Abstract
Objective
Negation is a linguistic phenomenon that marks the absence of an entity or event. Negated events are frequently reported in both biological literature and clinical notes. Text mining applications benefit from the detection of negation and its scope. However, due to the complexity of language, identifying the scope of negation in a sentence is not a trivial task.
Design
Conditional random fields (CRF), a supervised machine-learning algorithm, were used to train models to detect negation cue phrases and their scope in both biological literature and clinical notes. The models were trained on the publicly available BioScope corpus.
Measurement
The performance of the CRF models was evaluated on identifying the negation cue phrases and their scope by calculating recall, precision and F1-score. The models were compared with four competitive baseline systems.
Results
The best CRF-based model performed statistically better than all baseline systems and NegEx, achieving an F1-score of 98% and 95% on detecting negation cue phrases and their scope in clinical notes, and an F1-score of 97% and 85% on detecting negation cue phrases and their scope in biological literature.
Conclusions
This approach is robust, as it can identify negation scope in both biological and clinical text. To benefit text mining applications, the system is publicly available as a Java API and as an online application at http://negscope.askhermes.org.
Keywords: Natural language processing, negation detection, text mining
Negation is a linguistic phenomenon that marks the absence of an entity or event. Negated events are frequent in both biological literature and clinical notes; two examples are shown below:
(1) By contrast, the expression of other adherence-associated early genes, such as IL-8 and IL-1β, was not up-regulated in PBMC of tuberculous patients.
(2) No radiographic abnormality seen of the chest.
It is essential for a text mining application to identify negation.1 For example, in sentence (1), the author indicates that the upregulation of genes IL-8 and IL-1 in tuberculous patients did not occur. Similarly, in sentence (2), the report indicates that abnormalities in the chest of the patient are absent. To not account for negation in these sentences reverses the polarity of the information and can result in inaccurate and potentially harmful information.
Due to the importance of this issue, the task of negation detection is actively researched, but is ignored by many current biomedical text mining approaches. Negation detection is not an easy task. Although certain cue terms (eg, ‘not’, ‘no’ and ‘without’) are commonly used in negated statements, identifying negated statements merely based on the presence of the cue terms may lead to false results. Two examples are shown below:
(3) Thus, signaling in NK3.3 cells is not always identical with that in primary NK cells.
(4) This does not exclude the diagnosis of pertussis.
In sentence (3), although the negation cue term ‘not’ appears, it is not being used to negate the observation of signaling in NK3.3 cells being identical to that in primary NK cells. Rather, the authors apply the negation to argue that these signaling events are identical in both cells only sometimes (ie, not always). Similarly, in sentence (4), the clinician indicates that it is unclear if the patient was diagnosed with pertussis, but not that pertussis is necessarily absent.
Furthermore, the use of a negation cue in a sentence might not apply to the entire sentence; rather, its scope might be limited to only a part of the sentence. This can be seen in the following example sentences in which the negation scope is marked in square brackets and the negation cue is in boldface:
(5) PMA treatment, and [not retinoic acid treatment of the U937 cells] acts in inducing NF-κB expression in the nuclei.
(6) Less well defined opacity in the superior segment of the right lower lobe [which was not seen on prior studies].
In sentence (5), the proposition that retinoic acid treatment of U937 induces NF-κB expression in the nuclei is negated, whereas the proposition that PMA treatment induces NF-κB expression in the nuclei is not. Similarly, in sentence (6), the clinician indicates that the observation that opacity in the superior segment of the right lower lobe of lung is less well defined is present, but was not observed earlier. A system that identifies negation must thus identify the scope of negation or the results will be misleading.
Therefore, negation detection is a challenging research task, and we propose that the task of information extraction needs to add negation detection in addition to relation identification. We report here on the development of a supervised machine-learning system called NegScope that identifies biomedical sentences that contain negation and marks the scope of negation in such sentences.
Related work
Most existing work in biomedical and clinical negation detection classifies an entire sentence as negated or not and ignores negation scope. In the clinical domain, rule-based approaches have been developed for negation detection. For example, Chapman et al2 developed the NegEx system to identify the negation of target findings and diseases in narrative medical reports. The current version of NegEx utilizes 272 rules, which are matched using regular expressions. The reported recall of the system was 95.93% and precision was 93.27%, and it attained an accuracy of 97.73%. A similar system, Negfinder, was developed to identify negated concepts in medical narratives.3 The system first identifies negation markers in the sentence using regular expressions. These words are then passed to a parser that uses a single-token look-ahead strategy to identify negated concepts. The reported recall and precision of the system was 95.27% and 97.67%, respectively. Along the same lines, Elkin et al4 developed a system to identify the negation of concepts in electronic medical records. The system was built by identifying textual cues for negation in 41 clinical documents, and the reported recall and precision of the system was 97.2% and 91.2%, respectively.
Supervised machine-learning approaches have also been developed for negation detection. Auerbuch et al5 developed an algorithm to automatically learn negative context patterns in medical narratives. The algorithm uses information gain to learn negative context patterns.
A hybrid approach that classifies negations in radiology reports based on the syntactic categories of the negation signal and negation patterns was developed by Huang and Lowe.6 Thirty radiology reports were manually inspected to develop the classifier, and the classifier was validated on a set of 470 radiology reports. Evaluation was conducted on 120 radiology reports, and the reported recall and precision was 92.6% and 98.6%, respectively.
In the genomics domain, a rule-based system was developed by Sanchez-Graillet and Poesio7 to detect negated protein–protein interactions in the biomedical literature. The system was built using a full dependency parser. Hand-crafted rules were then used to detect negated protein–protein interaction. An example rule reads as follows: if cue verb, such as ‘interact’, is an object of ‘fail’, ‘Protein A’ is subject of fail, and ‘Protein B’ is object of interact, then there is no interaction between ‘Protein A’ and ‘Protein B’. Evaluation was conducted on 50 biomedical articles, and the best recall and precision reported was 66.27% and 89.15%.
Morante and Daelemans8 developed a two-phase approach to detect the scope of negation in biomedical literature. In the first phase, negation cues were identified by a set of classifiers, and in the second phase another set of classifiers was used to detect the scope of the negation. The system performed better than the baseline in identifying negation signals in text and the scope of negation. The percentage of correct scope for abstract, full-text and clinical articles was 66.07%, 41% and 70.75%, respectively.
Most systems reported above were developed to detect negation in either clinical notes or biomedical literature. In contrast, our system was trained on annotations from a large corpus of both clinical and biomedical texts, and therefore its ability to detect negations in both the medical and genomics domain is robust. Such a cross-domain negation detection system will also assist text mining systems that require the analysis of both clinical data and primary literature; an application example being the clinical question answering system AskHERMES9 10 that we are now developing. Furthermore, while the previous systems detect negation in a sentence, most of them do not detect the scope of negation. Ignoring the scope of negation can be potentially misleading, as only some clauses in the sentence might be negated, but other clauses offer non-negated information. The NegEx2 system makes use of simple rules to detect negation scope boundaries, but due to the complexity of language, the scope identified may not be correct. We thus report on a machine-learning approach to detect both negation and its scope here.
Finally, except for the NegEx system, none of the previous systems is available for general use. To our knowledge, NegScope is currently the only implemented system that is publicly available and detects both negation and its scope in both biological literature and clinical notes.
Methods
We briefly describe our methods in this section; the detailed version is available as supplementary material (available online only, at www.jamia.org). In general, our systems were built by training a supervised machine-learning algorithm known as conditional random fields (CRF) on the BioScope corpus.1 We created training and test data for biological and clinical sentences, and used them for training and evaluation, respectively. We also evaluated our systems on a test set used to evaluate NegEx.2
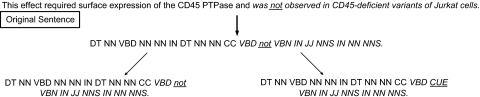
We trained CRF11–13 models on the BioScope training data to identify negation cue phrase and scope, and call them NegCue and NegScope, respectively. For NegScope, we also used a backoff smoothing model14 by replacing non-cue phrase words with part of speech tags15 (figure 1). Cue phrase words were either not replaced (ie, the original cue words were retained) or were replaced with a custom tag ‘CUE’ (figure 1).
Figure 1.
Example of a sentence used for training after it was replaced with its part of speech tags. The underlined word is the cue word in the sentence, while the words in italics represent the scope of negation. In the first step, all words except the cue word (underlined) were replaced with their part of speech tags. The cue word was either not replaced (bottom left) or replaced with a custom tag ‘CUE’ (bottom right). Other tags are CC, coordinating conjunction; DT, determiner; IN, preposition; JJ, adjective; NN, singular noun; NNS, plural noun; VBD, past tense verb; VBN, past participle verb.
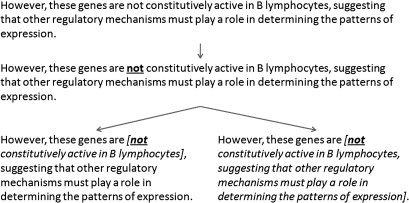
For comparison, we developed two baseline systems BaselineCue and BaselineScope, which detect negation cue phrase and scope, respectively. BaselineCue incorporates a negation cue phrase dictionary that was obtained by extracting negation cue phrases from the training data. It then looks up the dictionary for identifying cue phrases in the testing data. BaselineScope marks the scope from the beginning of a cue phrase identified by BaselineCue until the first occurrence of a comma or a period (figure 2). Another baseline system is NegEx,2 which is currently the only negation system made available to the public.
Figure 2.
An example showing the method in which BaselineScope marks the scope of the sentence. The negation cue is first identified using BaselineCue. BaselineScope then marks the scope of negation as the text from the negation cue to the first comma or period (left), or the first period (right).
We evaluate all systems by calculating recall, precision, F1-score and accuracy. For every word in the test sentence, if both the original annotation and tested system marked the word as a part of a cue phrase or scope, then the word was counted as a true positive; if the original annotation only marked the word as a part of the cue phrase, then the word was counted as a false negative; if only the tested system marked the word as a part of the cue phrase, then the word was counted as a false positive; and if neither the original annotation nor the tested system marked the word as a part of the cue phrase, then the word was counted as a true negative.
We also calculated the percentage of the correct scope (PCS) to evaluate the performance of scope predicting systems. If for a sentence, none of the words were marked as false positive or false negative, then we consider that the system had correctly predicted the scope of the sentence. For sentences with no negation, the system correctly predicted the scope of the sentence only if it indicated that there was no scope of negation.
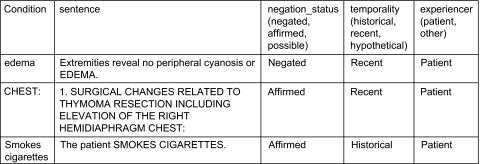
We also evaluated all systems on the NegEx test data. The NegEx test data do not mark the scope of negation, but mark the negation status of a target entity (figure 3). The publicly available version of the NegEx system can mark the negation status of a target entity in a sentence. To evaluate our system on the NegEx test data, we first marked the scope of negation of the sentence using the system and then checked if the target entity was a part of the negation scope. If it was, we marked the negation status target entity as ‘negated’; if not, we marked it as ‘affirmed’. We obtained the recall, precision, F1-score and accuracy for all systems.
Figure 3.
Examples of instances in the NegEx test set.
Results
Table 1 shows the performance of three systems—NegCue, BaselineCue and NegEx—for predicting negation cue phrases in the clinical subcorpus, biological subcorpus, and combination of both clinical and biological subcorpora. Table 2 shows the performance of three systems—NegScope, BaselineScope and NegEx—for negation scope detection.
Table 1.
Performance of NegCue, BaselineCue and the NegEx systems at identifying negation cue phrases in the BioScope testing set
| Clinical sentences | Biomedical sentences | Both clinical and biomedical sentences | |||||||
| Neg Cue | BaselineCue | Neg Ex | Neg Cue | BaselineCue | Neg Ex | Neg Cue | BaselineCue | Neg Ex | |
| Recall | 96.24±2.05 | 96.92±2.31 | 94.61±2.69 | 95.74±2.34 | 99.37±1.44 | 80.11±3.47 | 93.80±2.26 | 98.98±1.14 | 84.21±3.91 |
| Precision | 99.27±1.17 | 87.43±8.33 | 74.43±7.97 | 97.31±2.0 | 72.79±3.43 | 88.87±4.82 | 97.38±0.57 | 91.33±2.69 | 84.26±3.50 |
| F1-score | 97.72±1.48 | 91.76±5.03 | 83.11±5.34 | 96.50±1.7 | 83.98±2.28 | 84.20±3.28 | 95.55±1.38 | 94.99±1.84 | 84.17±2.77 |
| Accuracy | 99.74±0.16 | 98.99±0.58 | 97.79±0.63 | 99.86±0.07 | 99.20±0.11 | 99.37±0.13 | 99.80±0.07 | 99.76±0.09 | 99.29±0.12 |
Value after ‘±’ indicates SD. Values in bold represent the best performance.
Table 2.
Performance of NegScope and BaselineScope at predicting the scope of negation
| NegScope | BaselineScope | Negex | ||||||
| Features used | Words | Part of speech | Part of speech | Part of speech | Part of speech | Words | Words | Words |
| Cue identified using | – | NegCue | NegCue | BaselineCue | BaselineCue | BaselineCue | BaselineCue | – |
| Cue phrase replaced | – | No | Yes | No | Yes | – | – | – |
| Scope limited by | – | – | – | – | – | Comma and period | Period only | – |
| Both biological and clinical sentences used for training and testing | ||||||||
| Recall | 76.27±2.66 | 82.09±2.30 | 81.40±2.03 | 85.13±2.57 | 84.56±2.32 | 83.68±3.04 | 89.17±2.48 | 71.7±5.14 |
| Precision | 88.36±3.65 | 86.26±3.29 | 86.07±3.66 | 82.37±3.22 | 81.76±3.72 | 76.69±3.34 | 57.94±3.67 | 64.5±4.99 |
| F1-score | 81.79±1.64 | 84.08±1.94 | 83.63±2.12 | 83.68±1.90 | 83.08±2.09 | 79.99±2.59 | 70.16±2.70 | 67.79±3.0 |
| Accuracy | 94.31±0.64 | 94.79±0.77 | 94.66±0.82 | 94.42±0.94 | 94.22±0.96 | 92.97±1.19 | 87.31±1.35 | 88.62±1.26 |
| PCS | 84.21±1.74 | 83.79±2.25 | 83.93±2.22 | 82.65±2.41 | 82.65±2.27 | 80.89±2.50 | 76.0±2.36 | 74.62±2.46 |
| Biological sentences used for training and testing | ||||||||
| Recall | 73.72±2.39 | 84.07±3.84 | 81.76±3.59 | 87.27±3.82 | 85.94±3.70 | 84.62±3.74 | 90.36±2.65 | 72.67±3.84 |
| Precision | 85.53±5.02 | 84.74±3.47 | 82.74±4.03 | 71.76±4.04 | 69.69±3.33 | 67.50±4.58 | 51.12±3.66 | 63.81±4.69 |
| F1-score | 79.09±2.40 | 84.37±3.25 | 82.13±2.01 | 78.71±3.41 | 76.90±2.61 | 75.02±3.56 | 65.23±3.26 | 67.84±3.36 |
| Accuracy | 93.20±0.77 | 94.61±0.96 | 93.81±0.69 | 91.80±1.23 | 91.02±1.04 | 90.20±1.20 | 83.16±1.96 | 87.96±1.57 |
| PCS | 80.26±1.61 | 80.99±2.36 | 79.18±2.11 | 74.12±2.14 | 72.78±1.8 | 72.31±2.24 | 66.37±2.84 | 70.56±2.09 |
| Clinical sentences used for training and testing | ||||||||
| Recall | 93.68±2.40 | 94.99±3.02 | 95.18±2.74 | 95.30±3.01 | 95.49±2.71 | 93.75±2.23 | 96.00±2.42 | 85.94±6.93 |
| Precision | 95.25±4.61 | 94.74±4.23 | 94.07±4.91 | 85.98±9.11 | 85.37±8.78 | 85.73±10.10 | 82.14±8.97 | 90.81±5.26 |
| F1-score | 94.37±1.97 | 94.82±3.04 | 94.57±3.32 | 90.26±6.26 | 89.99±5.85 | 89.34±6.56 | 88.31±5.79 | 88.07±4.07 |
| Accuracy | 96.88±0.93 | 97.07±1.84 | 96.87±2.09 | 94.33±3.23 | 94.12±3.11 | 93.82±3.29 | 92.95±2.86 | 93.58±1.73 |
| PCS | 95.83±1.66 | 96.06±2.12 | 95.49±2.07 | 92.25±3.99 | 91.67±3.20 | 91.21±3.12 | 91.55±2.77 | 88.90±2.14 |
Value after ‘±’ indicates SD. Values in bold represent the best performance.
PCS, percentage of the correct scope.
The results show that NegScope performed better than both BaselineScope and NegEx and the differences were statistically significant (p<0.001%, t test, two-tailed). We calculated the micro-average, the sentence-level average performance of a category, of F1-score for every system. We explored two models for training and testing. In the first model, systems were trained and tested separately on biological and clinical data. This model has resulted in an F1-score of 87.60±2.35% and 79.44±3.0% for NegScope and BaselineScope, respectively. In the second model, we merged biomedical data with clinical data, and trained NegScope and BaselineScope on the merged data. This led to decreased performance for NegScope but increased performance for BaselineScope—84.08% and 79.99% F1-score, respectively. While the performance difference of BaselineScope between the two models was not statistically different (p=0.67), the difference of NegScope between two models was statistically significant (p=0.002).
The performance of our systems and the NegEx system in predicting the negation status of target conditions in the NegEx test set is shown in table 3.
Table 3.
Performance of NegScope, BaselineScope and the NegEx system on the Negex testing data. NegScope and BaselineScope were trained on both biological and clinical sentences
| NegScope | BaselineScope | NegEx | ||||||
| Features used | Words | Part of speech | Part of speech | Part of speech | Part of speech | Words | Words | – |
| Cue phrase identified using | – | NegCue | NegCue | BaselineCue | BaselineCue | BaselineCue | BaselineCue | – |
| Cue phrase replaced | – | No | Yes | No | Yes | – | – | – |
| Scope limited by | – | – | – | – | – | Comma and period | Period only | – |
| Both biological and clinical sentences used for trainining | ||||||||
| Recall | 75.3±6.86 | 67.89±7.30 | 67.11±8.34 | 81.13±5.25 | 83.83±4.93 | 75.94±5.79 | 85.24±4.77 | 96.27±3.08 |
| Precision | 90.48±3.57 | 97.21±2.87 | 96.48±3.77 | 90.54±2.68 | 87.16±3.51 | 78.9±4.88 | 76.22±4.39 | 95.43±2.02 |
| F1-score | 82.12±5.30 | 79.79±5.64 | 78.99±6.81 | 85.53±3.76 | 85.42±3.82 | 77.29±4.41 | 80.44±4.13 | 95.82±1.91 |
| Accuracy | 93.35±1.57 | 93.06±1.38 | 92.8±1.75 | 94.4±1.20 | 94.15±1.34 | 90.87±1.46 | 91.5±1.59 | 98.27±0.78 |
| Clinical sentences used for training | ||||||||
| Recall | 36.88±5.42 | 58.52±6.48 | 70.33±5.93 | 59.13±6.53 | 72.0±5.76 | 63.61±6.77 | 80.06±5.37 | 96.27±3.08 |
| Precision | 94.59±5.43 | 96.59±2.72 | 95.07±3.03 | 93.36±2.36 | 91.64±2.38 | 80.46±6.14 | 81.59±3.87 | 95.43±2.02 |
| F1-score | 52.92±6.10 | 72.73±5.32 | 80.72±4.41 | 72.22±4.99 | 80.55±4.17 | 70.99±6.28 | 80.78±4.39 | 95.82±1.91 |
| Accuracy | 86.53±1.79 | 91.04±1.48 | 93.18±0.94 | 90.7±1.46 | 92.93±0.94 | 89.35±2.06 | 92.21±1.45 | 98.27±0.78 |
Value after ‘±’ indicates SD. Values in bold represent the best performance.
When we tested the performance of NegScope and BaselineScope on the NegEx testing data, we observed that many false negative instances contained the words ‘denied’ and ‘denies’. This is because in the BioScope training data, there were no examples of negated sentences that used these words as a negation cue phrase. For example, in the test sentence ‘Denied any HEADACHES’, the tested entity ‘headaches’ is negated, but our training data contained no cases in which ‘denied’ appeared as the negation cue. In order to overcome the inability of our systems to identify cue phrases ‘denies’ and ‘denied’, we manually added these cue phrases to the set of cue phrases identified by the baseline system (BaselineCue). We could not manually add these cue phrases to the CRF system that identifies cue phrases (NegCue) because we would have to create specific training sentences for the system to learn these phrases. As only the baseline system could be modified, we were only able to test the performance of NegScope when the cue phrase was identified by BaselineScope. The performance of these systems is shown in table 4.
Table 4.
Performance of NegScope, BaselineScope and the NegEx systems on the NegEx testing set when additional cue words were added to the dictionary
| NegScope | BaselineScope | NegEx | |||
| Features used | Part of speech | Part of speech | Words | Words | – |
| Cue phrase identified using | BaselineCue | BaselineCue | BaselineCue | BaselineCue | – |
| Cue phrase replaced | No | Yes | – | – | – |
| Scope limited by | – | – | Comma and period | Period only | – |
| Recall | 92.73±3.08 | 95.85±2.73 | 85.79±3.60 | 97.48±2.22 | 96.27±3.08 |
| Precision | 91.66±2.25 | 88.64±2.89 | 80.95±3.89 | 78.61±3.65 | 95.43±2.02 |
| F1-score | 92.17±2.30 | 92.08±2.33 | 83.2±2.30 | 87.0±2.76 | 95.82±1.91 |
| Accuracy | 96.8±0.77 | 96.63±0.86 | 92.89±0.80 | 94.02±1.11 | 98.27±0.78 |
Value after ‘±’ indicates SD. Values in bold represent the best performance.
Discussion
We have developed CRF-based models to predict the scope of negation in biomedical sentences. We compared these models with baseline systems, which use dictionary lookup, and NegEx, which makes use of regular expressions and rules to mark the cue phrase and scope of negation in a sentence. Our results indicate that models using CRF for the detection of scope of negation in biomedical sentences perform better than models based on the use of dictionary lookup and NegEx (p<0.001, t test, two-tailed). Our system can be used to detect negation and its scope in both biological and clinical text.
Negation cue phrase detection
For detection of cue phrases, we observed that the F1-score and accuracy of NegCue is slightly better than BaselineCue; however, the difference was not statistically significant (F1-score p=0.45; accuracy p=0.28, t test, two-tailed). The recall of BaselineCue is significantly better than that of NegCue (p<0.0001, t test, two-tailed). This is because BaselineCue collects all phrases that have been seen as a cue for negation and marks any such phrase in the sentence as a negation cue, without considering the context in which it appears. BaselineCue thus achieves a lower precision than the CRF system, which lowers its F1-score and accuracy. NegCue's performance was significantly better than NegEx's performance. This was because NegEx's rules were generally longer that the cue phrases identified in the BioScope corpus. For example, in the sentence ‘No signs of tuberculosis’, the cue annotated in the BioScope corpus was ‘no’, whereas the NegEx rule that matched was ‘no signs of’. Moreover, certain cue terms such as ‘unable’ and ‘lacked’ were not present in NegEx's dictionary of negation triggers.
Negation scope detection
At the task of detecting negation scope, we noticed that the average F1-score of NegScope trained specifically for biological and clinical text was better than the F1-score of NegScope trained on the combination of biological and clinical text. This is because there are several differences in biological and clinical text. For example, biological sentences from articles published in journals are generally grammatically well formed, whereas many sentences from clinical notes are not (eg, ‘Normal two views of the chest without focal pneumonia.’).
We found that the NegScope system performed better than the BaselineScope system (F1-score p<0.001; PCS p=0.003, t test, two-tailed) and NegEx (p<0.001, t test, two-tailed). Within NegScope, we found that smoothing the data by replacing words with part of speech tags improved the F1-score (p=0.01, t test, two-tailed). However, the smoothing in which the cue words were replaced with ‘CUE’ did not improve the performance. Leaving the cue words as is resulted in slightly better performance than replacing them with the custom tag ‘CUE’, but the difference was not statistically significant (F1-score p=0.63, t test, two-tailed).
When comparing the BaselineScope systems, a better performance was observed when both the commas and periods were used to mark the end of scope. As expected, the baseline system that used only periods to mark the end of scope achieved a better recall (+5%) than when commas and periods were used; however, the decrease in precision was 19%, which decreased the overall F1-score by 9% (p<0.0001, t test, two-tailed).
In analyzing the cases that were not correctly annotated by NegEx, we found that many errors occurred because NegEx assumes that the cue appears at the beginning or the end of the scope. For example, in the sentence ‘In A20 B cells, the TNF-alpha gene is not regulated by NFATp bound to the kappa 3 element’, the cue ‘not’ was correctly identified by NegEx and it marked the scope from the cue to the end of the sentence. However, the scope in the gold standard was ‘the TNF-alpha gene is not regulated by NFATp bound to the kappa 3 element’, whereas the scope identified by NegEx was ‘not regulated by NFATp bound to the kappa 3 element’. False negative errors were seen when the cue phrase was not detected by NegEx. NegEx exhibits especially poor performance on biological sentences, which is expected because it was not designed for biological literature.
In analyzing the cases in which NegScope did not identify the scope of negation correctly, we found that the errors could be classified into three categories: (1) type I error (false positive): the model assigns scope when none exists (ie, it is a non-negative sentence); (2) type II error (false negative): the model assigns no scope when one does exist (ie, it is a negative sentence); and (3) boundary errors: the model correctly assigns negative polarity to the sentence, but it assigns a different boundary than that assigned in the testing data. In 2801 sentences, we observed 20, 49, and 386 type I, type II and boundary errors, respectively. We present an analysis of each error type along with examples in the supplementary material (available online only).
Despite these errors, our system achieved a strong performance in scope detection (84.08% F1-score), which makes it suitable to be used in conjunction with other text mining applications in both biological and clinical domains. We show two examples in which NegScope correctly determined the scope of negation whereas BaselineScope did not. In the first sentence, the correct scope is marked by square brackets, and in the second sentence there is no negation scope even though the sentence includes the frequently used negation cue phrase ‘not’:
In A20 B cells, [the TNF-alpha gene is not regulated by NFATp bound to the kappa 3 element].
These data suggest that interferon regulatory factor 1 not only triggers the activation of the interferon signal transduction pathway, but also may play a role in limiting the duration of this response by activating the transcription of IRF-2.
For the first example, the BaselineScope system incorrectly excluded the phrase ‘the TNF-alpha gene’ from the scope of negation, but the entire scope was correctly identified by the NegScope model. In the second example, the NegScope system did not mark the sentence, as there is no scope of negation, but the BaselineScope system marked the scope from ‘not’ to the end of the clause.
Performance of NegScope, BaselineScope and NegEx on the NegEx test data
As shown in table 4, NegEx2 performed best (F1-score 95.82%). NegScope had a F1-score of 85.53%. Adding two cue phrases (‘denied’ and ‘denies’) to the dictionary, the F1-score increased to 92.17% (difference with NegEx's F1-score was statistically significant at p=0.001, t test, two-tailed), which indicates that NegScope is robust and it can attain a good performance on different data without major adaptations. Although the F1-score of the modified NegScope system is still not as high as the F1-score of the NegEx system, it may still be useful for practical purposes. Details of all three system's performance are described in the supplementary material (available online only, at www.jamia.org).
Limitations
The NegScope was trained on the BioScope corpus, and therefore the quality and the size of the annotation impact NegScope's performance. In addition, like all other NLP systems, NegScope faces the complexity of natural language. An example is the cue term ‘allergy’. Consider a dummy sentence ‘the patient is allergic to DrugX’. If the task is to identify the medications that were given to the patient, then ‘allergic’ is a negation cue phrase. In other tasks, allergic may not be considered a negation cue phrase. For example, if the task is to determine if a clinical condition was negated, then allergy to DrugX is the clinical condition and it is not negated. NegScope cannot perform such task-specific negation detection, if the annotated data do not fall into the task requirement.
Comparison with another CRF-based approach
A CRF-based approach was used by Morante and Daelemans8 to identify scope in biomedical literature. Similar to our approach, their system was also trained on the BioScope data. In comparing their reported results with our results, we noticed that our system performed better than their system. This could be due to the difference in the training data used; Morante and Daelemans8 used only the abstract subcorpus for training. Surprisingly, our system's overall performance (PCS ∼84%) was also better than their system's performance on the abstract subcorpus (PCS ∼67%). This could be due to the difference in the size of the training data or the features used for selection. Unfortunately, the system of Morante and Daelemans8 is not publicly available, so we were unable to test the performance of their system on the same test sets as our system was tested on.
Conclusion and future work
We have created several CRF-based models that can automatically predict the scope of negation in biomedical literature. These models can also be used to predict the negation status of a target entity in the sentence. The choice of which model to use depends on the task at hand. For predicting the scope of negation, we recommend using a CRF-based model that identifies cue phrases using a CRF-based cue phrase identifier and replaces non-cue phrase words with their parts of speech. However, to test the negation status of a target entity in the sentence, we recommend using a CRF-based model that identifies cue phrases from a dictionary of possible cue phrases. More importantly, the models we have trained are robust and perform well in detecting negation in both biomedical and clinical documents. An online version of the negation scope detector is available at http://negscope.askhermes.org. The system is also available as a Java API from this link.
Any annotated corpus has size limitations, and unseen data encountered by a system trained on such a corpus will hurt the system's performance. In future work we may explore methods for automatically identifying negation cue phrases from a large corpus, including contextual similarity, which is commonly used for identifying semantically related words or synonyms.16 17 We may also explore bootstrapping18 or co-training approaches19 that partly overcome the limitations of training size.
Acknowledgments
The authors thank Dr Lamont Antieau for proofreading this manuscript. Any opinions, findings, or recommendations are those of the authors and do not necessarily reflect the views of the NIH.
Footnotes
Funding: This work received support from the National Library of Medicine, grant number 5R01LM009836 to HY and 5R01LM010125 to Isaac Kohane.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Szarvas G, Vincze V, Farkas R, et al. The BioScope corpus: annotation for negation, uncertainty and their scope in biomedical texts. In: Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing. Columbus, Ohio: Association for Computational Linguistics, 2008:38–45 [Google Scholar]
- 2.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 3.Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS. J Am Med Inform Assoc 2001;8:598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elkin P, Brown S, Bauer B, et al. A controlled trial of automated classification of negation from clinical notes. BMC Med Inform Decis Mak 2005;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auerbuch M, Karson TH, Ben-Ami B, et al. Context-sensitive medical information retrieval. Stud Health Technol Inform 2004;107:282–6 [PubMed] [Google Scholar]
- 6.Huang Y, Lowe HJ. A novel hybrid approach to automated negation detection in clinical radiology reports. J Am Med Inform Assoc 2007;14:304–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Graillet O, Poesio M. Negation of protein protein interactions: analysis and extraction. Bioinformatics 2007;23:i424–32 [DOI] [PubMed] [Google Scholar]
- 8.Morante R, Daelemans W. A metalearning approach to processing the scope of negation. In: Proceedings of the Thirteenth Conference on Computational Natural Language Learning. Boulder, Colorado: Association for Computational Linguistics, 2009:21–9 [Google Scholar]
- 9.Yu H, Lee M, Kaufman D, et al. Development, implementation, and a cognitive evaluation of a definitional question answering system for physicians. J Biomed Inform 2007;40:236–51 [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Cao Y. Automatically extracting information needs from ad hoc clinical questions. AMIA Annu Symp Proc 2008:96–100 [PMC free article] [PubMed] [Google Scholar]
- 11.Lafferty J, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and labeling sequence data. In: Proceedings of the Eighteenth International Conference on Machine Learning (ICML 2001). Williamstown, MA, USA, 2001:282–9 [Google Scholar]
- 12.Settles B. ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics 2005;21:3191–2 [DOI] [PubMed] [Google Scholar]
- 13.Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Pac Symp Biocomput 2008:652–63 [PubMed] [Google Scholar]
- 14.Katz S. Estimation of probabilities from sparse data for the language model component of a speech recognizer. IEEE Trans Acoust 1987;35:400–1 [Google Scholar]
- 15.Toutanova K, Manning C. Enriching the knowledge sources used in a maximum entropy part-of-speech tagger. In: Proceedings of the Joint SIGDAT Conference on Empirical Methods in Natural Language Processing and Very Large Corpora (EMNLP/VLC-2000). 2000:63–70 [Google Scholar]
- 16.Dagan I, Marcus S, Markovitch S. Contextual word similarity and estimation from sparse data. In: Proceedings of the 31st Annual Meeting on Association for Computational Linguistics. Columbus, Ohio: Association for Computational Linguistics, 1993:164–71 [Google Scholar]
- 17.Yu H, Agichtein E. Extracting synonymous gene and protein terms from biological literature. Bioinformatics 2003;19:340–9 [DOI] [PubMed] [Google Scholar]
- 18.Weiss SM, Kapouleas I. An empirical comparison of pattern recognition, neural nets, and machine learning classification methods. In: Proceedings of the Eleventh International Joint Conference on Artificial Intelligence. Detroit, MI: International Joint Conferences on Artificial Intelligence (IJCAI) 1989:781–7 [Google Scholar]
- 19.Blum A, Mitchell T. Combining labeled and unlabeled data with co-training. In: Proceedings of the Eleventh Annual Conference on Computational Learning Theory. Madison, Wisconsin, United States: ACM, 1998:92–100 [Google Scholar]