parameters following a defined protocol and in a given environment. This information is typically stored in a database or explained in an article, and needs to be extracted from these data sources to be made available to researchers in a form useful for their research. However, these available data are not systematized, which creates problems in data management, retrieval and accessibility. The CO comprises the computational architecture to structure these data and thus provide researchers and end users with a user-friendly tool to facilitate the use of comparative biology infrastructure for crops such as rice, maize and wheat. In doing so, the CO facilitates the use of biological information to accelerate the crop improvement efforts under way in institutions around the world.
Materials and methods
Crop selection
In 2006, GCP scientists developed and applied a new method for identifying areas where poverty and production of drought-prone crops coincide. This analysis of global spatial data sets identified five farming systems in South Asia, five in sub-Saharan Africa, four in East Asia and one in Mesoamerica where drought coincides with a high level of poverty. The analysis also examined a global database of harvested areas and production data to determine which crops poor farmers rely on the most. This analysis identified 12 crops as covering at least 5 % of the cultivated area in each of the 15 farming systems most stricken by poverty and drought (Website 1). Among 12 GCP mandate crops, banana (Musa spp.), chickpea (Cicer arietinum), maize (Zea mays), potato (Solanum tuberosum), rice (Oryza sativa), sorghum (Sorghum spp.) and wheat (Triticum spp.) were selected for the CO because information on these crops is well characterized in research databases.
CO resources
The team identified the sources of trait names for the CO and ways to validate them; each source was crop specific. CGIAR databases including the International Maize Information System (IMIS; Website 2), the International Rice Information System (Website 3), the International Wheat Information System (Website 4), the Musa Germplasm Information System (Website 5), the International Crop Research Institute for the Semi-Arid Tropics (ICRISAT; Website 6) information system for sorghum and chickpea, and the International Potato Center (CIP; Website 7) information system for potato were used as resources for developing the CO. The data generated by GCP-funded projects and deposited in the GCP Central Registry were another source. For several years, Bioversity International, in collaboration with crop experts, has been developing a controlled vocabulary, called descriptors, for characterizing crop varieties in the field. These descriptors are available as downloadable files (Website 8) as well as a series of booklets available online from the System Wide Genetic Resources Information System (SINGER; Website 9). The descriptors for the priority crops formed the first core of concepts for the CO. This vocabulary was enriched by the extraction of numerous trait names from breeders' databases and the literature. Public-domain ontologies or equivalent concept catalogues including the Gene Ontology (GO) (Ashburner and Lewis, 2002), Plant Ontology Consortium (POC; Jaiswal et al., 2002), MIAME-Plant (Zimmermann et al., 2006) and the Food and Agriculture Organization FAO/Bioversity Multi-Crop Passport Descriptors (FAO/IPGRI, 2001) were used as references for building the CO.
Ontology landscape for the trait concept
The GO is the most well-known bio-ontology; it describes the gene and gene products in several model organisms with a controlled vocabulary. The new Environment Ontology (EnvO) is bringing similar benefits through consistent annotation grounded in an ontological framework, with the ability to facilitate the semantic retrieval of any biological record anchored to EnvO. The Plant Ontology (PO) mainly describes structure, anatomy and growth stages of plants. The Gramene Cereal Plant Trait Ontology (TO), Phenotype and Trait Ontology (PATO) and Sol Genomics Network (SGN) Ontology have been developed mainly for accessing plant-science information. Gramene, a database of grass genomes providing comparative genomics tools for grasses, is being used for the development of TO in collaboration with the POC (Jaiswal et al., 2002). The PATO has been constructed to capture qualitative and quantitative information about phenotypes. This ontology of phenotypic qualities can be used to capture the differences between wild and mutant phenotypes of all organisms. The SGN has recently developed an ontology for Solanaceae phenotype information (Menda et al., 2008).
Tools
For CO development and curation, OBO-Edit (version 2.0, Day-Richter et al., 2007) was the tool of choice due to its simplicity and the generation of text output (OBO ontology format) that is readable by biologists. Each term in the ontology was assigned: a globally unique CO identifier composed of the prefix CO_ followed by a three-digit number denoting the index of the ontology from which the term was adopted; a decimal point; and finally an alphanumeric suffix, which is the specific identifier for that term or concept. The scope of the CO indices is provided in Table 1, and the fully indexed inventory of CO is published at Website 10.
Table 1.
Scope and index ranges for subclasses of crop ontology.
| CO prefix | Subclasses of crop ontology |
|---|---|
| 010–089 | General Germplasm and Passport Ontology |
| 90–099 | Taxonomic Ontology |
| 100–299 | Plant Anatomy and Development Ontology |
| 300–499 | Phenotype and Trait Ontology |
| 500–699 | Structural and Functional Genomic Ontology |
| 700–799 | Location and Environment Ontology |
| 800–899 | General Science Ontology |
| 900–999 | Other |
At each level of the CO, specific trait-type information (e.g. synonyms, trait description, note of application for a crop) was included for each term. The Open Biomedical Ontologies (OBO) format allows various types of relationship between terms. The most common relationships are (i) ‘is_a’ (e.g. the plant height is a vigour trait or shoot anatomy and morphology-related trait); (ii) ‘part_of’ (e.g. the stem length is part of the plant height) as in GO; and (iii) ‘derived_from’ (e.g. abscisic acid content to sugar content ratio is derived from parent terms ‘abscisic acid content’ and ‘sugar content’). The additional relationship ‘has_a’, which is more common in web ontology language (OWL), is also used between the terms in CO.
Additional software tools, primarily in the Java and Perl programming languages, were used to facilitate ontology management. These included tools to parse and convert ontology term metadata to Web Language for Ontology or OBO, and tools for storing the ontology catalogues in the Chado-controlled vocabulary schema of the Genetic Model Organism Database project. These Java and Perl tools include GCPModelToOwl, GPCOBOParser, OBOWriter, OboToChadoLoader and OwlToChadoWriter (they can be found at Website 11). Perl scripts are also available to convert a source of data in a certain format into an OBO-formatted file.
Validation of trait descriptions and definitions
For each crop, a group of crop experts including breeders, physiologists, agronomists and pathologists validated trait descriptions and other trait information. Crop breeders' networks forming communities of practice were also identified within GCP challenge initiatives to exchange information and build further the CO.
Term-submission process
Plant structure, anatomy, growth and development terms were mapped with PO terms, and traits of selected crops were mapped with TO terms when applicable. A map was made as a dbxref in an OBO-Edit-formatted file. Terms in the crop-specific trait ontology without an equivalent term or synonym in PO or TO were considered crop-specific terms and nominated for submission to PO and TO. Regular meetings with the POC were held for discussing new terms on the term-submission page of the PO. An automated tracking system for new term submission is being developed at Website 12 for CO collaborators and researchers involved in the project.
Using Terminizer for assisted mark-up of literature with CO terms
Ontologies can assist in both searching for documents and data sets by enabling smarter matching and automatic generation of search terms, and interpreting them, since the unambiguous nature of ontological annotation leads to improved comprehension.
Terminizer (Website 13), developed by the Department of Computer Science, University of Manchester, is an easy-to-use tool that promotes the inclusion of ontologies in scientific data by assisting in the detection of ontological terms found in free text. The resulting terms are overlaid on the original text or displayed in a list organized by ontology and frequency. Users can accept or reject each match, or try to find a more appropriate match by exploring the network of ontology concepts.
Terminizer offers the full set of ontologies from the OBO Foundry, a collection of over 40 general-purpose biological ontologies. Since the system is implemented as a web service, both the term-detection service and the interactive-presentation layer can be easily incorporated within other websites or programs.
A version of Terminizer that uses the GCP ontologies (Website 14) has been implemented by the Terminizer team, enabling mark-up of GCP ontology terms in previously published crop science articles.
Results
Inventory of CO
Most breeders and germplasm users would like to select directly for the most stress-resistant, high-yielding, drought-tolerant, early-maturing, bright-green accessions. However, information on these accessions is not often readily available and, if it is, it may be difficult to interpret since a phenotype is largely shaped by its environment. Another problem is the lack of standardization of trait names, methods of measuring scale (continuous, cm/m vs. categorical, 1–9, 0–9, 0–5 %), experimental design (number of replicates), treatment (inoculation, irrigation, fertilizer), growth stages for treatment or expression of traits, and other factors that influence scores and their reliability. Other types of information, such as molecular marker data and information about quantitative trait locus (QTL) and genes, are simpler to provide and should be made available to users soon.
In order to cover all important domains of crop science, nine subclasses were created in the CO (Table 1). The General Germplasm and Passport Ontology Subclass is one of the most important subclasses of the CO. It was adapted from common concepts relating to genetic resources, especially crop descriptors. Passport information is extremely useful for genebank management, particularly for discovering duplicates and ensuring diversity in collections. This subclass therefore includes passport information on germplasm, management-related data and specific attributes, including technical terms used by genebanks to describe, for example, a single seed sample or plant clone. These vocabularies are derived from the Multi-Crop Passport Descriptors developed by the FAO of the United Nations and Bioversity International (FAO/IPGRI, 2001). Controlled vocabularies from the International Crop Information System (ICIS; Website 15) model have also been incorporated into this subclass.
The Taxonomic Ontology Subclass is a collection of plant taxonomy ontologies adopted from external taxonomy databases such as the United States Department of Agriculture's Genetic Resources Information System (GRIN), the National Center for Biotechnology Information (NCBI), UniProt and PO. The POC (Jaiswal et al., 2002) has provided the PO database for plant structure, anatomy, morphology and developmental stages (Ilic et al., 2007; Avraham et al., 2008). The Plant Anatomy and Development Ontology Subclass contains ontologies for maize (Z. mays), banana (Musa spp.; Fig. 1) and wheat (Triticum spp.).
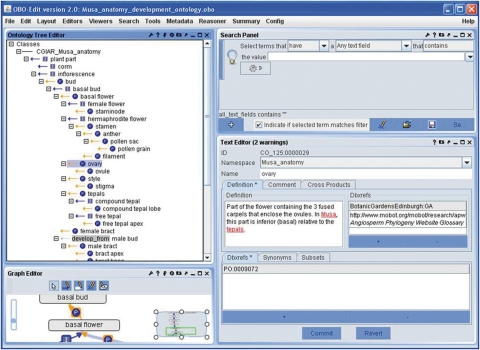
Fig. 1.
A screen capture of the Musa anatomy and developmental stages ontology, as seen within the OBO-Edit tool, displaying all the information associated with the term ‘ovary’.
Another focused subclass is the PATO, which includes the GCP crop-specific trait ontology. The goal of developing a crop-specific anatomy, development and trait ontology is to provide exact meanings of terms that are related to phenotypes that are described by crop physiologists, plant breeders and other crop scientists. Crop-specific trait ontologies have been developed for chickpea (C. arietinum), maize (Z. mays), potato (S. tuberosum), rice (O. sativa), sorghum (Sorghum spp.) and wheat (Triticum spp.). Development of a banana-specific trait ontology is ongoing. The rice mutant ontology is also integrated into this subclass of the CO. The OBO-formatted ontology files for these crops are publicly available online and described on the Pantheon website (Website 10).
The Structural and Functional Genomics Ontology Subclass consolidates many of the cellular and molecular level process ontologies, including the GO (Ashburner and Lewis, 2002) and the Ontology for Biomedical Investigations. The Location and EnvO Subclass includes location metadata such as country lists, geographical information system metadata and environmental descriptors. Included in this category are public efforts such as the EnvO project. The General Science Ontology Subclass contains physical and chemical property data for chemical species such as the Chemical Entities of Biological Interest (CHEBI)-controlled vocabularies.
To date, several crop-specific traits for quality and disease resistance have been submitted by the CO team and added to the Cereal Plant Trait Ontology (TO). Trait names for quality and disease resistance remain crop specific compared with the names of morphological and agronomic traits, which can be generic for all crops. The submitted traits can be viewed at Website 19. For example, several chickpea-specific trait names like ‘chickpea pod borer’, ‘chickpea botrytis grey mold resistance’, ‘chickpea fusarium wild resistance’, or sorghum-specific traits such as ‘sorghum stem borer resistance’, ‘sorghum shoot fly resistance’, ‘sorghum downy mildew resistance’ were added to TO along with other necessary generic trait names such as ‘seed texture’, ‘seed coat spots’, ‘grain weight to panicle weight ratio’ and ‘shoot potassium content’. In the case of wheat, ‘glume length’, ‘glume width’, ‘glume pubescence density’, ‘glume pubescence’ and ‘glume waxiness’ were added to the TO database. The wheat quality-related traits such as ‘crum structure’, ‘flour falling number’, ‘flour protein content’, ‘grain falling number’, ‘gluten type’, ‘semolina protein content’, several disease and pest-related traits have already been submitted by the CO team to TO. This trait submission process to TO will be continued for maize, Musa and potato. The CO is developed by the GCP community of practice and enriches the global public ontologies related to plants with additional concepts and definitions that were lacking.
CO browser—online ontology look-up service
A web-based GCP CO lookup service is available online at Website 16. This service, at the moment, is primarily for developers or/and curators to search for ontology terms or browse specific ontology hierarchies—users can browse a complete ontology or a subset by clicking the ‘browse’ button on the main page. A user-friendly interface will be available for end-users to browse ontology hierarchies, query annotated data with CO terms. The root terms of the ontology or subsets are shown, and users can navigate an ontology dynamically by clicking on a term to load its children. Selecting a term will display the term name, accession, definition, synonyms and annotation, if any. The browser utilizes files maintained in OBO-Edit (Website 17) to provide updated synchronized ontologies, using the Code Versioning System and Subversion version-control system repositories. Updated local copies of OBO files are being loaded into the system, changing ontology files into a database. Or, on demand, entire ontologies can be reloaded. The database searches are enhanced by text searching based on a technology known as Lucene indexes (Website 18).
A CO embedded Terminizer
In collaboration with Manchester University, controlled vocabularies of the CO have been embedded in Terminizer (Website 14), an open-source software developed by the Department of Computer Science, University of Manchester. The ontological terms are stored in an omixed resource, which provides a convenient platform for managing this kind of data. The omixed server also handles navigation functions and graph generation for the ontology browser (Website 13). The dictionary builder takes all of the terms from the omixed server and builds an in-memory dictionary, which can be used to quickly retrieve information about a specific term. When presented with input text, the look-up engine discovers which of the terms in the dictionary appear in that text and replies with an XML document listing those terms. Finally, the result formatter converts this information into an HTML representation with attendant Javascript code, which can be handled by a web browser.
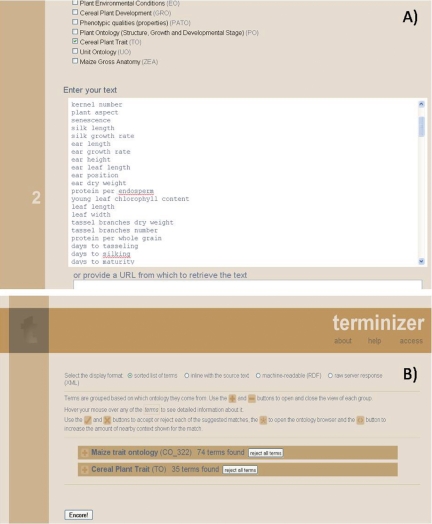
This tool has been useful for analysing traits that are common in several crops. For example, the tool found that plant height, days to flowering, days to maturity, harvest index and seed weight are common traits in chickpea, maize, sorghum, rice and wheat. In addition, this tool has been used to find crop-specific and new traits, and as a test case, 150 traits of the IMIS database were analysed using the subset maize trait ontology (CO_322) and TO; 74 traits were matched to those of maize trait ontology and 35 to those of TO (Fig. 2). Since the maize trait ontology did not include all IMIS traits, Terminizer could not show all matching traits. Based on the trait query result, new traits are being submitted to TO as a generic ontology for plant traits.
Fig. 2.
A screen capture of the tool ‘Terminizer’ for GCP crop ontology at http://crops.terminizer.org/. (A) List of IMIS traits entered for analysing common and new traits in the maize trait ontology of GCP CO and Cereal Plant Trait Ontology (TO). (B) The query result showing the numbers of IMIS terms (traits) that matched to the maize trait ontology and TO.
Another potential application of the crop Terminizer includes marking up crop science-related publications in a library collection using the Terminizer web service, which then outputs an XML file containing the ontologies found in each article (this exercise would be extremely tedious if done manually). After the library collection is marked up, the XML output for each article can then be searched instead of the entire article, offering a faster and more intelligent search result.
Discussion
Free-text searching forms the basis of information mining and retrieval, but is extremely limited because of an inherent lack of accuracy and specificity (Gkoutos et al., 2004). For example, complex free-text descriptions used for phenotypes are almost impossible to index and retrieve in a useful way. The use of bio-ontologies to describe phenotypes, coupled with advanced search tools, enables researchers to fully exploit and realize the potential of these data.
It is noteworthy that many bio-ontologies are available publicly. This begs the question why the CO is needed. The reason is that the CGIAR centres have accumulated a huge amount of data over the past decades, including those related to germplasm, breeding, disease, phenotypes and agronomically important traits. In addition, the GCP Central Registry contains data related to both genotype and phenotype. The PO includes controlled vocabularies that are related to plant structure, plant growth and development, and anatomy. In the case of TO, it describes plant traits but essential information on each trait such as method, scale and scale value is missing. Moreover, these existing ontologies do not cover controlled vocabularies that are required for managing germplasm and passport information in genebanks. Therefore, CO was needed to create not only for traits, but also for managing all crop information that is required for integrating agricultural data as a whole. The CO is then the responsibility of a community of practice that curates the terms and adds additional concepts and definitions that are lacking in the global public ontologies related to plants.
The International Crop Information System (Website 15) is a database system that is being developed by agricultural scientists and information technicians in several CGIAR centres, in Advanced Research Institutions, and in National Agricultural Research Systems to address the problem of ambiguous germplasm identification, difficulty in tracing pedigree information, and lack of integration between genetic resources, breeding, evaluation, utilization and management data. This provides integrated management of global information on crop improvement and management both for individual crops and for farming systems. To make the ICIS a more user friendly and knowledge-intensive crop research information system, integration of ontologies with ICIS version 6 is ongoing. This new version of ICIS would demonstrate the importance of ontologies in databases.
Genesys, a new gateway to information on the crop varieties conserved in genebanks worldwide, is being developed by Bioversity International in collaboration with the Global Crop Diversity Trust and the Treaty for Plant Genetic Resources (PGR) for Food and Agriculture. This gateway aims at releasing available data sets about the germplasm in the interest of breeders. The second development phase will integrate the terms of the CO, ensuring that trait names in use are compatible.
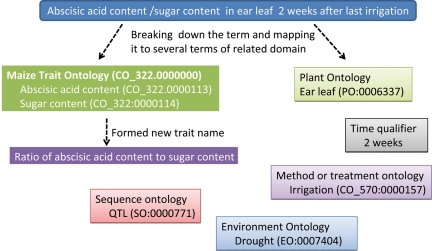
It has already been mentioned that traits were extracted from several CGIAR centres' databases for each crop and GCP central registry to develop the current CO. For example, the trait ‘abscisic acid content/sugar content in ear leaf 2 weeks after the last irrigation’ is difficult to understand by simple reading of the trait name (this name was given to a QTL-based drought-tolerant maize trait). The exact meaning of the term is the ratio of abscisic acid to sugar content in ear leaf, which is measured after 2 weeks of irrigation (J.-M. Ribaut, pers. comm.). In this case, it is clear that two traits or attributes, ‘abscisic acid content’ and ‘sugar content’, are linked together. The entity ‘ear leaf’, time qualifier ‘2 weeks’ and treatment method are written along with the traits. It is obvious that such complex terms need to be broken down into a simpler form and annotated separately (Fig. 3). In addition, a suggestion can be made for a term to cover a new trait: ‘ratio of abscisic acid content to sugar content’. In order to capture all information on traits such as this, TO alone was not enough. Therefore, CO was developed to cover other ontology domains as well.
Fig. 3.
Schematic diagram representing a method for dissecting a complex phenotyping trait term and mapping it to terms of respective ontology domains. The new trait was formed using two parents ‘Abscisic acid content'and ‘Sugar content’. The trait was also mapped to drought of EO and QTL of SO because the experiment involved QTL-based phenotyping in drought conditions.
Research has increasingly been focused at the molecular level, with QTLs, molecular breeding and simple sequence repeat (SSR) tools used extensively for crop phenotyping. Different alleles in one gene can have effects on many important traits. For example, the dwarf1 gene is associated with several phenotypic traits in rice, such as plant height, stem length, leaf colour and panicle length. Capturing such information using a controlled vocabulary allows researchers to compare data that are stored in and between databases. With the inception of the CO project, the rice mutant ontology is being integrated as a CO resource, and each mutant phenotype controlled vocabulary is now an ontology term in the rice mutant ontology. To facilitate smooth data exchange across databases and data annotation, a controlled vocabulary will be used in GCP data templates to help control data quality.
The term ‘trait’ is used in the wide range of research fields, including both plant and animal biology. A trait is defined as any morphological, physiological or phenological feature measurable at the individual level, from the cell to the whole-organism level, without reference to the environment or any other level of organization (Violle et al., 2007). Trait-based approaches are widely used in agriculture, especially in the field of plant breeding for producing stress-resistant, high-yielding varieties and cultivars. Recently, the plant research community has also been using trait-based approaches in ecological and evolutionary research to address questions from organisms to ecosystems and beyond. Plant research communities working on ecology are interested in the concept of ‘functional trait’—any trait that impacts fitness indirectly via its effects on growth, reproduction and survival (Violle et al., 2007). Trait-based approaches have been used in various research communities, but information related to plant traits needs to be brought together in order to facilitate knowledge sharing. With this concept, the CO project is working with the PO Consortium to harmonize plant traits globally. Recently, the CO project has initiated collaboration with the integrated breeding platform (IBP; Website 20) of the GCP for developing a crop trait dictionary based on crop-specific traits of the CO. This platform aims at servicing the plant breeders and the use of the CO derivated trait dictionaries will enable researchers to construct their field book and submit new terms. The inclusion of CO will enable multi-crop searching for traits in common on the platform. The dictionary will provide elaborated standard protocols that will explain scale, scale value, scoring guidelines and growth stages of the traits. The information for the wheat trait ‘stem rust’, for example, with CO term ID (CO_321:0000118) can be viewed at Website 21. The use of such traits in the development of field books and exporting them (or parts of them) to hand-held devices will provide a standard and clear specification of the traits to be measured in the field. Moreover, field books or hand-held devices will be more effective and user friendly for breeders or/and researchers.
The creation of a CO is an important first step for crop scientists who wish to integrate information from various electronic sources. Online documents can be marked up with CO terms, which in turn enable semantic web technology to work with them; documents with common ontology terms can be cross-referenced unambiguously. Furthermore, documents that do not appear to share common key words, but are properly tagged for ontological terms, can still be cross-referenced as long as there is a common parent ontological term.
Data sources and data availability
Webpage and downloads
To facilitate development of the CO, a site for curators and collaborators has been developed on the Pantheon project website (Websites 11 and 12). The site contains the complete indexed inventory of the CO and a ‘best practices’ methodology for CO curation. A project created for the CO on the CropForge project management site (Websites 9 and 10) complements the Pantheon website. This site provides both the latest releases and previous versions of ontology flat files, which describe terms, relationships, definitions and software tools. The site also provides forums and mailing lists for communication among collaborators. All the ontology flat files can be downloaded for local use.
Conclusions and forward look
Crop science and PGR practitioners span diverse research communities, each with its favourite internet communication protocols (BioMoby, TAPIR, SSWAP, GDPC, etc). The CO, along with the GCP domain model (Bruskiewich et al., 2006, 2008) and associated Pantheon software, provides a common semantic and software framework for global integration of data across diverse community protocols. Generation Challenge Programme's semantics are designed to be extensible, and to allow for the addition of new semantics and novel data types as the needs of crop researchers evolve.
The development of crop-specific ontologies for anatomy and plant traits is a collaborative process, involving the CGIAR institutions, GCP communities of practice and the POC. In developing the CO, priority will be given to ontology terms that describe crop drought-tolerance experiments, a primary focus of the GCP. The POC best practices will be followed for specifying ontology term nomenclature, definition and semantics; phenotype terms will be defined as cross-products of PO, PATO and other related public ontologies. The CO is a global public good and will be promoted for wide use by other data sources, ontology or semantic web applications. GCP will work at expanding CO with additional priority crops, the next being cassava (Manihot esculenta), and reinforce collaboration with similar partners' initiatives for crops in common. The use of ontology terms to describe agronomic phenotypes, and the ability to accurately map these descriptions into other resource databases and literature, will be an important step in gene discovery.
Sources of funding
This crop ontology development was supported by the Generation Challenge Programme [G4005.22, G4009-03] and the Terminizer development was jointly funded via the UK NERC contract F3/G13/18/04 to the NERC Environmental Bioinformatics Centre and the EU Framework 6 ActinoGEN project (FP6-5224).
Contributions by the authors
R.S. is a coordinator of the crop ontology team. She is involved in developing and curating crop-specific plant and trait ontology for maize and wheat, and maintaining GCP crop ontology web documentation. E.A. is leading the crop ontology project from 2009 as principal investigator (PI). She is involved in facilitating crop ontology project work and interacting with collaborators. She also supervises the Musa team for developing Musa plant structure, anatomy and trait ontology. R.S. and E.A. have contributed equally in preparing this manuscript. R.M. contributed in developing rice mutant ontology and coordinated embedding crop ontology in Terminizer with D.H. and N.M. of Manchester University. M.S. is a principal designer of the GCP core domain models, of the Pantheon software framework and the Ontology look-up service. G.D. is a principal designer of the Pantheon framework and software support for the GCP domain model, and is leading the implementation of CO in version 6 of ICIS. D.H. and N.M. contributed in developing crop Terminizer. R.B. was a PI of the project from 2007 to 2008. He was a principal designer of the GCP core and GCP Scientific Domain Model and Ontology, and a core contributor to the Pantheon framework. G.M. contributed by supervising overall project work.
Conflict of interest statement
None declared.
Acknowledgements
We would like to thank Genevieve Mae Aquino and Jeffrey Detras of IRRI for their contribution in indexing of GCP crop ontology. G.M.A. contributed to GCP platform project management and to elements of GCP germplasm domain model and ontology. J.D. converted several public controlled vocabularies into formal OBO managed ontology files. We would also like to thank the collaborators Jayashree Balaji (ICRISAT; chickpea and sorghum), Reinhard Simon (CIP; potato) and Stephanie Channeliere (Bioversity International; Musa) for working on crop-specific plant and trait ontology. We would like to thank Adriana Alercia, Bioversity International, for continuously providing the latest revisions of the crop descriptors that she coordinates, Chih-Wei Tung (Cornell University) for her valuable suggestions during the crop ontology meeting in Rome for its development, Kevin Manansala (IRRI) for designing and implementing most GCP specific ontology management software utilities, Milko Skofic (Bioversity International) and Franjel Consolacion (IRRI) for installing the mirror of the Ontology look-up service in Bioversity and CIMMYT, respectively, and for developing web services as well as providing feedback on crop ontology as users. We also thank Theo van Hintum (Wageningen University) for spearheading development of core GCP germplasm and passport ontology, and Tom Hazekamp (Bioversity International) for the early development of the Passport data ontology.
References
- Ashburner M, Lewis SE. On ontologies for biologists: the Gene Ontology—uncoupling the web. Novartis Foundation Symposium. 2002;247::66–80. (discussion 80–83, 84–90, 244–252) [PubMed] [Google Scholar]
- Avraham S, Tung C-W, Ilic K, Jaiswal P, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, Schaeffer M, Stein L, Stevens P, Vincent L, Zapata F, Ware D. The Plant Ontology database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Research. 2008;36::D449–D454. doi: 10.1093/nar/gkm908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruskiewich R, Davenport G, Hazekamp T, Metz T, Ruiz M, Simon R, Takeya M, Lee J, Senger M, McLaren G, van Hintum T. The Generation Challenge Programme (GCP)-Standards for Crop Data. OMICS. 2006;10::215–219. doi: 10.1089/omi.2006.10.215. [DOI] [PubMed] [Google Scholar]
- Bruskiewich R, Senger M, Davenport G, Ruiz M, Rouard M, Hazekamp T, Takeya M, Doi K, Satoh K, Costa M, Simon R, Balaji J, Akintunde A, Mauleon R, Wanchana S, Shah T, Anacleto M, Portugal A, Ulat VJ, Thongjuea S, Braak K, Ritter S, Dereeper A, Škofič M, Rojas E, Martins N, Pappas G, Alamban R, Almodiel R, Barboza LH, Detras J, Manansala K, Mendoza MJ, Morales J, Peralta B, Valerio R, Zhang Y, Gregorio S, Hermocilla J, Echavez M, Yap JM, Farmer A, Schiltz G, Lee J, Casstevens T, Jaiswal P, Meintjes A, Wilkinson M, Good B, Wagner J, Morris J, Marshall D, Collins A, Kikuchi S, Metz T, McLaren G, van Hintum T. The generation challenge programme platform: semantic standards and workbench for crop science. International Journal of Plant Genomics. 2008 doi: 10.1155/2008/369601. Article ID 369601, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day-Richter J, Harris MA, Haendel M, Lewis S. Obo-Edit—an ontology editor for biologists. Bioinformatics. 2007;23::2198–2200. doi: 10.1093/bioinformatics/btm112. [DOI] [PubMed] [Google Scholar]
- FAO/IPGRI. Multi-crop passport descriptors. Rome, Italy: FAO and IPGRI.; 2001. [Google Scholar]
- Gkoutos GV, Green ECJ, Mallon A-M, Blake A, Greenaway S, Hancock JM, Davidson D. Ontologies for the description of mouse phenotypes. Comparative Functional Genomics. 2004;5::545–551. doi: 10.1002/cfg.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic K, Kellogg EA, Jaiswal P, Zapata F, Stevens PF, Vincent LP, Avraham S, Reiser L, Pujar A, Sachs MM, Whitman NT, McCouch SR, Schaeffer ML, Ware DH, Stein LD, Rhee SY. The plant structure ontology, a unified vocabulary of anatomy and morphology of a flowering plant. Plant Physiology. 2007;143::587–599. doi: 10.1104/pp.106.092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal P, Ware D, Ni J, Chang K, Zhao W, Schmidt S, Pan X, Clark K, Teytelman L, Cartinhour S, Stein L, McCouch S. Gramene: development and integration of trait and gene ontologies for rice. Comparative and Functional Genomics. 2002;3::132–136. doi: 10.1002/cfg.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N, Buels RM, Tecle I, Mueller LA. A community-based annotation framework for linking Solanaceae genomes with phenomes. Plant Physiology. 2008;147::1788–1799. doi: 10.1104/pp.108.119560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R, Davenport GF, Bruskiewich R, Arnaud E. Development of crop ontology for sharing crop phenotypic information. In: Monneveux P, Ribaut JM, editors. Drought phenotyping in crops: from theory to practice. Mexico: Generation Challenge Programme (GCP), c/o CIMMYT; 2010. pp. 167–176. [Google Scholar]
- Violle C, Navas M-L, Vile D, Kazakou E, Fortunel C, Hummel I, Garnier E. Let the concept of trait be functional! Oikos. 2007;116::882–892. [Google Scholar]
- Zimmermann P, Schildknecht B, Craigon D, Garcia-Hernandez M, Gruissem W, May S, Mukherjee G, Parkinson H, Rhee S, Wagner U, Hennig L. MIAME/Plant—adding value to plant microarrray experiments. Plant Methods. 2006;9: doi: 10.1186/1746-4811-2-1. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Website 1. Generation Challenge Programme Phase II. http://www.generationcp.org/gen.php?da=09128234#gcp/ . (19 February 2010)
- Website 2. International Maize Information System. http://imis.cimmyt.org . (23 April 2010)
- Website 3. International Rice Information System. http://iris.irri.org/ . (23 April 2010)
- Website 4. International Wheat Information System. http://iwis.cimmyt.org . (7 May 2010)
- Website 5. Musa Germplasm Information System. http://www.crop-diversity.org/banana/ . (23 February 2010)
- Website 6. International Crops Research Institute for the Semi-Arid Tropics. http://www.icrisat.org/ . (23 February 2010)
- Website 7. International Potato Centre. http://www.cipotato.org/ . (23 February 2010)
- Website 8. Crop descriptors lists. http://www2.bioversityinternational.org/Themes/Germplasm_Documentation/Crop_Descriptors/ . (7 May 2010)
- Website 9. SINGER, System-wide information system on Genetic Resources. http://www.singer.cgiar.org/ . (7 May 2010) [Google Scholar]
- Website 10. Pantheon. http://pantheon.generationcp.org/ . (7 May 2010) [Google Scholar]
- Website 11. CropForge Domain Models. http://cropforge.org/projects/gcpmodels . (23 February 2010) [Google Scholar]
- Website 12. CropForge Project Info. http://cropforge.org/projects/gcpontology/ . (February 2010) [Google Scholar]
- Website 13. Terminzer. http://terminizer.org/ . (22 February 2010) [Google Scholar]
- Website 14. Terminizer search page. http://crops.terminizer.org/ . (22 February 2010) [Google Scholar]
- Website 15. International Crop Information System. http://www.icis.cgiar.org/ . (23 February 2010) [Google Scholar]
- Website 16. GCP—Crop Ontology Lookup Service. http://cropontology.org/ . (22 February 2010) [Google Scholar]
- Website 17. OBO-Edit. http://oboedit.org/ . (23 February 2010) [Google Scholar]
- Website 18. Lucene. http://lucene.apache.org/ . (22 February 2010) [Google Scholar]
- Website 19. Tracker: Plant Trait Ontology (TO) requests. http://sourceforge.net/tracker/?group_id=76834&atid=835557 . (23 April 2010) [Google Scholar]
- Website 20. Molecular breeding platform. http://ibp.generationcp.org/ . (23 February 2010) [Google Scholar]
- Website 21. Trait dictionaries—stem rust. http://ibp.generationcp.org/confluence/display/MBP/CO_321+0000118 . (23 February 2010) [Google Scholar]