Abstract
Objective
Active drug safety surveillance may be enhanced by analysis of multiple observational healthcare databases, including administrative claims and electronic health records. The objective of this study was to develop and evaluate a common data model (CDM) enabling rapid, comparable, systematic analyses across disparate observational data sources to identify and evaluate the effects of medicines.
Design
The CDM uses a person-centric design, with attributes for demographics, drug exposures, and condition occurrence. Drug eras, constructed to represent periods of persistent drug use, are derived from available elements from pharmacy dispensings, prescriptions written, and other medication history. Condition eras aggregate diagnoses that occur within a single episode of care. Drugs and conditions from source data are mapped to biomedical ontologies to standardize terminologies and enable analyses of higher-order effects.
Measurements
The CDM was applied to two source types: an administrative claims and an electronic medical record database. Descriptive statistics were used to evaluate transformation rules. Two case studies demonstrate the ability of the CDM to enable standard analyses across disparate sources: analyses of persons exposed to rofecoxib and persons with an acute myocardial infarction.
Results
Over 43 million persons, with nearly 1 billion drug exposures and 3.7 billion condition occurrences from both databases were successfully transformed into the CDM. An analysis routine applied to transformed data from each database produced consistent, comparable results.
Conclusion
A CDM can normalize the structure and content of disparate observational data, enabling standardized analyses that are meaningfully comparable when assessing the effects of medicines.
Keywords: Pharmacoepidemiology; product surveillance, Postmarketing; drug safety, medical records systems, computerized; Epidemiologic methods; drug Toxicity; databases, Factual; medical Informatics Computing
Introduction
Drug safety scientists have traditionally relied on information from clinical trials and post-market individual patient case reports of adverse drug reactions to identify potential safety issues in marketed medicinal products.1 More recently, data mining methods have been applied to large collections of patient case reports to detect patterns that may not be obvious by individual case review.2 The limitations of the analytic methods and the current sources of safety information are well documented.3–7 There is emerging interest in the use of observational databases, including administrative claims and electronic medical records, to augment post-approval drug safety surveillance activities. However, data recorded in observational databases were collected for purposes other than drug safety research; administrative claims data support insurance reimbursement processes, while electronic medical records are aimed at supporting clinical practice at the point of care. In addition, differences between the data organization, format, and terminologies used among individual observational data sources have historically made safety analyses utilizing multiple observational data sources time consuming and expensive, and comparisons among results of studies utilizing disparate databases difficult.
Recent drug safety initiatives have begun to explore the use of a common data model (CDM) to enable systematic analysis of observational databases. The concept behind this approach is to transform data contained within disparate databases into a common format (data model), and then perform systematic analyses using a library of standard analytic routines that have been written based on the common format. If the approach proves successful, large numbers of records in disparate data sources could be analyzed rapidly and efficiently to identify and evaluate potential drug safety signals. In this paper, we examine the results of the transformation of two disparate, observational databases into a CDM that was specifically developed for the purpose of supporting drug safety research. We evaluate the effects of the transformation on the data itself, as well as performance characteristics of the CDM for supporting drug safety analyses.
Background
Gaps in the current post-approval drug safety system
In 2004, the highly visible recall of rofecoxib focused the attention of the industry, public, government, and press on the issue of drug safety.8 Subsequent drug safety issues and recalls have kept the topic of drug safety in the public eye and have highlighted well-documented shortcomings in the current drug safety monitoring system. In a 2006 report, the Institute of Medicine provided an assessment of the current system for evaluating and ensuring drug safety post-approval. Among the findings documented in this report are recommendations for improvement of the current drug safety system including the increased use of automated healthcare databases for formulation and testing of drug safety hypotheses.3 In the following year, the US Congress passed the Food and Drug Administration Amendments Act of 2007, which in part, mandated the “…development of validated methods for the establishment of a post-market risk identification and analysis system to link and analyze safety data from multiple sources.”9 In May 2008, partially in response to the Institute of Medicine report and Food and Drug Administration Amendments Act, the United States Food and Drug Administration (FDA) issued a report describing its intention to establish a national, integrated, electronic system for monitoring medical product safety using multiple, existing electronic medical record systems and claims databases to augment the agency's current capability. The Sentinel Initiative was launched to instantiate this vision.10 Related to this initiative, several organizations have been established to focus on research into the use of observational data for drug safety research, including the Observational Medical Outcomes Partnership (OMOP).11 Interest in the use of observational data for drug safety research is not limited to the United States; EU-ADR12 and IMI-PROTECT13 are two European consortia currently working on the development of an innovative computerized system to detect safety signals in observational data to supplement spontaneous reporting systems.
The observational data landscape
A large number of observational databases are already being utilized for medical research and could potentially be implemented as part of a national drug safety monitoring system. De-identified patient claims, pharmacy, laboratory, and electronic medical records are available for license through vendors such as GE Healthcare, IMS Health, Thomson Reuters, and i3 Ingenix. Large repositories of identifiable patient claims, pharmacy, medical record, hospital, and laboratory data are owned and curated by public, private, and not-for-profit managed healthcare organizations such as Kaiser-Permanente, WellPoint, and United Health. Although patient privacy issues make access to these databases more difficult, many are routinely used for medical research through collaborations with the data owners. In addition, government agencies such as the Veterans Administration and the Department of Defense administer large repositories of patient data for US veterans and service members; both of these agencies routinely participate in clinical studies utilizing these databases.
The need for a CDM
The use of observational healthcare databases in support of drug safety and health outcomes studies is not new.14 15 However, disparate observational databases have different logical organizations and physical formats, and the terminologies used to describe the medicinal products and clinical conditions vary from source to source. Therefore, data analyses performed in support of these ad hoc safety studies are typically accomplished by developing custom programs that conform to a specific observational data format and incorporate source-specific assumptions. These programs are time-consuming to develop and validate, and cannot be systematically reproduced on other observational data sources. In addition, transformation rules and assumptions applied to the data are often embedded within the programs and not clearly documented. This complicates the interpretation of results for anyone not familiar with the program code, and makes meaningful comparisons among results from disparate databases more difficult.
As an example, consider the association between rofecoxib and acute myocardial infarction mentioned previously. Rofecoxib was withdrawn from the US market in 2004 following intense evaluation of disparate information from clinical trials, spontaneous adverse event reporting, and observational healthcare databases.16–21 Many have cited the rofecoxib example when highlighting concerns about the current pharmacovigilance process and the increasing need to establish a national active surveillance system.5 8 22–24 Since 2002, dozens of observational database studies have been published on the subject,25–35 covering a wide array of different data sources utilizing a variety of study designs. The results among these studies have varied significantly; a meta-analysis of some observational studies highlighted the heterogeneity in these results.36
While it should be expected that data sources will have unique limitations with inherent bias that can influence results, a CDM can be used to minimize variability and enable common interpretation within the context of underlying source data. This is accomplished by standardizing the data structure, creating one set of transparent data transformation rules for each data source, developing a common terminology to define exposures, outcomes, and covariates, and establishing a common library of analytic routines for such things as characterizing the populations, identifying new safety signals, and producing drug–outcome effect estimates.
A recent report developed for the FDA recommends the adoption of a CDM for the Sentinel Initiative.37 In this report, several conceptual data models are described, including: encounter based patient-level, patient-level summary data, drug and condition eras, and summary data models. The report summarizes the ability of each of these models to meet the Sentinel system needs based on the data content comprising each model. However, it does not include operational information regarding the process and effects of transforming source data into each type of model nor performance characteristics of each model for the subsequent support of drug safety research.
As part of ongoing research in this area, the OMOP has recently published a detailed specification for a CDM38 39 that includes characteristics of both the encounter based and drug and condition era data models described in the FDA report. Research into the transformation of data into the OMOP Common Data Model and performance of different types of analysis methods utilizing this model is currently underway.
Although there is a great deal of discussion and activity, little has been published to date about the operational consequences of utilizing a CDM to enable drug safety research.
Model description
Overview
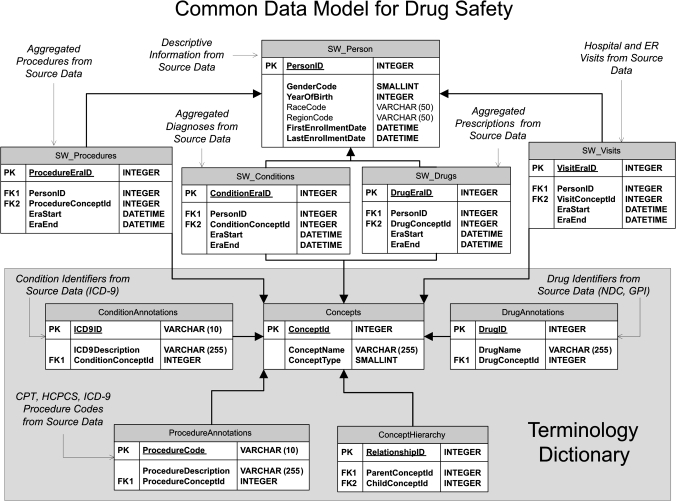
To accomplish the research described in this paper, we developed a CDM designed to support drug safety research. Our CDM, which is similar to the OMOP model, contains the basic data elements required for drug safety analysis. Figure 1 describes the CDM schema as an entity-relationship diagram; the remainder of the paper provides details of the development and validation of the model.
Figure 1.
CDM schema.
PHARMetrics Choice, a claims database from IMS Health and the GE Commercial Data Set, a database of electronic medical records from GE Healthcare, were selected for transformation into our CDM and subsequent analysis. Both datasets contain anonymized and de-identified person-level information from 2000 through 2008. Each database was first transformed into the CDM format, and then the results were analyzed in two ways. First, a series of descriptive statistics were produced on both the raw and transformed data to better understand the effects of the transformation process on the data itself. The second goal was to understand the performance characteristics of utilizing a CDM for drug safety analyses. To accomplish this goal, a program conforming to the CDM format was developed. This program was executed on the transformed data from both databases examining the results of two clinical cohorts: persons who were exposed to the drug rofecoxib and persons with a diagnosis of acute myocardial infarction.
CDM for drug safety
Person Timeline
The format of the CDM is person-centric, organizing the healthcare encounters for each person into a ‘Person Timeline’ to facilitate longitudinal analysis. Within the CDM, de-identified information stored about each person includes a unique Person Identifier and descriptive characteristics found within most observational databases which may be important for drug safety analysis. These characteristics include date of birth, gender, race, and geographic region; the data model has been developed in a way that additional characteristics can be added. For this research, two types of healthcare encounters are recorded for each person: exposure to medications and occurrences of conditions, which are represented as ‘Drug Eras’ and ‘Condition Eras’ and are associated with each person via the Person Identifier.
Drug Eras and Condition Eras
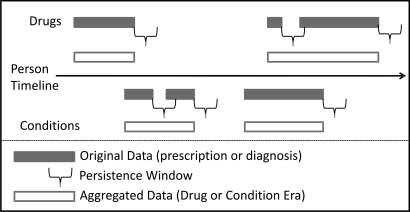
Drug Eras represent a span of time that a given person has been persistently exposed to a given Drug Concept, which can include a generic drug name (such as rofecoxib), a brand name (such as Vioxx), or a drug class or group (such as Cox-2 inhibitors or NSAIDs). Within the CDM, each Drug Era is represented by a unique drug identifier and a start and end date, so that the period of time of drug exposure for a drug can be calculated by (Era end—Era start). The information stored about a Drug Era is derived based on the data elements available for drug prescriptions and medications contained within the source data. The model takes into account the fact that recurring prescriptions for the same product may actually represent one continuous period of drug use. Independent prescriptions are combined into a single Drug Era through the use of a persistence window, which is the allowable span of time after a prescription is scheduled to be completed within which another prescription of the same drug needs to be filled in order to maintain persistence. This persistence window accounts for such things as the logistics of getting a new prescription refilled and the fact that many patients are not 100% compliant in taking their medication every day.
Condition Eras represent an episode of care for a given condition concept, which can include individual conditions such as acute myocardial infarction, or groups of related conditions such as ischemic coronary artery disorders. Within the CDM, each Condition Era is also represented by a unique Condition Concept and a start and end date so the period of time of the episode of care for a condition can be calculated by (Era end—Era start). The information stored about a particular Condition Era is derived based on the data elements available for each diagnosis healthcare encounter contained within the source data. Like Drug Eras, Condition Eras can be aggregated using a persistence window. In this case, a persistence window represents an allowable span of time occurring between recorded diagnoses of the same condition in order to maintain persistence of the episode of care for that condition.
Figure 2 provides a schematic view of a Person Timeline, including the creation of Drug and Condition Eras using a persistence window.
Figure 2.
Schematic of the Person Timeline Common Data Model.
Standardized Terminology Dictionary
The Person Timeline described above enables the standardization of observational data into a format suitable for drug safety analyses; however, it does not address standardization of the data content itself. Drugs and conditions can be coded using various source vocabularies across disparate observational databases. For example, drugs may be recorded using NDC, GPI or Multilex, while conditions may be documented as ICD-9, ICD-10, SNOMED, MedDRA, READ-OXMIS, or any other local codes. In addition, to fully enable robust querying, searching, and analysis of observational data, it is desirable to have the capability of aggregating related drugs and conditions for certain analyses. For example, we may want to analyze the individual brand name Vioxx, the generic name rofecoxib, or all products contained within the drug groups Cox-2 inhibitors or NSAIDs. For conditions we may want to analyze acute myocardial infarctions, coronary artery disorders, or more broadly cardiac disorders.
A Terminology Dictionary provides the capability for drugs and conditions represented within the source data to be mapped into a standard set of hierarchical terminologies, enabling robust analyses and making possible a common interpretation of results.40 The CDM Terminology Dictionary was created by storing the individual concepts from selected medical terminologies, as well as the hierarchical relationships among concepts, within the CDM structure. Our Terminology Dictionary includes a Drug Terminology based on the SNOMED-CT Drug and Medicament hierarchy as well as a Condition Terminology based on the MedDRA hierarchy. Creation of the Terminology Dictionary was accomplished utilizing publicly available data and procedures provided by the Unified Medical Language System (UMLS) from the National Library of Medicine41 as well as procedures developed internally.42 43 The MedDRA was selected for inclusion in the Terminology Dictionary as it is the current standard vocabulary for adverse event reporting used by the FDA.44
Data transformation
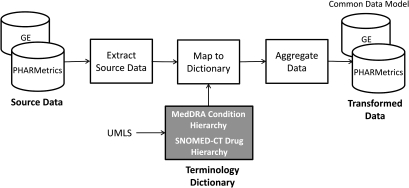
The PHARMetrics and GE databases have different underlying formats. In PHARMetrics, all administrative claims, including pharmacy records and inpatient and outpatient medical claims are captured in one large file; data within this file are differentiated using a ‘type code’. The GE patient encounter data is distributed across 15 tables, each representing a different aspect of a patient encounter. While PHARMetrics captures diagnoses using ICD-9 codes on medical claims, GE records conditions on a problem list and maps conditions to corresponding ICD-9 diagnosis codes. Drug exposure can be inferred from PHARMetrics using pharmacy dispensing information (coded in NDC), while GE drug information is inferred from medication history records and prescriptions written (as coded in GPI). The process of transforming native data from each data source into the CDM involves several steps which are illustrated in figure 3 and described in more detail below.
Figure 3.
Data transformation process.
Extract data
The initial import transforms source data from the native schemas into the general CDM format. The process for each data source is similar and includes the following steps:
Extract Person data for each unique Person
Extract Drug Eras and Condition Eras for each Person, including drug and condition identifiers as found within the data source, an Era start date, and an Era end date
Create Drug and Condition Reference files of all source-specific, unique drugs and conditions. To uniquely identify each drug, Product Identifiers are constructed consisting of the Product name+Strength for each drug in the reference file. The ICD-9 codes are used to uniquely identify each condition.
The extraction details for each data source are different due to the differences in the organization and format of vendor data. Source specific rules for transforming the diagnosis, medication, and prescription data into the CDM are built into the extract programs; development of these rules requires significant data content expertise for each source dataset being transformed.
Map to Terminology Dictionary
The goal of this step is to map or annotate each of the drugs and conditions found in the Drug and Condition Reference files to the appropriate Concept in the Terminology Dictionary that was previously created.
To map drug data, the drugs found within the source data Drug Reference files are annotated to the appropriate SNOMED-CT Drug Concepts found in the Terminology Dictionary. The goal of the annotation is to associate each unique drug reference found in the native source data with one or more Drug Concepts. The fundamental approach for accomplishing the annotation is to match the string representation of the product names found in the Drug Reference file to product name Drug Concepts in the Drug Hierarchy. Although string normalization tools are available within the UMLS, the specific requirements of this project necessitated the development of a string normalizer which is tuned to the needs of a clinical drug vocabulary.43
To map condition data, the conditions found within the source data Condition Reference files are annotated to the MedDRA Condition Concepts in the Terminology Dictionary. The process for annotating conditions is somewhat simpler than for drugs, and the UMLS Metathesaurus is utilized to map ICD-9 codes to MedDRA Preferred Term Condition Concepts using equivalence between Concept Unique Identifiers found in the Metathesaurus. The ICD-9 codes and MedDRA Preferred Terms that have equivalent Concept Unique Identifiers are assumed to refer to the same medical condition.42
Aggregate Drug and Condition Eras
At this point in the process, Person data from each observational data source has been transformed into the Person Timeline format including Drug and Condition Eras, and drug and condition references found within the Person data have been mapped to a common Terminology Dictionary. The final step in the data transformation process is to aggregate Drug and Condition Eras that occur within the allowable persistence windows. During this process, Drug and Condition Eras that represent the same Drug or Condition Concept from the Terminology Dictionary are aggregated if the start of the second Era occurs within 30 days of the end of first Era. Multiple Eras can be merged into one using this method. Thirty days was selected as the persistence widow for aggregation based on a conservative approach that may be more appropriate for chronic drugs and acute conditions; this value is an input parameter which can be modified.
Consider the following example: a Condition Era representing ICD-9 code 41001 (AMI ANTEROLATERAL WALL, INITIAL EPISODE) would be aggregated to a Condition Era representing ICD-9 code 41041 (AMI INFERIOR WALL, INITIAL EPISODE) occurring within 30 days as both of these ICD-9 codes annotate to the same Condition Concept, Acute Myocardial Infarction, within the MedDRA hierarchy. However, a Condition Era representing the ICD-9 code 412 (INFARCTION, MYOCARDIAL OLD) would not be aggregated to either of the Condition Eras above since ICD-9 code 412 annotates to a different Condition Concept, Myocardial Infarction.
The end result of the aggregation process is that drugs and conditions with the same Concept and occurring within 30 days of each other are aggregated into one Era. When this process is complete, the data from each database have been fully transformed into the CDM format.
Load transformed data into a production database
The transformed data in the CDM format for each data source are loaded into a commercial relational database as normalized relational tables (figure 1 describes the CDM tables). A relational structure was chosen due to the large size of the data and the need for data access and execution efficiency. Data from each source are not integrated, but maintained in separate, identical table structures. After the relational tables have been successfully loaded, the transformed data are ready for analysis.
Data transformation statistics
To assess the effect of the data transformation on the content of the data, a variety of statistics were calculated at each step of the data transformation process. From each native data source, the overall number of persons, the number of persons with at least one rofecoxib prescription or medication record, and the number of persons with at least one diagnosis code representing an acute myocardial infarction were counted.
To evaluate the performance of the drug and condition mapping process, the number of unique drugs (by Product Identifier) and Conditions (by ICD-9 code) within the Drug and Condition Reference files produced for the native data sources, as well as the total number of Drug and Condition Concepts found within our Terminology Dictionary, were counted. Drug Concepts include brand names, generic names, and drug groups at all levels of the SNOMED-CT hierarchy, and Condition Concepts include MedDRA Preferred Terms, High Level Terms, High Level Groups, and System Organ Classes. These numbers were used to calculate two metrics: Distinct Annotation Proportion and Instance Annotation Proportion. The Distinct Annotation Proportion is the proportion of unique drugs and conditions from the Source Reference files that were successfully annotated to Drug and Condition Concepts in the Terminology Dictionary. The Instance Annotation proportion is the proportion of drug and condition references found within the Person data that were successfully annotated. For conditions, the proportion of condition references that are ICD-9 E and V codes was also calculated. Finally, the specific mapping results for Concepts representing the drug rofecoxib and the condition acute myocardial infarction were analyzed.
To understand the consequences of the data aggregation process, several metrics were calculated based on the number and average length of prescriptions/medications and diagnoses records found within the native source data, and the number and average length of Drug and Condition Eras within the transformed data. These metrics were calculated for the overall data, as well as for rofecoxib and acute myocardial infarction.
CDM performance statistics
To assess the capability of the CDM to support systematic drug safety analyses, a program based on the CDM format was written to analyze clinical cohorts. This program was executed against each transformed database for two test cases: persons exposed to the drug rofecoxib and persons with a diagnosis of acute myocardial infarction. The program produces descriptive statistics describing the persons comprising the cohort, including a demographic summary and a summary of concomitant medications occurring either during the use of the cohort drug (rofecoxib), or at the same time as the occurrence of the cohort condition (acute myocardial infarction) for each person within that cohort.
Model validation
Data transformation summary
A total of 43 096 800 person records across both databases were included in this analysis; of this total, 76.1% are PHARMetrics records. The number of persons remained unchanged during the transformation process and is the same in the native and transformed data. The rofecoxib cohort comprises 1.07% of the total PHARMetrics persons, and 1.82% of GE persons. For the acute myocardial infarction cohort, 0.67% of PHARMetrics persons and 0.52% of GE persons are included. Table 1 provides summary statistics regarding the number of persons found within each native and transformed source file.
Table 1.
Person counts in native and transformed data for each database
| PHARMetrics | GE | |
| Number of persons | 32818355 | 10278445 |
| Number of persons in rofecoxib cohort | 349929 (1.07%) | 186763 (1.82%) |
| Number of persons in acute myocardial infarction cohort | 221437 (0.67%) | 53063 (0.52%) |
Terminology Dictionary mapping
In total, 97.91% of PHARMetrics and 96.68% of GE drugs associated with prescription or medication records found within the Person data mapped to Concepts in our Terminology Dictionary (instance annotations). This represents 79.39% of PHARMetrics and 69.79% of GE unique Product Identifiers found within the Drug Reference files (distinct annotations). A review of the unmapped Product Identifiers reveals items such as ‘100 CC SYRINGE’ and ‘ATTENDS BRIEFS LARGE’ contained within the medication files of the source data; it is appropriate that these are not annotated for our purposes.
In both PHARMetrics and GE, multiple Vioxx Product Identifiers correctly annotate to the Drug Concept Vioxx within the Terminology Dictionary and GE references to the generic Product Identifier Rofecoxib correctly annotate to the Rofecoxib NOC Concept.
For Conditions, 84.56% of PHARMetrics and 84.11% of GE ICD-9 codes found within actual Person data (instance annotations) annotate to MedDRA Concepts, representing 75.41% of PHARMetrics and 85.47% of GE unique ICD-9 codes in the Condition Reference files (distinct annotations). The ICD-9 E and V codes account for approximately 12.01% and 13.80% of unmapped, unique codes in the Condition Reference file for PHARMetrics and GE, respectively, since there is no corresponding concept within MedDRA for a majority of these health and service indicator codes. The remaining 12.58% (PHARMetrics) and 0.73% (GE) of unmapped, unique ICD-9 codes found in the Condition Reference files are not represented in the current ICD-9 coding dictionary; this may be due to transcription errors during data entry or represent local modifications to the official ICD-9 coding scheme. These non-standard and/or invalid ICD-9 codes represent only 0.16% and 0.11% of the conditions found within the actual PHARMetrics and GE Person data, respectively. All ICD-9 codes starting with ‘410’ appropriately annotated to the Condition Concept Acute Myocardial Infarction.
Table 2 provides detailed statistics produced from the annotated Terminology Dictionary.
Table 2.
Standardized Terminology Dictionary mapping performance
| Total Drug Concepts in Terminology Dictionary | 16269 | |
| PHARMetrics | GE | |
| Unique Product Identifiers in reference file | 22454 | 28501 |
| Distinct annotation proportion | 79.39% (17827) | 69.79% (19892) |
| Drug References in Person data | 718590080 | 231492741 |
| Instance annotation proportion | 97.91% (703584362) | 96.68% (223799256) |
| Source drug annotations to Terminology Dictionary Drug Concepts Vioxx and rofecoxib |
|
|
| Total Condition Concepts in Terminology Dictionary | 20666 | |
| PHARMetrics | GE | |
| Unique ICD-9 codes in reference file | 29630 | 14972 |
| Distinct annotation proportion | 75.41% (22345) | 85.47% (12797) |
| Unmapped, unique ICD-9 E and V codes | 12.01% (3559) | 13.80% (2066) |
| Unmapped, unique invalid codes | 12.58% (3726) | 0.73% (109) |
| Condition references in Person data | 3627194305 | 81693914 |
| Instance annotation proportion | 84.56% (3067136827) | 84.11% (68713570) |
| ICD-9 E and V codes in Person data | 15.28% (554089276) | 15.78% (12894228) |
| Invalid codes in Person data | 0.16% (5968202) | 0.11% (86116) |
| PHARMetrics | GE | |
| Source ICD-9 annotations to Condition Terminology Dictionary Concept ‘Acute Myocardial Infarction‘ (AMI) |
|
|
Data aggregation
Data aggregation results for drugs are consistent with the transformation rules which aggregate multiple occurrences of the same Drug Concept within the allowable persistence window of 30 days. The aggregation process reduces the overall number of Person drug records in both databases. For PHARMetrics, the total number of Drug Eras found in the transformed data is only 43.11% of the total number of Person drug records within the native data; for GE this number is 41.74%. The reduction for rofecoxib Drug Eras is similar, at 35.84% and 40.58% of the number of native rofecoxib drug records for PHARMetrics and GE, respectively.
Within the native data, the average number of drug records per person is 21.9 in PHARMetrics and 22.52 in GE. The aggregation process reduces this number to an average of 9.44 Drug Eras per person in PHARMetrics and 9.40 Drug Eras per person in GE. The consistency of results across both database types is also seen for rofecoxib, although the average number of rofecoxib drug records per person is lower in both the native and transformed data.
The average length of a Drug Era in the transformed data is longer in GE than in PHARMetrics, although the length of rofecoxib exposure in GE is only slightly longer. In PHARMetrics, the overall average length of a transformed Drug Era is 2.55 times larger than the average length of a prescription record in the native data (as measured by days supply); a rofecoxib Drug Era is on average 2.91 times larger than a native prescription. Length of exposure is not directly obtainable from the GE raw data but requires a defined set of rules to infer utilization, such as those applied here to enable estimation.
Condition Eras within the CDM represent the concept of an ‘episode of care’, which does not exist in the native data but is constructed from diagnoses information during the transformation process. The Condition aggregation process produces different results between the two types of databases, reflecting differences in the underlying motivation for recording a condition in each data source. For PHARMetrics the number of Condition Eras in the transformed data is only 22.19% of the number of Conditions found in the native data; for GE this number is 75.64%, representing less aggregation of Condition Eras. For Acute Myocardial Infarctions, these aggregation reductions were consistent (Acute Myocardial Infarction Condition Eras represent only 5.28% and 62.01% of the number of original diagnoses of acute myocardial infarction found within PHARMetrics and GE, respectively).
Within the native data sources, the average number of diagnosis records per person is 110.52 in PHARMetrics, and 7.95 in GE. The aggregation process reduces this number to 24.53 Condition Eras per person for PHARMetrics and 6.01 Condition Eras per person for GE. Condition aggregation also reduces the number of Acute Myocardial Condition Eras per person in the transformed data in both databases.
The average length of a Condition Era within the transformed data is significantly longer in GE than in PHARMetrics, both in the overall data and for Condition Eras representing acute myocardial infarctions.
Table 3 presents the results of the data aggregation steps for the entire database as well as for rofecoxib and acute myocardial infarction.
Table 3.
Impact of data aggregation on drugs and conditions
| PHARMetrics | GE | |||
| Native | Transformed | Native | Transformed | |
| Total persons | 32818355 | 10278445 | ||
| Drug aggregation | ||||
| Total Drug Eras | 718590080 | 309797580 | 231492741 | 96614637 |
| Eras per person | 21.90 | 9.44 | 22.52 | 9.4 |
| Average exposure length | 31 days | 79 days | NA | 108 days |
| Rofecoxib | ||||
| Total persons | 349929 | 186763 | ||
| Total rofecoxib Eras | 1423273 | 510078 | 652234 | 264654 |
| Eras per person | 4.07 | 1.46 | 3.49 | 1.42 |
| Average exposure length | 34 days | 99 days | NA | 102 days |
| Condition aggregation | ||||
| Total Condition Eras | 3627194305 | 804977267 | 81693914 | 61795940 |
| Eras per person | 110.52 | 24.53 | 7.95 | 6.01 |
| Average Condition Era length | NA | 6 days | NA | 88 days |
| Acute myocardial infarction (AMI) | ||||
| Total persons | 221437 | 53063 | ||
| Total AMI Eras | 5547711 | 292980 | 85940 | 53290 |
| AMI Eras per person | 25.05 | 1.32 | 1.62 | 1.00 |
| Average Condition Era length | NA | 11 days | NA | 175 days |
CDM performance
The CDM performance metrics were produced from a single analysis program executed against the transformed data in both databases. Table 1 provides summary statistics produced by the analysis program, regarding the total number of persons within each transformed source file, as well as the count of persons in the rofecoxib and acute myocardial infarction cohorts. The transformed person counts match the statistics produced from the native data.
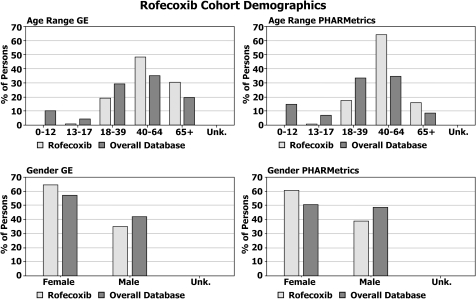
Figures 4 and 5 provide a summary of the cohort demographics for the rofecoxib (figure 4) and acute myocardial infarction (figure 5) cohorts compared to the database background for the transformed GE and PHARMetrics data. Figures 6 and 7 compare the concomitant medications found within each transformed database for the rofecoxib (figure 6) and acute myocardial infarction (figure 7) cohorts. Although the underlying data within each source database were recorded in different ways and for different reasons, the CDM has allowed us to execute one systematic analysis across both databases utilizing standard definitions and assumptions and to present the results in a consistent format enabling common interpretation.
Figure 4.
Demographic summary of rofecoxib cohort in each transformed database.
Figure 5.
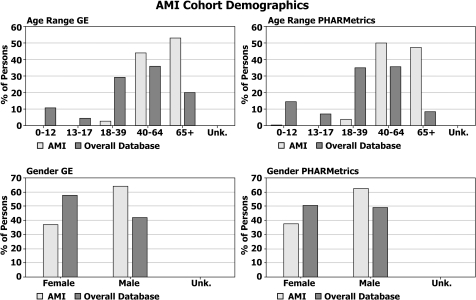
Demographic summary of the acute myocardial infarction (AMI) cohort in each transformed database.
Figure 6.
Comparison of concomitant medications within each transformed database for the rofecoxib cohort.
Figure 7.
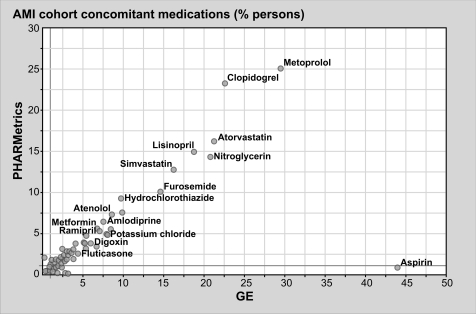
Comparison of concomitant medications occurring during acute myocardial infarction Condition Eras, within each transformed database.
These results highlight many similarities, as well as a few differences, in the underlying populations captured by administrative claims versus electronic medical records databases. For instance, 19.9% of the persons in the GE database are 65 years old or older, while only 8.8% within the PHARMetrics database are 65 or older. This is a characteristic of claims databases reflecting the fact that the elderly population in the USA is eligible for healthcare coverage provided by the government versus private coverage. And, although the concomitant medications reported among comparable cohorts are strikingly similar across both databases, one major discrepancy—concomitant aspirin use in both cohorts—highlights differences in the underlying data capture purposes of the two data sources. Over-the-counter medications such as aspirin are not typically reimbursed by health insurance providers but their use is recorded by healthcare providers in an electronic medical record (EMR).
Discussion
The results of this research confirm that a CDM is a feasible and useful approach to enable systematic analyses of disparate healthcare data sources, including administrative claims and EMR data. We have successfully demonstrated the implementation of a Person Timeline CDM including Drug and Condition Eras derived from prescriptions and diagnoses found within the source data; this same methodology could be extended to other types of healthcare encounters such as procedures, laboratory results, and hospital visits. Although this research demonstrated the successful use of this approach for claims and EMR data, we believe that the approach is extensible to other types of longitudinal healthcare data such as patient and disease registry data. Because data from individual data sources are normalized but not integrated, this approach is viable for both a centralized data warehouse as well as a distributed network of healthcare databases.
While each type of database captures healthcare encounters in different ways, to different degrees, and for different underlying reasons, the use of a CDM enforces a series of standardized, transparent rules and assumptions to be applied during the data preparation process rather than at analysis time. Transformation rules embody assumptions that are unique to the underlying data in each data source and an understanding of these rules is critical to properly interpret any analysis performed on the data. The consistent application of data transformation rules and analysis procedures enables results to be meaningfully comparable within the context of the underlying sources. Transforming data into a common model requires a significant amount of work and validation up front, however this work is re-used for each analysis that utilizes the transformed data. This approach is different from the current paradigm for the analysis of observational data, where the assumptions and rules are generally embedded within individual analysis programs and validated at analysis time. It is important for source data experts to participate in the development and validation of the transformation rules for each source database to provide insights that cannot be gleaned from the data itself. And the transformation rules should be reviewed and evaluated each time a new version of the source data is received to ensure that they are still valid.
We believe the use of a standard Terminology Dictionary is a critical component in the development of a CDM for drug safety, and this approach is extensible to any codes found within source observational data. The MedDRA was selected as the initial reference condition terminology for our data model because it is a standard vocabulary used by drug safety scientists and the FDA and it comprises conditions that are drug adverse events. Therefore, analyses can be done in a language that is most familiar to our target user population. It is feasible that vocabularies other than MedDRA and SNOMED-CT could be selected as reference vocabularies; key factors for vocabulary selection are a correct and uniform classification of drug and condition concepts within the domain being studied and comprehensive coverage relative to the source data. A key limitation of utilizing a terminology dictionary is that the mapping from the source data may not always be consistent with a specific clinician's expectations and should always be reviewed prior to analysis. In addition, the selection of terminology restricts any analysis performed to the concepts found within that terminology. For example, the SNOMED-CT hierarchy we selected for this research does not include concepts for drug strength, so analyses by drug strength are not supported using this data model. Because most source vocabularies change over time, including a process for updating and versioning the terminology dictionary is important for maintaining currency.
The descriptive analyses of cohorts of rofecoxib and myocardial infarction patients described in this paper illustrate the use of the CDM to enable systematic exploration of the similarities and differences in patient characteristics recorded in disparate databases. Apart from this project, the CDM described here has also been utilized for systematic signal detection45 46 and risk evaluation studies.47 48 These studies demonstrate that the CDM is of sufficient fidelity to support drug safety research.
There are some additional considerations when performing an analysis utilizing a CDM. The transformation rules and assumptions applied to the drugs and conditions may not be appropriate for all circumstances. For our research above we selected a persistence window of 30 days for the aggregation of both drugs and conditions. This is more appropriate for acute conditions such as acute myocardial infarction. Depending on the type of analysis being performed, it may be less appropriate for a chronic condition such as a malignancy and would require adjustment at analysis time. In addition, if the source database contains data that are not supported by the data model, the unsupported data will not be available for analysis using the data model. For example, the CDM we used for our research did not include laboratory results at the time of this analysis, so performing analysis of an outcome defined by a particular laboratory result would not be possible even though the native GE database contains that information. The CDM was designed to be extensible for this reason—as additional source databases are incorporated the data model can be extended to include coverage of additional types of data.
Conclusion
In conclusion, we have studied the real-world impact of utilizing a CDM to support drug safety analyses and found that we were able to successfully execute a systematic descriptive analysis of cohorts across two disparate observational data sources using a Person Timeline CDM developed for drug safety analysis. We have reviewed the impact of the data transformation process on the content of each source database and found that the characteristics of transformed data from disparate databases are consistent with underlying data capture motives and the transformed data exhibit many similar characteristics despite the fact that underlying data organization and formats are different. Areas of future research include extending the model to incorporate additional types of healthcare encounters, incorporating additional reference vocabularies into the Terminology Dictionary, including additional sources of observational data such as patient and disease registries, and further exploring analytic techniques that are best suited for systematic pharmacovigilance.
The rofecoxib–acute myocardial infarction association, as well as other known drug–outcome pairs, have been used as exemplars to illustrate the performance of analysis methods,45 47 49–52 but substantial work is required to determine the appropriate methods and selection of data sources that can meaningfully contribute to a national active surveillance system.53 Several efforts, including OMOP and EU-ADR, should provide research to inform this broader debate. In this paper we do not argue for a particular analysis approach or for the use of either the PHARMetrics or GE databases. However, we do assert that a CDM can serve as a necessary foundation to facilitate the integration of the appropriate databases and methods into a coordinated system. The demonstration of transforming both an administrative claims and electronic health record database into this CDM offers promise that data from other health plans, providers, and clinical systems could also be implemented in either a centralized or distributed network within a broader national active surveillance system. The results of applying the CDM show how differences in drug and condition coding can be successfully resolved, how periods of persistent exposure and episodes of care for outcomes can be systematically and consistently estimated, and how population-level characteristics that could meaningfully impact an analysis can be identified and defined through a standardized process. This demonstration shows how a CDM can be a valuable tool to significantly contribute to our collective ability to better understand the effects of medical products.
Footnotes
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Anonymous. Managing the Risks From Medical Product Use: Creating a Risk Management Framework. Rockville, MD: US Department of Health and Human Services, Food and Drug Administration, 1999. Contract No.: Document Number. [Google Scholar]
- 2.Almenoff J, Tonning JM, Gould AL, et al. Perspectives on the use of data mining in pharmacovigilance. Drug Saf 2005;28:981–1007 [DOI] [PubMed] [Google Scholar]
- 3.Baciu A, Stratton K, Burke S. The Future of Drug Safety: Promoting and Protecting the Health of the Public. Institute of Medicine, 2006. Contract No.: Document Number. [Google Scholar]
- 4.FDA The future of drug safety—promoting and protecting the health of the public, FDA's Response to the Institute of medicine's 2006 report, 2007. http://www.fda.gov/downloads/Drugs/DrugSafety/PostmarketDrugSafetyInformationforPatientsandProviders/UCM171627.pdf (accessed 28 Nov 2009).
- 5.Furberg CD, Levin AA, Gross PA, et al. The FDA and drug safety: a proposal for sweeping changes. Arch Intern Med 2006;166:1938–42 [DOI] [PubMed] [Google Scholar]
- 6.Goldman SA. Limitations and strengths of spontaneous reports data. Clin Ther 1998;20(Suppl C):C40–4 [DOI] [PubMed] [Google Scholar]
- 7.Couzin J. Drug safety. Gaps in the safety net. Science 2005;307:196–8 [DOI] [PubMed] [Google Scholar]
- 8.Psaty BM, Furberg CD. COX-2 inhibitors–lessons in drug safety. N Engl J Med 2005;352:1133–5 [DOI] [PubMed] [Google Scholar]
- 9.Public Law 110-85: Food and Drug Administration Amendments Act of 2007, 2007.
- 10.FDA The sentinel initiative: a national strategy for monitoring medical product safety, 2008. http://www.fda.gov/Safety/FDAsSentinelInitiative/ucm089474.htm (accessed May 2008).
- 11.Observational medical outcomes partnership, 2009. http://omop.fnih.org (accessed 28 Nov 2009).
- 12.EU-ADR. 2009. http://www.alert-project.org/ (accessed 28 Nov 2009).
- 13.IMI-PROTECT. 2009. http://www.imi-protect.eu/ (accessed 28 Nov 2009).
- 14.Strom B. Pharmacoepidemiology. 4th edn Chichester, UK: Wiley, 2005 [Google Scholar]
- 15.Hartzema AG, Tilson HH, Chan KA. Pharmacoepidemiology and Therapeutic Risk Management. Cincinnati, OH: Harvey Whitney Books, 2008 [Google Scholar]
- 16.FDA Merck Withdraws Vioxx; FDA Issues Public Health Advisory. http://www.fda.gov/fdac/features/2004/604_vioxx.html (accessed 26 Apr 2009). [PubMed]
- 17.Juni P, Nartey L, Reichenbach S, et al. Risk of cardiovascular events and rofecoxib: cumulative meta-analysis. Lancet 2004;364:2021–9 [DOI] [PubMed] [Google Scholar]
- 18.Velentgas P, West W, Cannuscio CC, et al. Cardiovascular risk of selective cyclooxygenase-2 inhibitors and other non-aspirin non-steroidal anti-inflammatory medications. Pharmacoepidemiol Drug Saf 2006;15:641–52 [DOI] [PubMed] [Google Scholar]
- 19.Abraham NS, El-Serag HB, Hartman C, et al. Cyclooxygenase-2 selectivity of non-steroidal anti-inflammatory drugs and the risk of myocardial infarction and cerebrovascular accident. Aliment Pharmacol Ther 2007;25:913–24 [DOI] [PubMed] [Google Scholar]
- 20.Chen LC, Ashcroft DM. Risk of myocardial infarction associated with selective COX-2 inhibitors: meta-analysis of randomised controlled trials. Pharmacoepidemiol Drug Saf 2007;16:762–72 [DOI] [PubMed] [Google Scholar]
- 21.Scott PA, Kingsley GH, Smith CM, et al. Non-steroidal anti-inflammatory drugs and myocardial infarctions: comparative systematic review of evidence from observational studies and randomised controlled trials. Ann Rheum Dis 2007;66:1296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Psaty BM, Furberg CD. The record on rosiglitazone and the risk of myocardial infarction. N Engl J Med 2007;357:67–9 [DOI] [PubMed] [Google Scholar]
- 23.Avorn J. Evaluating drug effects in the post-Vioxx world: there must be a better way. Circulation 2006;113:2173–6 [DOI] [PubMed] [Google Scholar]
- 24.Ray A. Beyond debacle and debate: developing solutions in drug safety. Nat Rev Drug Discov 2009;8:775–9 [DOI] [PubMed] [Google Scholar]
- 25.Arellano FM, Yood MU, Wentworth CE, et al. Use of cyclo-oxygenase 2 inhibitors (COX-2) and prescription non-steroidal anti-inflammatory drugs (NSAIDS) in UK and USA populations. Implications for COX-2 cardiovascular profile. Pharmacoepidemiol Drug Saf 2006;15:861–72 [DOI] [PubMed] [Google Scholar]
- 26.Garcia Rodriguez LA, Varas-Lorenzo C, Maguire A, et al. Nonsteroidal antiinflammatory drugs and the risk of myocardial infarction in the general population. Circulation 2004;109:3000–6 [DOI] [PubMed] [Google Scholar]
- 27.Garcia Rodriguez LA, Gonzalez-Perez A. Long-term use of non-steroidal anti-inflammatory drugs and the risk of myocardial infarction in the general population. BMC Med 2005;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mamdani M, Rochon P, Juurlink DN, et al. Effect of selective cyclooxygenase 2 inhibitors and naproxen on short-term risk of acute myocardial infarction in the elderly. Arch Intern Med 2003;163:481–6 [DOI] [PubMed] [Google Scholar]
- 29.Moore RA, Derry S, McQuay HJ. Cyclo-oxygenase-2 selective inhibitors and nonsteroidal anti-inflammatory drugs: balancing gastrointestinal and cardiovascular risk. BMC Musculoskelet Disord 2007;8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray WA, Stein CM, Hall K, et al. Non-steroidal anti-inflammatory drugs and risk of serious coronary heart disease: an observational cohort study. Lancet 2002;359:118–23 [DOI] [PubMed] [Google Scholar]
- 31.Ray WA, Stein CM, Daugherty JR, et al. COX-2 selective non-steroidal anti-inflammatory drugs and risk of serious coronary heart disease. Lancet 2002;360:1071–3 [DOI] [PubMed] [Google Scholar]
- 32.Schmidt H, Woodcock BG, Geisslinger G. Benefit-risk assessment of rofecoxib in the treatment of osteoarthritis. Drug Saf 2004;27:185–96 [DOI] [PubMed] [Google Scholar]
- 33.Solomon DH, Schneeweiss S, Glynn RJ, et al. Relationship between selective cyclooxygenase-2 inhibitors and acute myocardial infarction in older adults. Circulation 2004;109:2068–73 [DOI] [PubMed] [Google Scholar]
- 34.Solomon DH, Glynn RJ, Levin R, et al. Nonsteroidal anti-inflammatory drug use and acute myocardial infarction. Arch Intern Med 2002;162:1099–104 [DOI] [PubMed] [Google Scholar]
- 35.van Staa TP, Smeeth L, Persson I, et al. What is the harm-benefit ratio of Cox-2 inhibitors? Int J Epidemiol 2008;37:405–13 [DOI] [PubMed] [Google Scholar]
- 36.Hernandez-Diaz S, Varas-Lorenzo C, Garcia Rodriguez LA. Non-steroidal antiinflammatory drugs and the risk of acute myocardial infarction. Basic Clin Pharmacol Toxicol 2006;98:266–74 [DOI] [PubMed] [Google Scholar]
- 37.Brown J, Lane K, Moore K, et al. Database Models to Implement the FDA Sentinel Initiative FDA, 2009. http://www.regulations.gov/search/Regs/home.html#documentDetail?R=090000648098c282 (accessed 21 Sep 2010).
- 38.Ryan PB, Griffin D, Whittenburg L, et al. Points to consider in developing a common semantic data model and terminology dictionary for observational analyses. Observational Medical Outcomes Partnership, 2009. http://omop.fnih.org/CDMandTerminologies (accessed 28 Nov 2009). [Google Scholar]
- 39.Ryan PB, Griffin D, Reich C, et al. OMOP common data model (CDM) Specifications, 2009. http://omop.fnih.org/CDMandTerminologies (accessed 28 Nov 2009).
- 40.Reich C. OMOP standard vocabulary specifications, 2009. http://omop.fnih.org/CDMandTerminologies (accessed 28 Nov 2009).
- 41.National Library of Medicine Unified Medical Language System (UMLS), 2009. http://www.nlm.nih.gov/research/umls/ (accessed 28 Nov 2009).
- 42.Ryan PB, Painter JL, Merrill GH. Defining Medical Conditions by Mapping ICD-9 to MedDRA: A Systematic Approach to Integrating Disparate Observational Data Sources for Enabling Enhanced Pharmacovigilance Analyses. Boston, MA, USA: Drug Information Association, 2008 [Google Scholar]
- 43.Merrill GH, Ryan PB, Painter JL. Using SNOMED to Normalize and Aggregate Drug References in the SafetyWorks Observational Pharmacovigilance Project. Phoenix, AZ, USA: KR-MED, 2008 [Google Scholar]
- 44.Postmarketing Safety Reports for Human Drug and Biological Products; Electronic Submission Requirements. Proposed Rule. Federal Register 74 (21 August 2009), 42184–220 [PubMed] [Google Scholar]
- 45.Ryan PB, Powell GE, Patishall EN, et al. Performance of Screening Multiple Observational Databases for Active Drug Safety Surveillance. Providence, RI, USA: International Society of Pharmacoepidemiology, 2009 [Google Scholar]
- 46.Powell G, Ryan PB, Patishall E. Comparison of Quantative Signal Detection Using Observational and Spontaneous Adverse Event Data. Brighton, UK: International Society of Pharmacoepidemiology, 2010:49 [Google Scholar]
- 47.Beach K, Le HV, Powell G, et al. Performance of a Semi-Automated Process for Estimation of Risk using Observational Databases, 2. Providence, RI, USA: International Society of Pharmacoepidemiology, 2009 [Google Scholar]
- 48.Mera R, Beach KJ, Powell G, et al. Semi-automated risk estimation using large databases: quinolones and clostridium difficile associated diarrhea. Pharmacoepidemiol Drug Saf 2010;19:610–17 [DOI] [PubMed] [Google Scholar]
- 49.Brown JS, Kulldorff M, Petronis KR, et al. Early adverse drug event signal detection within population-based health networks using sequential methods: key methodologic considerations. Pharmacoepidemiol Drug Saf 2009;18:226–34 [DOI] [PubMed] [Google Scholar]
- 50.Brown JS, Kulldorff M, Chan KA, et al. Early detection of adverse drug events within population-based health networks: application of sequential testing methods. Pharmacoepidemiol Drug Saf 2007;16:1275–84 [DOI] [PubMed] [Google Scholar]
- 51.Norén G, Bate A, Hopstadius J, et al. Temporal pattern discovery for trends and transient effects: its application to patient records. Paper presented at: Proceeding of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Las Vegas, Nevada, USA, 2008 [Google Scholar]
- 52.Curtis JR, Cheng H, Delzell E, et al. Adaptation of Bayesian data mining algorithms to longitudinal claims data: coxib safety as an example. Med Care 2008;46:969–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryan PB, Welebob E, Hartzema AG, et al. Surveying US observational data sources and characteristics for drug safety needs. Pharmaceut Med 2010;24:1–8 [Google Scholar]