Abstract
Objective
To propose a centralized method for generating global unique identifiers to link collections of research data and specimens.
Design
The work is a collaboration between the Simons Foundation Autism Research Initiative and the National Database for Autism Research. The system is implemented as a web service: an investigator inputs identifying information about a participant into a client application and sends encrypted information to a server application, which returns a generated global unique identifier. The authors evaluated the system using a volume test of one million simulated individuals and a field test on 2000 families (over 8000 individual participants) in an autism study.
Measurements
Inverse probability of hash codes; rate of false identity of two individuals; rate of false split of single individual; percentage of subjects for which identifying information could be collected; percentage of hash codes generated successfully.
Results
Large-volume simulation generated no false splits or false identity. Field testing in the Simons Foundation Autism Research Initiative Simplex Collection produced identifiers for 96% of children in the study and 77% of parents. On average, four out of five hash codes per subject were generated perfectly (only one perfect hash is required for subsequent matching).
Discussion
The system must achieve balance among the competing goals of distinguishing individuals, collecting accurate information for matching, and protecting confidentiality. Considerable effort is required to obtain approval from institutional review boards, obtain consent from participants, and to achieve compliance from sites during a multicenter study.
Conclusion
Generic unique identifiers have the potential to link collections of research data, augment the amount and types of data available for individuals, support detection of overlap between collections, and facilitate replication of research findings.
Keywords: Patient identification systems, database management systems, registries, medical record linkage, epidemiologic studies, confidentiality, privacy, computer security
Introduction
Autism spectrum disorder (ASD) is a heterogeneous syndrome characterized by atypical social behavior, disrupted communication, and repetitive behaviors. The complexity of this disorder requires the development of large, well-characterized collections of affected individuals and their families to facilitate genotype–phenotype studies.1 Because replication is crucial for confirming the validity of such research, it is important to consider collaborations among multiple independent studies; this can yield greater generalizability of findings across the diversity of populations and exposures.2
Current collections of autism data and biospecimens include Autism Genetic Resource Exchange,3 Autism Genome Project,4 the Autism Tissue Program,5 National Database for Autism Research (NDAR),6 and Simons Foundation Autism Research Initiative (SFARI) Simplex Collection.7 There is no universal method for combining data across these collections, even when patient consent would allow it. Similarly, it is difficult to assess when collections overlap.
In this paper, we propose a centralized method for generating global unique identifiers (GUIDs) to link autism collections. The approach uses identifying information that is known to subjects or easily accessible. Each item of identifying information is invariant over a subject's lifetime (eg, name and location at the time of birth), so the method will always generate the same identifier for a subject despite the passage of time or movement across locations. Multiple items are gathered and combined in different ways, facilitating matching even in the face of variation across collection sites. The identifying information undergoes one-way encryption before being shared with the central system, so that personal identifying information (PII) is never transmitted or stored outside collection sites.
Background
In the present state of epidemiologic research, investigators often rely on a ‘patchwork quilt’ of record linkage techniques to bring separate datasets together.8 A solution on a national scale could greatly improve linkage efficiency. A proposed architecture for the National Health Information Network seeks to link public health research to routine patient care, but provides no robust mechanism for identifying a single patient across multiple locations.9 The Connecting for Health initiative proposes a Record Locator Service that aggregates records from all locations where a patient has received care, but does not explicitly address the needs of researchers.10 An architecture for disease registries proposes a method for secure submission of patient information, but relies on probabilistic methods to carry out record linkage.11
Other approaches to linking research datasets involve the services of a third party to carry out the matching process on identifying information.12 Cryptographic methods can be used to perform the merging of two sets of data without a human agent.13 A similar method combines hashing and encryption to allow secure linkage across studies, but is limited to the numeric identifiers assigned in patient care.14 A broader approach combines multiple pieces of identifying information (such as name and date of birth), but does not address how variant forms from different sources are handled.15 There are a number of algorithms that preserve privacy that are robust to variation in data.16 These record linkage approaches attempt to address the problem after subjects have been enrolled and the study data have been collected.
This work is a collaboration between the SFARI and the NDAR. The latter was initially using Biomedical Informatics Research Network in which each participating site maintains a table mapping local identifiers to a central identifier.17 Discussions with members of the Autism Genome Project suggested an approach based on hashing, but noted challenges in getting local sites to adopt the method.13
The SFARI and NDAR team required a method that would work on a large scale. From our collective experience, we had strong signals from institutional review boards that any approach that involved transmission of PII outside of data collection sites would be unacceptable, in particular, sharing such information with a central authority or storage in a central system. We decided to pursue a new method based on hashing to balance the competing demands of detecting the identity and protecting the privacy of subjects. In contrast with record linkage approaches, we sought a method that would enable assignment of a GUID to each subject before data collection begins in a study. The SFARI group developed a prototype algorithm, refined it with input from the NDAR team, and then tested the production code developed by NDAR.
Methods
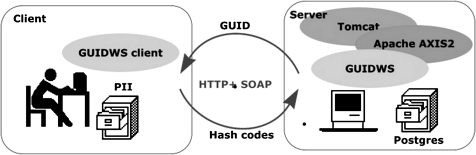
A GUID for research purposes is a random sequence of characters that is unique to each research participant, regardless of the study. The process for generating a GUID for a participant is implemented here as a web service (GUIDWS), involving a client application and a server application (figure 1). The two applications communicate via hypertext transfer protocol and simple object access protocol. The following steps provide a broad outline of the process, with details described in the following sections.
Using the client application, the researcher enters a specific set of PII obtained from the research participant, such as name, date, and city at birth. The client application processes the identifiers into several intermediary codes using a one-way hash function, and transmits the codes in a secure manner to the server application.
The server application compares the transmitted hash codes against an internal database. If there is no match, the server application generates a random GUID according to a particular format, and stores the association between the hash codes and the GUID for future use. If the codes match those from a previous transmission, the associated GUID is obtained. In either case, the server returns the resulting GUID to the client application.
The researcher obtains the GUID from the client application, and stores it locally to establish an association between the GUID and the participant's research data.
In order to share the participant's research data with other investigators, the researcher removes all identifying information except the GUID. This anonymized dataset can now be linked with other datasets that follow this process (assuming appropriate consent agreements are in place).
Figure 1.
System architecture. GUIDWS, global unique identifier web service; PII, personal identifying information; Postgres, an open-source Relational Database Management System.
Implementation of client application
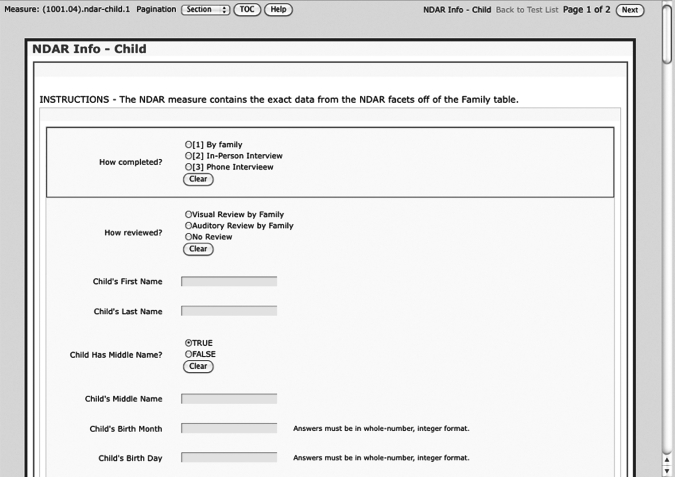
The client application requires Java JRE 1.5 to be installed on the local machine, running the UNIX, LINUX, Apple MacOS X, or Microsoft Windows operating system. A researcher can interact with the client application as a graphical user interface. Figure 2 shows an example screen for data entry to be used after the information is captured on a paper form.
Figure 2.
Portion of online graphical interface for entering fields of identifying information for a child.
Alternatively, a researcher can run the client application from the command line, allowing efficient generation of GUIDs for multiple subjects using information stored in a simple text file.
The client program can also be invoked through an application programming interface (API), which enables software developers to integrate their data processing applications directly with the GUIDWS. For example, we used the API method in the SFARI project to launch the client from SFARI Outpost, a clinical site data-entry and study management system developed for SFARI by our software partner, Prometheus Research, LLC.
Securing client and server communication
Security in the GUIDWS architecture is provided at the application level and at the network level. At the application level, Web Services Security supports a secure connection between a client and server through encryption and authentication. Encryption is implemented using a key system in which each client program identifies itself to the server using a particular pair of public and private keys. This reduces the opportunity for interception of messages and unauthorized access to the server.
At the network level, access to the GUIDWS server is restricted by the client's internet protocol (IP) address. Each site that wishes to use the service must submit the fully qualified domain name (eg, somehost.example.com) and static IP address of the machine that will run the client application. If the client machine has a dynamic IP address, the site must provide a range of addresses with a subnet mask. In addition, the server audits all access and requests and reports unusual activity to the system administrator. For additional security, the Java code of the client application is obfuscated to reduce the potential for the algorithm to be decompiled and analyzed.
Data fields and hash code formation
The fields of PII employed in GUIDWS were chosen to be relatively easy to acquire, yet invariant over the lifetime of the subject. Most can be obtained from a birth certificate. For each field, table 1 lists the name, abbreviation, inverse probability, and whether the field is required in order to generate a GUID. The inverse probability is a very rough estimate of the unlikelihood of two different individuals drawn randomly from the subject population sharing the same value for that field.
Table 1.
Fields of identifying information used in the generation of the hash codes
| Index | PII field name | Abbreviation | Inverse probability | Required |
| 1. | Government issued ID or national ID | GIID | 1000000000 | No |
| 2. | Complete legal given name of subject at birth | FN | 200 | Yes |
| 3. | Complete legal family name of subject at birth | LN | 3000 | Yes |
| 4. | Complete additional legal name or names at birth, if any, such as a middle name | MN | 100 | Yes |
| 5. | Day of month of birth | DOB | 30 | Yes |
| 6. | Month of birth | MOB | 12 | Yes |
| 7. | Year of birth | YOB | 15 | Yes |
| 8. | Physical sex of subject at birth (M/F) | Sex | 2 | Yes |
| 9. | Name of city/municipality in which subject was born | COB | 300 | Yes |
| 10. | Mother's complete legal given name at her birth | MFN | 200 | No |
| 11. | Mother's complete legal family name at her birth | MLN | 3000 | No |
| 12. | Father's complete legal given name at his birth | FFN | 200 | No |
| 13. | Father's complete legal family name at his birth | FLN | 3000 | No |
| 14. | Mother's day of month of birth | MDOB | 30 | No |
| 15. | Mother's month of birth | MMOB | 12 | No |
| 16. | Father's day of month of birth | FDOB | 30 | No |
| 17. | Father's month of birth | FMOB | 12 | No |
PII, personal identifying information.
The key feature of the GUID system is that the fields of identifying information (table 1) are combined in several different ways to form hash codes (table 2). Each field is normalized to have only uppercase letters and numbers, no spaces, and no punctuation. For example, the city of birth ‘St Paul’ is rendered STPAUL; the two middle names for ‘Sally Emma Clark Richards’ are rendered ‘EMMACLARK’, the hyphenated last name ‘Port-Wetherby’ is ‘PORTWETHERBY’. A special flag is included to indicate whether the subject has a middle name or not; this distinguishes between having a middle name with an unknown value and not having a middle name at all (see Discussion for explanation of usage).
Table 2.
Combination of fields used to form the hash codes
| Hash Code | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Prob | L | U |
| 1 | YOB* | DOB* | GIID | Sex* | 9E+12 | 0 | 1 | ||||
| 2 | FN* | MN* | LN* | COB* | DOB* | MOB* | 6E+12 | 0 | 0 | ||
| 3 | FN* | YOB* | MFN | MLN | FFN | FLN | 5E+15 | 1 | 3 | ||
| 4 | FN* | LN* | MDOB | MMOB | FDOB | FMOB | COB* | Sex* | 4E+13 | 1 | 3 |
| 5 | FN* | MN* | MOB* | MFN | FFN | MLN | 3E+13 | 1 | 3 |
Indicates required fields.
FN, first name; MN, middle name; LN, last name; YOB, year of birth; MOB, month of birth; DOB, day of birth; COB, city of birth; GIID, government issued identifier; MFN, mother's first name; MLN, mother's last name; MMOB, mother's month of birth; MDOB, mother's day of birth; FFN, father's first name; FLN, father's last name; FMOB, father's month of birth; FDOB, father's day of birth; Prob, estimated inverse probability of combined fields; L, lower threshold for number of missing values in hash code; U, upper threshold.
The fields are concatenated into the five patterns shown in table 2. Each combination of fields is converted to a hash code using a one-way hash algorithm. An additional byte is added to each resulting code to indicate the number of missing values for that code. The combination of PII fields was chosen such that each combined inverse probability of each hash code is sufficiently high to confidently delineate subjects.
Matching hash codes on the server
When the GUIDWS server application receives a request, it must determine whether the hash codes match those of an individual already in the database. The server can make a match using a subset of the five hash codes, provided they exceed a certain level of quality. When the quality conditions are not met, a new GUID is returned and stored in the database with its associated hashes. The quality of the match is based on the number of values missing in each hash code. This number is compared with a lower threshold (L) and an upper threshold (U) specific to each hash code (table 2). Each hash code is assessed as follows:
Perfect hash code: the number of missing values that form the particular hash is equal to or less than L.
Good hash code: the number of missing values that form the particular hash is greater than L, but equal to or less than U.
Bad hash code: the number of missing values that form the particular hash is greater than U.
The assessment of hash codes is applied to the new hashes in the current request as well as to the old hashes that are stored in the database. A ‘good match’ results when a good, new hash code is identical with a good, old hash code. A ‘perfect match’ results when a perfect, new hash code is identical with a perfect, old hash code. The server finds a match whenever there is at least one perfect match, or at least two good matches.
Generating a GUID
When the server determines that a set of hash codes represents a new individual, it generates a new GUID. The server produces a human-readable identifier in a particular format—for example, NDARCJ743PV3. In our implementation, a GUID consists of a prefix, an alphanumeric pattern, and a check character. For research on ASD, we have chosen the prefix NDAR (National Database for Autism Research). The alphanumeric pattern is AANNNAA (CJ743PV in the current example), where A represents an alphabetic character and N represents a numeric character. The server uses a nondeterministic random number generator to generate the pattern, excluding the letters I, O, Q, and S, because of their similarity to numeric characters. A check character (the final character ‘3’ in the current example) is calculated from the pattern portion of the identifier, and is used to verify the accuracy of the other characters—for example, when typed in during data entry.
Simulation study
The purpose of the GUIDWS system is to generate identifiers for research participants such that no two individuals are assigned the same identifier (false identity) and no individual is assigned more than one identifier (false split). To evaluate this, we conducted a simulation study using mailing list information on one million individuals18 consisting of first name (FN), last name (LN), middle name (MN), and city of residence. The city of residence was used to simulate city of birth (COB). Values for date of birth (YOB, MOB, and DOB) and government issued identifier were generated randomly. Individuals were randomly grouped into 250 000 families of four members each. (Many autism collections consist of families consisting of parents, one or more affected children, and zero or more unaffected siblings.) Individuals were assigned parents' names (MFN, MLN, FFN, and FLN) and dates of birth (MDOB, MMOB, FDOB, FMOB) to be logically consistent with the family structure.
The 17 fields in table 1 (with no missing values) were transmitted to the GUIDWS server using the API version of the client application. The same individuals were then loaded again using only the eight required fields (FN, MN, LN, DOB, MOB, YOB, SEX, and COB), with no missing values. The GUIDs generated on the second round were compared with the first round, in terms of false splits and false identity.
Field study
In order to support research, researchers must be able to successfully collect the identifying information used by the GUIDWS system. We evaluated this in the SFARI Simplex Collection, which is gathering phenotype and genotype information on 3000 families in North America, in which each family has only one child affected with an ASD and at least one unaffected sibling. At present, over 2000 families have been recruited at 13 university-based sites.
The SFARI team developed a paper form that participants completed as part of enrolling in the study. Form data were reviewed for completeness by site coordinators, and follow-up was conducted by phone if necessary. Form data were entered by site staff with privileges to see PII into a corresponding electronic form in SFARI Outpost, an integrated data entry and study management system for SFARI clinical sites. The electronic form (figure 2) supported live field-level validation rules (eg, date fields must contain valid dates) and dual-data entry. The SFARI Outpost submitted data to the GUIDWS client application via the API, and recorded the returned identifiers in the clinical database. The NDAR team provided access to the GUIDWS application hosted at the National Institutes of Health.
Results
Simulation study
The goal of this study was simple: individuals loaded on the second round had to produce GUIDs identical with those in the first round. In earlier versions of the algorithm, errors generated by such testing helped confirm certain decisions about required fields, lower bounds, and upper bounds. For example, first name needs to be a component of hash code 2; without it there is no way to distinguish identical twins. Testing also revealed the importance of distinguishing an unknown middle name from a middle name that is known to be empty. The Discussion provides more details on these design choices. The current version of hashes shown here was successful in matching individuals, with no false identities and no false splits. This provides face validity that the algorithm can distinguish large numbers of individuals.
Batch requests to the GUIDWS for several hundred GUIDs are usually processed in less than 10 s. This suggests that large-volume uses of the system are feasible, whether for testing such as this study or for other kinds of linkage applications.
Field study
Our experience with using the client application in the field revealed that significant design work is required to obtain a usable graphical user interface. Field names must be carefully chosen to make absolutely clear what information is being requested. For example, it is challenging to clearly communicate that the field should contain the legal name at birth of the proband's (affected child's) mother. The time required to fill out the paper form for NDAR GUID is ∼3 min. The time needed for dual entry and validation using our SFARI Outpost software is ∼4 min. The response time to receive a GUID from the server depends on network conditions, but is usually no more than 3 s.
Research coordinators experienced significant resistance from subjects in collecting PII, particularly mother's maiden name and government issued identifier (usually social security number). Many sites experienced considerable difficulty obtaining approval from their local institutional review boards, causing delays in deploying the form for the study. Several sites had low rates of compliance, even after making the form a mandatory part of the study (it was not mandatory at the start). Initially, only about one half of subjects in the study were assigned identifiers (table 3). Compliance increased to 72% after a series of announcements to collection sites requesting remediation. A substantial reclamation effort was required to raise compliance to the current levels, and involved recontacting families whose forms lacked information by email, US Mail and phone. This effort was focused primarily on probands, 97% of whom now have valid identifiers. Siblings are close to this rate at 96%, while parents lag behind a bit at 84%. (The fall in valid identifiers for parents in the last quarter is due to recent data submissions that have not been cleaned, such as families that are still undergoing validation.)
Table 3.
Percentage of valid global unique identifiers for each type of family member in the SFARI Simplex Collection, over the previous four quarters
| Proband | Sibling | Mother | Father | |
| Jul-09 | 56.2 | 51.6 | 54.2 | 53.3 |
| Oct-09 | 83.0 | 80.2 | 79.1 | 78.5 |
| Nov-09 | 88.5 | 86.5 | 84.7 | 84.1 |
| Apr-10 | 96.6 | 95.8 | 77.9 | 77.4 |
Table 4 shows statistics on the availability of specific fields (for subjects for whom we were able to obtain completed forms). As in table 3, results are somewhat better for probands and siblings than for mothers and fathers. The required fields (first eight rows) show somewhat higher availability than the remaining fields (pertaining to information about parents). Social security number has the poorest acquisition rate, and is missing for about half of parents and more than half of probands and siblings. When composing these fields into hashes, it is not surprising that hash code 2 performs the best (formed from only required fields), and hash code 1 performs the worst (containing social security number), with the hashes using parental information falling in between.
Table 4.
Percentage of available fields and hashes for subjects in total, probands, siblings, mothers, and fathers
| Total | Proband | Sibling | Mother | Father | |
| MOB | 0.995 | 0.996 | 0.994 | 0.995 | 0.996 |
| FN | 0.995 | 0.996 | 0.994 | 0.995 | 0.996 |
| YOB | 0.995 | 0.996 | 0.994 | 0.995 | 0.996 |
| DOB | 0.995 | 0.996 | 0.994 | 0.995 | 0.996 |
| Sex | 0.995 | 0.996 | 0.994 | 0.995 | 0.994 |
| LN | 0.994 | 0.996 | 0.994 | 0.991 | 0.996 |
| MN | 0.975 | 0.982 | 0.981 | 0.965 | 0.973 |
| COB | 0.971 | 0.987 | 0.981 | 0.967 | 0.950 |
| MFN | 0.890 | 0.993 | 0.993 | 0.802 | 0.789 |
| FLN | 0.889 | 0.994 | 0.993 | 0.792 | 0.793 |
| FFN | 0.887 | 0.994 | 0.993 | 0.788 | 0.789 |
| MLN | 0.885 | 0.990 | 0.990 | 0.798 | 0.782 |
| MMOB | 0.855 | 0.993 | 0.993 | 0.758 | 0.698 |
| MDOB | 0.849 | 0.993 | 0.993 | 0.758 | 0.673 |
| FMOB | 0.834 | 0.994 | 0.993 | 0.719 | 0.655 |
| MYOB | 0.831 | 0.993 | 0.993 | 0.729 | 0.636 |
| FDOB | 0.822 | 0.994 | 0.993 | 0.707 | 0.624 |
| FYOB | 0.811 | 0.994 | 0.993 | 0.680 | 0.609 |
| GIID | 0.503 | 0.447 | 0.420 | 0.571 | 0.558 |
| Hash2 | 0.952 | 0.974 | 0.968 | 0.936 | 0.933 |
| Hash3 | 0.872 | 0.988 | 0.987 | 0.769 | 0.762 |
| Hash5 | 0.854 | 0.975 | 0.974 | 0.744 | 0.745 |
| Hash4 | 0.792 | 0.984 | 0.978 | 0.672 | 0.566 |
| Hash1 | 0.502 | 0.447 | 0.420 | 0.571 | 0.557 |
| Perfect | 0.803 | 0.875 | 0.867 | 0.753 | 0.728 |
| Good | 0.129 | 0.111 | 0.116 | 0.138 | 0.148 |
| Bad | 0.045 | 0.000 | 0.000 | 0.080 | 0.093 |
| Incomplete | 0.023 | 0.013 | 0.017 | 0.029 | 0.031 |
For hashes with fields missing, ‘perfect’ have minimum missing, ‘good’ up to maximum, ‘bad’ more than maximum, and ‘incomplete’ have required fields missing.
FN, first name; MN, middle name; LN, last name; YOB, year of birth; MOB, month of birth; DOB, day of birth; COB, city of birth; GIID, government issued identifier; MFN, mother's first name; MLN, mother's last name; MMOB, mother's month of birth; MDOB, mother's day of birth; FFN, father's first name; FLN, father's last name; FMOB, father's month of birth; FDOB, father's day of birth.
Even with missing fields, the hash codes still perform very well. On average, four out of five (80%) of an individual's hashes are perfect (missing values are less than or equal to the lower bound in table 2), and the remaining hash is usually good (missing values are less than or equal to the upper bound in table 2). Bad hashes (missing values exceed the upper bound) occur in roughly one out of five parents (none for probands or siblings), and incomplete hashes (missing a required field) occur in roughly one out of 10 individuals.
Discussion
The GUIDWS system must strike a balance among three goals. To prevent false identity, a sufficient number of identifying fields must be available to distinguish one individual from another. To prevent false splits, it must be feasible to collect the fields with sufficient accuracy to enable matching. To protect confidentiality, it must be very difficult to guess the original values of the hashes stored on the server.
These goals compete in a number of ways. To ensure that two individuals are different, we would like to know as much about them as possible. However, this makes data collection more difficult. Protecting identity requires combining fields in fixed ways, which reduces the combinations available to establish unique identity. Because fields in a hash are packaged together, an error in any one field blocks matching in the database. The more missing values that we allow in a hash, the more potential there is to confuse two individuals.
Distinguishing individuals
One can think about the fields in table 1 as descriptors that ‘locate’ an individual in time (eg, YOB), space (eg, COB), and parentage (eg, MLN). A ‘perfect’ hash must adequately discriminate individuals using the set of fields that have known values. For example, hash code 2 must always be perfect, because all the fields are required. If the values are all entered correctly, a match will be found. The inverse probability of false identity is approximately 6E+12, which occurs when two persons are born in the same city, in the same month, on the same day with the same complete name.
Two ‘good’ hashes must adequately discriminate individuals with their combined set of known values. For example, when only the required fields of hash codes 1 and 5 are available, both hash codes qualify as good. The union of the fields in these two hashes (FN, MN, MOB, DOB, YOB, Sex) results in a combined inverse probability of approximately 2.16E+8.
Twins are an important case requiring discrimination, because most of the identifying information for twins will be the same. In the general population, the rate is about 1.2% (including both dizygotic and monozygotic twins). Studies that focus on twins will have higher rates. Hash code 2 adequately separates twins, since the first names (and middle names, if present) are almost certainly distinct. For this reason, first name is also a component of hash codes 3, 4, and 5.
Because the GUIDWS system always returns a GUID, it cannot be used to detect cases of false identity. This function requires a study database that records the GUIDs that have been assigned to participants in the study of interest, and ideally across many studies. When a new participant is being enrolled, the research coordinator uses the client application to generate a GUID, and checks the study database to see whether the identifier has been seen before. If so, the coordinator asks the participant about prior participation, and may also contact relevant collection sites to resolve any discrepancies. Depending on the study, the participant may be disqualified from enrolling because of previous participation, or may be eligible for follow-up. In the case of true confusion of identity, attempts should be made to correct identifying information for the two individuals to obtain distinct GUIDs. If this fails, the coordinator cannot assign a GUID, and may choose not to enroll the new participant.
Feasible collection of accurate data
For the GUIDWS system to work properly, the identifying information collected must be readily available and have a high chance of accurate entry. Items such as government issued identifier and information about parents cannot be required fields. If there are many errors in the items that are captured, none of the hashes will match, causing a false split. Matching can be made more robust if each field is hashed separately, but this conflicts with the goal of making hashes hard to guess by an attacker.
All the fields for hash code 2 are required: FN, MN, LN, MOB, DOB, COB. An error in any value will cause matching to fail. For example, there may be inconsistent entry for LN or COB, particularly when transliterating from foreign languages. In either case, we can fall back on hash codes 1 and 5 as described above. However, an error in the middle name (MN) is fatal. Without it, only hash code 1 is good, which is not enough to identify the subject. For this reason, it is essential to distinguish between an unknown middle name and a middle name that is known to be empty.
Variants in first name are common, and even more so in middle names. Asking participants for ‘official’ versions of their names does not solve the problem, because their memory of what is printed on a birth certificate may be unreliable (eg, when one spouse completes the form for another). Most studies will not be able to exclude participants who do not bring a birth certificate, but subjects could be strongly encouraged to do so.
One way to avoid dependence on middle name is to add a sixth hash code: FN, LN, Sex, DOB, MOB, YOB, COB. This would allow the situation in which for some city, two individuals with the same first and last names are born on the same day. Other techniques to consider include: computing additional fields that are more stable than their sources (eg, first letter of last name), phonetic representation of strings, and advanced string hashing techniques.16 These methods have not been investigated in the current implementation.
The approach presented here is pragmatic: we use double data entry of required PII items that are highly reproducible and invariant over the life time of the subject. If all items are entered, then the GUID system is tolerant to errors of the PII fields because there are five hash codes and only one needs to be correct to match a subject. Our experience to date indicates that first name accuracy is actually very high.
False splits are of little practical concern for a single study, such as the SFARI Simplex Collection, which involves 13 geographically disparate sites over a period of 3 years. For follow-up studies, participants who return to the clinical sites where they initially enrolled will already have GUIDs on file, and there will be no need to access the GUID server again. The only need to look up GUIDs will be for subjects who enroll in a follow-up study through a different site than they originally registered with.
False splits could have negative consequences for studies involving genetic screens, which generally seek rare events. Seeing a particular gene twice rather than once has the potential to make a random effect appear to be a new discovery. Researchers must follow-up any such events with genetic checks for identity on subjects culled from such screens. In studies conducting large-scale genomic analysis like the SFARI Simplex Collection, it may be possible to confirm identity by comparing pairs of subjects. The GUID system should therefore provide an easy way to record any such instances where two identifiers are found to refer to the same person, and permanently merge them into a single identifier.
Protecting confidentiality
As noted above, the GUIDWS system cannot store fields of identifying information as separate items. This would open the system to a dictionary attack. With access to the database, an attacker could hash all common first names, all common last names, all common cities, and so forth and then read off a huge fraction of the entries. Therefore, each hash has to be a combination of multiple fields. Since hashing is cheap for the attacker, there must be at least billions of possibilities for the contents of each hash. In the proposed scheme, each hash code has an approximate data value range of more than 1012 (when all of the required fields are supplied). These probabilities are only rough approximations, because not all fields are truly independent (eg, sex is highly correlated with first name). Even so, the magnitude should make dictionary attacks highly infeasible.
A crucial feature of the GUIDWS system is that it only returns a GUID, and does not provide any information as to whether it is a new or existing identifier. Even if someone had access to the system and accurate identifying information on an individual, it would not be possible to use the GUID server to determine whether that individual was previously enrolled in a study. As noted above, the ability to detect prior enrollment is an important function, but is only available to research coordinators who already are permitted access to identifying information, because of direct contact with the participant. On a larger scale, verification of enrollment across multiple studies could be supported by research registries, but only by authorized personnel, with prior consent from participants.
These protections must be well understood by investigators, and made absolutely clear to institutional review boards and to participants. For example, institutional review boards appeared concerned about collection of unusually sensitive identifying information (social security numbers, mother's maiden name). They were understandably concerned that this information would not be stored securely, and had some difficulty understanding that the PII would never be transmitted off-site.
It is crucial to communicate that the GUIDWS database does not store any identifying information. In addition, after a GUID is obtained for a participant, the local database need only store a minimum set of identifying information needed by coordinators for future identity checks. The intention to use GUIDs to link collections and share data through public, anonymized databases must be made absolutely clear when obtaining informed consent.
Conclusion
Generic unique identifiers have the potential to link collections of research data, augmenting the amount and types of data available for individuals, supporting detection of overlap between collections, and facilitating replication of research findings. The proposed system balances three goals: distinguishing individuals from each other, collecting information with sufficient accuracy for matching, and protecting confidentiality. Of these, the greatest risk is assigning an individual more than one identifier (false split). This can be mitigated by developing study protocols that involve use of birth certificates and working closely with communities and institutional review boards to gain approval for collecting such information. Collections involving genomic scans can also help to verify identity.
Footnotes
Funding: This work was supported by the Simons Foundation Autism Research Initiative (SFARI) and the National Database for Autism Research (NDAR), National Institute of Mental Health. Other Funders: NIH; Simons Foundation Autism Research Initiative; National Database for Autism Research; National Institute of Mental Health.
Ethics approval: This study was conducted with the approval of Columbia University.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet 2008;9:341–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanock SJ, Manolio T, Boehnke M, et al. Replicating genotype-phenotype associations. Nature 2007;447:655–60 [DOI] [PubMed] [Google Scholar]
- 3.Geschwind DH, Sowinski J, Lord C, et al. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet 2001;69:463–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu-Lince D, Craig DW, Huentelman MJ, et al. The Autism Genome Project: goals and strategies. Am J Pharmacogenomics 2005;5:233–46 [DOI] [PubMed] [Google Scholar]
- 5.Haroutunian V, Pickett J. Autism brain tissue banking. Brain Pathol 2007;17:412–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Database for Autism Research; National Institute of Mental Health. September 28, 2009. http://ndar.nih.gov
- 7.Simons Foundation Autism Research Initiative; Simons Foundation. September 28, 2009. http://sfari.org
- 8.Leufkens HJ, van Delden JJ. Ethical aspects of epidemiological research. In: Ahrens W, Pigeot I, eds. Handbook of epidemiology. Berlin: Springer, 2005 [Google Scholar]
- 9.Mcmurry AJ, Gilbert CA, Reis BY, et al. A self-scaling, distributed information architecture for public health, research, and clinical Care. J Am Med Inform Assoc 2007;14:527–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Record Locator Service The Connecting for Health Common Framework. September 28, 2009. http://www.connectingforhealth.org
- 11.Churches T. A proposed architecture and method of operation for improving the protection of privacy and confidentiality in disease registers. BMC Med Res Methodol 2003;6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruse RL, Ewigman BG, Tremblay GC. The Zipper: a method for using personal identifiers to link data while preserving confidentiality. Child Abuse Negl 2001;25:1241–8 [DOI] [PubMed] [Google Scholar]
- 13.Segre AM, Wildenberg A, Vieland V, et al. Privacy-Preserving Data Set Union. In: Domingo-Ferrer J, Franconi L, eds. Privacy in Statistical Databases. Berlin: Springer-Verlag, 2006:266–76 [Google Scholar]
- 14.Quantin C, Fassa M, Coatrieux G, et al. Combining hashing and enciphering algorithms for epidemiological analysis of gathered data. Methods Inf Med 2008;47:454–8 [DOI] [PubMed] [Google Scholar]
- 15.Wjst M. Anonymizing personal identifiers in genetic epidemiologic studies. Epidemiology 2005;16:131. [DOI] [PubMed] [Google Scholar]
- 16.Schnell R, Bachteler T, Reiher J. Privacy-preserving record linkage using Bloom filters. BMC Med Inform Decis Mak 2009;9:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keator DB, Grethe JS, Marcus D, et al. A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN). IEEE Trans Inf Technol Biomed 2008;12:162–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingsbridge Marketing Partners, Inc. 23801 Calabasas Road, Suite 102, Calabasas, CA 91302.