Abstract
Objective
To study existing problem list terminologies (PLTs), and to identify a subset of concepts based on standard terminologies that occur frequently in problem list data.
Design
Problem list terms and their usage frequencies were collected from large healthcare institutions.
Measurement
The pattern of usage of the terms was analyzed. The local terms were mapped to the Unified Medical Language System (UMLS). Based on the mapped UMLS concepts, the degree of overlap between the PLTs was analyzed.
Results
Six institutions submitted 76 237 terms and their usage frequencies in 14 million patients. The distribution of usage was highly skewed. On average, 21% of unique terms already covered 95% of usage. The most frequently used 14 395 terms, representing the union of terms that covered 95% of usage in each institution, were exhaustively mapped to the UMLS. 13 261 terms were successfully mapped to 6776 UMLS concepts. Less frequently used terms were generally less ‘mappable’ to the UMLS. The mean pairwise overlap of the PLTs was only 21% (median 19%). Concepts that were shared among institutions were used eight times more often than concepts unique to one institution. A SNOMED Problem List Subset of frequently used problem list concepts was identified.
Conclusions
Most of the frequently used problem list terms could be found in standard terminologies. The overlap between existing PLTs was low. The use of the SNOMED Problem List Subset will save developmental effort, reduce variability of PLTs, and enhance interoperability of problem list data.
Introduction
The problem-oriented approach of organizing information in a medical record was first advocated by Weed almost 40 years ago.1 Central to this approach is a problem list which is ‘a complete list of all the patient's problems, including both clearly established diagnoses and all other unexplained findings that are not yet clear manifestations of a specific diagnosis, such as abnormal physical findings or symptoms.’ According to Weed, this list should also cover ‘psychiatric, social and demographic problems.’ Although the adoption of Weed's problem-oriented approach to the whole medical record has been limited, the use of problem lists is widespread in both paper- and computer-based medical records. Furthermore, many sanctioning bodies and medical information standards organizations consider the problem list to be an important element of the electronic health record (EHR), including the Institute of Medicine, Joint Commission, American Society for Testing and Materials and Health Level Seven.2–6 An encoded problem list is also one of the core objectives of the ‘meaningful use’ regulation of EHR published by the Department of Health and Human Services.7 In a recent national survey on the use of the EHR in US hospitals, an expert panel considers the problem list an essential component of both a basic and comprehensive EHR.8
Problem lists have value beyond clinical documentation. Common uses include the generation of billing codes and clinical decision support. To drive many of these functions, an encoded problem list (as opposed to data entered as free-text) is often required. This probably explains why the problem list is often the first (if not the only) location within an EHR with encoded clinical statements. This paper explores the use of controlled terminologies in electronic problem lists and their associated problems.
In an ideal world, everybody will be using a single, standardized problem list terminology (PLT). In reality, most institutions use their own local terminologies. In the USA, even though SNOMED CT is the terminology designated for problem lists by the Consolidated Health Informatics Initiative,9 10 the adoption of SNOMED CT has not been widespread.11 We started the UMLS-CORE Project in 2007 to study PLTs. As a flagship terminology product of the US National Library of Medicine (NLM), the Unified Medical Language System (UMLS) is a valuable resource for terminology research.12 The mnemonic CORE stands for ‘clinical observations recording and encoding,’ which refers to the capture and codification of clinical information in the summary segments of the EHR such as the problem list, discharge diagnosis, and reason for encounter. The UMLS-CORE Project has two goals:
To study and characterize the PLTs of large healthcare institutions in terms of their size, pattern of usage, mappability to standard terminologies, and extent of overlap.
To identify a subset of concepts based on standard terminologies that occur with high frequency in problem list data to facilitate the standardization of PLTs.
This paper describes the methods, findings, and implications of this project.
Methods
We asked large-scale healthcare institutions to submit their problem list terms together with the actual frequency of usage in their clinical databases. We also requested any mapping from the local problem list terms to standard terminologies (eg, ICD-9-CM, SNOMED CT) if available.
We mapped the local terms to the UMLS Metathesaurus using the 2008AA release. We only used exact maps that captured the full meaning of the local terms (ie, no inexact or broader/narrower maps). Only existing UMLS concepts were used and no attempt was made to create new meaning by combining existing concepts (post-coordination).
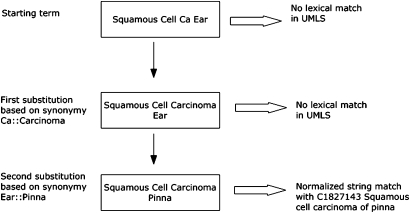
The UMLS mapping was carried out in three sequential steps. The first step was lexical matching. We first looked for exact (case-insensitive) and normalized matches using all English terms in the UMLS. Normalization was performed using the ‘norm’ function of the UMLS lexical tools, in order to abstract away from differences due to word inflection, case, word order, punctuation, and stop words.13 For example, ‘Perforating duodenal ulcers’ and ‘Duodenal ulcer, perforated’ both normalize to the same string ‘duodenal perforate ulcer’. We further enhanced lexical matching by synonymous word or phrase substitution.14 Local terms that did not have exact or normalized matches were scanned for words or phrases listed in a synonymy table (used internally in UMLS quality assurance). If found, we replaced the word or phrase with their synonyms and reapplied the matching algorithm. Empirically, we allowed a maximum of two substitutions to avoid unintentional meaning drift. Figure 1 shows an example of lexical matching with synonymous word substitution.
Figure 1.
Lexical matching augmented by synonymous word or phrase substitution.
Terms that could not be mapped by lexical matching were passed through to the second step, which made use of local maps to standard terminologies (either ICD-9-CM or SNOMED CT) if they were available. Not all local maps were exact maps. Only those maps that were explicitly labeled as exact maps by the institutions were used to map automatically (ie, without manual review) to the UMLS. For example, the local term ‘Adrenal insufficiency’ was mapped to the SNOMED CT concept ‘111563005 Adrenal hypofunction’ by one institution and it was labeled as an exact map. This map was used to map the local term to the UMLS concept C0001623 containing that SNOMED CT concept. Local maps that were not labeled explicitly as exact maps were manually reviewed and only those that were considered exact maps were used.
The final step was manual mapping. All terms that remained unmapped after the first two steps were manually mapped to the UMLS, using the RRF browser included with the UMLS as the searching tool.15 All manual mapping was carried out or validated by one of the authors (KWF). No manual validation was performed on terms that were algorithmically mapped in steps one and two. All terms that were ultimately unmapped were analyzed for reasons of failure by KWF.
All subsequent analysis was based on those terms that could be mapped to the UMLS. Comparison between PLTs was carried out using the UMLS concept (identified by its concept unique identifier (CUI)) as the proxy for the local term, so it was performed at the concept (not term) level. We defined the pairwise overlap between institutions A and B as follows:
We also compared the usage statistics of concepts shared among institutions with concepts occurring only in one institution.
Results
Characteristics of the institutions and datasets
We obtained datasets from the following six healthcare institutions: Kaiser Permanente (KP), Mayo Clinic (MA), Intermountain Healthcare (IH), Regenstrief Institute (RI), University of Nebraska Medical Center (NU) and Hong Kong Hospital Authority (HA). The HA was the only institution outside the USA. The RI data came from the Wishard Hospital Project. All the US datasets covered both ambulatory care and hospital patients. The HA dataset represented the discharge diagnoses of hospital inpatients. The characteristics of these institutions and their datasets are summarized in table 1.
Table 1.
Characteristics of the institutions and their datasets
| HA | IH | KP | MA | NU | RI | |
| Type of service | Inpatient | Mixed | Mixed | Mixed | Mixed | Mixed |
| Inpatient percentage | 100 | 10 | No data | 20 | 15 | 50 |
| Patient count (million) | 1.3 | 0.36 | 10 | 1.5 | 0.5 | 0.16 |
| Period of data retrieval | 3 years | All current patients | All current patients | 3 years | All current patients | 1 year |
| Count of problem terms | Patient-based | Patient-based | Patient-based | Encounter-based | Patient-based | Patient-based |
| Average problem terms per patient | 3.1 | 3.0 | 5.2 | 6.8 | 5.3 | 4.2 |
| Total unique terms | 12449 | 5685 | 26890 | 14921 | 13126 | 3166 |
| Unique terms covering 95% of total usage | 2635 | 1077 | 2961 | 3610 | 3320 | 792 |
HA, Hong Kong Hospital Authority; IH, Intermountain Healthcare; KP, Kaiser Permanente; MA, Mayo Clinic; NU, University of Nebraska Medical Center; RI, Regenstrief Institute.
The institutions were of relatively large scale and provided service in all major medical specialties including internal medicine, general surgery, pediatrics, obstetrics & gynecology, psychiatry, and orthopedics. The problem lists in these institutions were generated and maintained by physicians from all specialties as part of the care process, and the information was also available to other care givers. All institutions were using, or planning to use, the encoded problem list data for purposes beyond clinical documentation. These uses included: clinical decision support (eg, pharmacy alerts, best practice alerts, suggestion of order sets), generation of administrative codes (eg, ICD-9-CM codes and diagnosis related groups (DRG) for billing, ICD-10 codes for public health reporting), clinical research (eg, identification of study subjects, enrollment to research protocols), and compilation of management statistics.
The six datasets together covered 14 million patients. For HA and MA, the datasets represented all patients encountered by the system in 3 years. The RI data were collected over 1 year. For IH, KP, and NU, the datasets included all patients currently registered in their systems. Most of the usage data were patient-based, meaning that a given problem in one patient would be counted only once, although it could be documented multiple times in different encounters. Only the MA usage data were encounter-based. The average number of problem terms per patient varied from three to seven. The size of the PLTs varied considerably across institutions, ranging from just over 3000 to almost 27 000 unique terms.
Usage pattern of terms
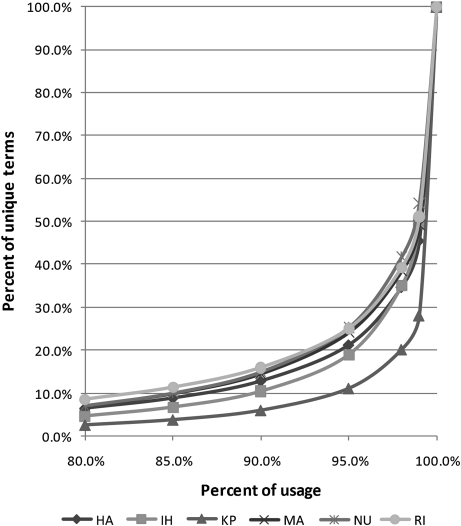
For each institution, we looked at the statistical distribution of usage. Figure 2 shows the percentage of unique terms (vertical axis) required in each institution to cover a certain percentage of total usage (horizontal axis). We used the percentage (instead of the absolute number) of unique terms to facilitate cross-institution comparison, because there was significant variation in the size of the PLTs. About 10% of unique terms already covered 85% of usage. To cover 95% of usage, the average percentage of terms required was 21%. The same skewed distribution—that is, a small proportion of commonly used terms and a long tail of rarely used ones—was found in all institutions.
Figure 2.
Usage pattern of the problem list terms. HA, Hong Kong Hospital Authority; IH, Intermountain Healthcare; KP, Kaiser Permanente; MA, Mayo Clinic; NU, University of Nebraska Medical Center; RI, Regenstrief Institute.
Mapping to the UMLS
We performed algorithmic lexical mapping for all 76 237 terms (65 678 terms unique across institutions) in the six datasets. For pragmatic reasons, we only carried out exhaustive mapping (including manual mapping) for the most frequently used 14 395 terms (10 081 terms unique across institutions), which was the union of the terms that covered 95% of usage in each institution. The following analysis and percentages were based on this subset of terms.
Results of UMLS mapping
Lexical mapping yielded maps for 10 812 terms (75% of 14 395 exhaustively mapped terms, same below). Among them, exact string match (case-insensitive) found 8102 terms (56%), normalized string match found 2035 terms (14%), and synonymous word or phrase substitution found an additional 675 terms (5%). The next mapping step made use of local maps when available. Some maps to ICD-9-CM were available for HA, MA, and RI, while maps to SNOMED CT were available for IH, KP, and NU. Only maps from HA, IH, and KP were explicitly labeled for the degree of exactness. Altogether, these local maps yielded UMLS maps for 1007 terms (7%). We manually reviewed the remaining 2576 terms and mapped 1442 (10%) terms, leaving 1134 terms (8%) ultimately unmapped. The mapping results are summarized in table 2. Altogether, 13 261 local terms were mapped to 6776 UMLS concepts, constituting what we called the UMLS-CORE Subset, representing the most commonly used UMLS concepts in problem lists. As for the nature of these concepts, altogether they were assigned 6938 UMLS semantic types (67 unique semantic types), with 91% of the semantic types belonging to the semantic group ‘Disorders’ and 7% to ‘Procedures’.16
Table 2.
Results of mapping to the UMLS
| Number of terms (%) | |
| Lexical matching | 10812 (75) |
| Mapping based on local maps provided by sources | 1007 (7) |
| Manual mapping | 1442 (10) |
| Terms not mapped to UMLS | 1134 (8) |
| Total | 14395 (100) |
UMLS, Unified Medical Language System.
Reasons for unmappability
We assigned each unmapped term to one of eight categories which were derived empirically (table 3). The most common category (53% of 1134 unmapped terms, same below) contained terms that included a high level of detail. One example was ‘Benign prostatic hyperplasia with age-related prostate cancer risk and obstruction’. Although ‘Benign prostatic hyperplasia’ and ‘Benign prostatic hyperplasia with obstruction’ both existed in the UMLS, the further qualification with cancer risk was not found. Many of these terms could be considered subtypes of existing terms with additional specificity. On the other hand, 11% of the unmapped terms were very general—for example, ‘Abnormal blood finding’. Seven percent of terms conveyed administrative rather than clinical information—for example, ‘Other Mr. # exists’—presumably to alert the healthcare provider about another patient with the same name. Another 7% of terms contained laterality information, which was not usually captured in standard terminologies. Again, like the highly specific terms, these terms could be considered subtypes of existing terms. Terms conveying negative findings and composite concepts each made up 3% of unmapped terms. A small number of terms (2%) were ambiguous and could not be mapped without clarifying their meaning. For example, red conjunctiva can refer to either acute conjunctivitis or conjunctival hemorrhage, which are distinct clinical entities. Finally, there were 13% of miscellaneous terms not easily classifiable to the other categories. For example, while ‘Sinusitis’, ‘Acute sinusitis’ and ‘Chronic sinusitis’ existed in the UMLS, ‘Subacute sinusitis’ was not found. Many of the unmapped terms, particularly those in the highly specific, laterality and composite concept categories would be mappable if post-coordination was allowed (see discussion).
Table 3.
Categorization of terms that could not be mapped to the UMLS
| Category | % | Example |
| Highly specific | 53 | Benign prostatic hyperplasia with age-related prostate cancer risk and obstruction |
| Very general | 11 | Abnormal blood finding |
| Administrative | 7 | Other Mr. # exists |
| Laterality | 7 | Renal stone, right |
| Negative finding | 3 | No urethral stricture |
| Composite concept | 3 | Diarrhea with dehydration |
| Meaning unclear | 2 | Conjunctiva red |
| Miscellaneous | 13 | Subacute sinusitis |
UMLS, Unified Medical Language System.
Relationship of usage to mapping
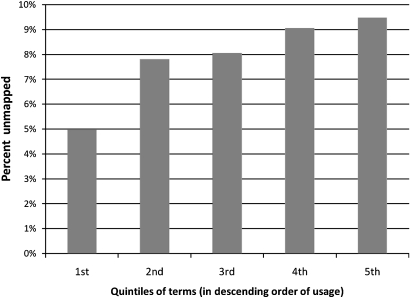
To study the relationship between frequency of usage and mappability to the UMLS, we arranged all 14 395 terms in descending order of usage. Starting from the most frequently used terms, we divided the terms into five equal parts and calculated the percentage of unmapped terms for each quintile (figure 3). The most frequently used quintile had 5% terms that were unmapped, which gradually increased to almost 10% for the last quintile. This showed that more frequently used terms were also more mappable. This finding is not a surprise because frequently used terms are more likely to have already made their way into standard terminologies and thus the UMLS.
Figure 3.
Relationship of term usage to mappability to the Unified Medical Language System.
Overlap between institutions
We calculated pairwise overlap between PLTs based on the terms that could be mapped to the UMLS, using the CUI as the basis for comparison. The pairwise overlap showed considerable variability (table 4). The lowest pairwise overlap was between HA and RI (11%), and the highest between KP and NU (29%). HA had the lowest mean pairwise overlap (15%) with all other institutions, while NU had the highest (24%). The overall mean pairwise overlap for all six institutions was 21% (median 19%).
Table 4.
Pairwise overlap between the PLTs
| Institution | Pairwise overlap with | Mean pairwise overlap | ||||
| IH | KP | MA | NU | RI | ||
| HA | 13% | 17% | 17% | 18% | 11% | 15% |
| IH | 25% | 19% | 25% | 27% | 22% | |
| KP | 29% | 29% | 17% | 23% | ||
| MA | 31% | 14% | 22% | |||
| NU | 19% | 24% | ||||
| RI | 18% | |||||
HA, Hong Kong Hospital Authority; IH, Intermountain Healthcare; KP, Kaiser Permanente; MA, Mayo Clinic; NU, University of Nebraska Medical Center; PLTs, problem list terminologies; RI, Regenstrief Institute.
Of the 6776 concepts (CUIs), 4201 concepts (62%) were unique to only one institution, while 2575 concepts (38%) were shared (table 5). When concept sharing was correlated with usage data, the unique concepts were found to be used much less frequently than the shared ones. Even though unique concepts made up 62% of total concepts, they accounted for only 15% of mean usage (average usage over six institutions). On the other hand, the 38% of shared concepts accounted for 77% of mean usage. We also calculated a Usage Index (mean usage × 1000/number of CUIs) for each group, which was the theoretical mean usage that 1000 concepts in that group would cover. The Usage Index increased exponentially with the number of sharing institutions, almost doubling at each step (tripling at the last step). There was a 50-fold difference in the Usage Index between the unique concepts and those shared by all six institutions. If we pooled all shared concepts together, the average Usage Index of shared concept was eight times that of a unique concept.
Table 5.
Distribution of concept unique identifiers (CUIs) among datasets and the corresponding usage coverage
| CUI appearing in | Number of CUIs (%) | Mean usage | Usage Index |
| 1 dataset | 4201 (62) | 15% | 0.035 |
| 2 datasets | 1130 (17) | 9% | 0.079 |
| 3 datasets | 607 (9) | 9% | 0.149 |
| 4 datasets | 391 (6) | 11% | 0.271 |
| 5 datasets | 282 (4) | 17% | 0.594 |
| 6 datasets | 165 (2) | 30% | 1.84 |
| Total | 6776 (100) | 90% | 0.133 |
Discussion
Despite the efforts to standardize medical terminologies, PLTs are still very much products of independent creation and evolution. In the beginning, institutions created their own terminologies, which could be derived from some pre-existing term lists or mined from clinical data.17–19 The subsequent changes of these terminologies are driven by a multitude of factors related to the various uses of the problem list data. The primary use of the problem list is clinical documentation. The problem list is ‘a table of content and an index’ which provides a convenient summary of the patient's problems and significant co-morbidities. This information helps to facilitate the continuity of care, formulation of plan of treatment or further investigations, and management of risk factors. To facilitate clinical documentation, the terms in the PLT need to resemble clinical parlance closely so that users can find their terms easily. Most institutions allow users to request terms that they cannot find. These requests are usually vetted through an editorial process and added to the terminology if necessary.20 Factors such as patient mix, level of care (primary or specialty care), care setting (eg, inpatient versus ambulatory), specialty distribution, user preferences, term request process, and editorial policy all affect the scope and granularity of the PLT.
Problem list data are often used to drive functions other than clinical documentation—for example, generation of billing codes, supporting clinical research, and quality assurance. The type and extent of these additional uses vary significantly across institutions,21 and they pose their own requirements on the PLT. It is not uncommon that different requirements are in conflict (eg, clinical documentation versus billing requirements), and the result is often a compromise between the usability of the PLT, primarily for the purpose of clinical documentation, and the usefulness of the data captured for other purposes.
This ‘letting a thousand flowers bloom’ scenario has two problems. Firstly, every institution that creates its own PLT is duplicating work that has been done by others. Given that there may be special needs unique to a particular institution, many uses and requirements of the PLT are common and a lot of the duplicated efforts could be avoided. Moreover, from the data interoperability perspective, the lack of a common set of problem list terms is an impediment to data sharing.
Our study shows that the average pairwise overlap of existing PLTs is a meager 21%, hardly encouraging for the sharing of problem list data. However, the overlap figure only includes cases in which exactly the same concept is used across institutions. It does not cover cases of different but related concepts (eg, hierarchically related concepts). Content overlap would be higher if these relationships are taken into account. One piece of good news is that actual usage is concentrated heavily on relatively few terms, which makes the problem more tractable because we only have to standardize a relatively small proportion of terms to reap large benefits in data interoperability. Another encouraging finding is that terms that are shared are the heavily used ones. This reduces the effort and increases the yield of standardization.
We think the UMLS-CORE Subset identified in this study can help to alleviate the above problems. On the basis of the UMLS-CORE Subset, we further identified a CORE Problem List Subset of SNOMED CT (the SNOMED Problem List Subset), which has been available for download by UMLS licensees since June 2009.22 The SNOMED CT can be used freely in the USA and any member country of the International Health Terminology Standards Development Organisation (IHTSDO).9 Details about the principles, creation, and characteristics of the SNOMED Problem List Subset will be covered in a separate paper. We recommend the use of the SNOMED Problem List Subset as a ‘starter set’ for PLTs. Based on the scale of the institutions and the scope of the datasets used in the creation of this Subset, we believe that it can support a high proportion of data entry requirements in healthcare institutions providing comprehensive medical services. The Subset will save considerable time and effort compared with starting from scratch. It can enhance data interoperability in two ways. Firstly, by pre-selecting a set of terms, one can avoid the arbitrary (unintentional) variations in the creation of PLTs that are not based on clinical necessity or importance. For example, one could have chosen either of the two terms ‘Infectious colitis, enteritis, and gastroenteritis’ or ‘Infectious gastroenteritis’ (both are actual terms from standard clinical terminologies) to populate a PLT. Semantically, the two terms can be considered different as the former explicitly states ‘colitis’ whereas the latter does not. However, in most clinical situations, the diagnosis of infectious gastroenteritis is made without radiological or endoscopic studies. Whether the colon is involved is usually neither known nor clinically important, because it does not affect the patient's management or prognosis. Limiting the choice to either one term (but not both) will reduce unnecessary variation in problem list data. A further way in which the SNOMED Problem List Subset can enhance data interoperability is that, if existing problem list terms can be mapped to concepts in the Subset, it can become the lingua franca for data exchange.
Among the most frequently used 14 395 terms, 92% could be found in standard terminologies in the UMLS. It is encouraging to see that existing terminologies already cover the majority of the frequently used terms in problem lists. There was a previous study, also by NLM, called the Large Scale Vocabulary Test (LSVT) Study, which evaluated the extent to which existing medical terminologies covered the terms needed for health information systems.23 It is interesting to compare the results of the LSVT with the present study. The LSVT found that only 64% of terms used in problem lists were covered by the medical terminologies in the UMLS. The lower percentage in LSVT could be explained by two reasons. Firstly, the LSVT mapped all submitted terms, whereas the present study only focused on the frequently used ones. As we have shown, more frequently used terms were more likely to be found in the UMLS, and therefore a higher percentage of mappable terms was found in the present study. Secondly, the UMLS used in LSVT (1996 version with some additions) was considerably smaller, containing about 250 000 concepts and 600 000 terms compared with 1.5 million concepts and 6.4 million terms in the 2008AA version used in this study. It is likely that this has contributed to the higher coverage in the present study.
One important finding that is common in both LSVT and the present study is that a significant proportion of terms that did not exist in standard terminologies could be derived from existing terms by the addition of modifiers. In LSVT, two-thirds of the terms that did not have exact matches were narrower in meaning than an existing concept in the controlled terminologies, and most of these terms could be represented with the addition of modifiers to the broader concept. In the present study, more than half of the terms that were not found in the UMLS were highly specific concepts or contained laterality information (table 3). Many of these terms could be represented by adding modifiers to existing concepts. The implication is that post-coordination will be a good way to ‘fill in the gaps’—that is, to provide the concepts missing from a PLT. The advantage of post-coordination (compared with just adding completely new terms de novo) is twofold. Firstly, if everybody follows the same rules to combine concepts to generate new meanings, at least theoretically one can determine computationally whether any two new concepts are equivalent even if they are created independently. Secondly, the new concepts will maintain their links to existing concepts. For example, if the new concept ‘Left kidney stone’ is created by adding the qualifier ‘Left’ to the existing concept ‘Kidney stone’ in the SNOMED Problem List Subset, the system will be able to recognize that the new concept is a subtype of kidney stone, and meaningful aggregation of existing and new concepts can still occur.
Finally, we note that this study is based on a convenience sample of datasets from six large-scale healthcare institutions. We have not performed an exhaustive survey of all healthcare institutions. Based on the size of the patient population and medical specialties covered, we postulate that the results would be generalizable to most large healthcare institutions providing broad-spectrum services. Whether the SNOMED Problem List Subset can be used in other institutions with adequate coverage will be the subject of further study. In future, if we obtain additional good quality datasets, we can employ the same methodology to expand and improve the Subset. The analysis of overlap between PLTs is only based on a subset of frequently used terms that can be mapped to the UMLS, because there is no straightforward way to detect equivalence of terms if they cannot be mapped to a common terminology system. Therefore the degree of overlap across institutions should be interpreted as a lower bound.
Conclusion
The problem list has been widely embraced as an efficient way to organize clinical information in EHRs. Encoded problem list information is required to invoke many of the intelligent functions of an EHR. A high proportion of commonly used terms in PLTs are already found in standard terminologies. There is only modest overlap between existing PLTs from large healthcare institutions. The terms that are shared are used more heavily than terms that are not shared. The lack of a publicly available set of problem list terms results in duplication of effort in PLT development and impaired data interoperability. A SNOMED Problem List Subset of frequently used problem list concepts has been identified. The use of this Subset will save effort in terminology development, reduce variability, and facilitate sharing of problem list data.
Acknowledgments
We thank Christopher Chute and David Mohr of Mayo Clinic, Robert Dolin of Kaiser Permanente, Stanley Huff and Naveen Maram of Intermountain Healthcare, Vicky Fung of Hong Kong Hospital Authority, James Campbell of University of Nebraska Medical Center, and Marc Overhage and Jeff Warvel of Regenstrief Institute for providing their institution's datasets for this study. Thanks also to John Kilbourne for helping to map the local terms to the UMLS, and the anonymous reviewers of JAMIA for their useful feedback and suggestions.
Footnotes
Funding: This work was supported by the Intramural Research Program of the National Institutes of Health and the National Library of Medicine.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Weed LL. The problem oriented record as a basic tool in medical education, patient care and clinical research. Ann Clin Res 1971;3:131–4 [PubMed] [Google Scholar]
- 2.Dick RS, Steen EB, Detmer DE. The computer-based patient record: an essential technology for health care. Revised edn Washington, DC: National Academy Press, 1997 [PubMed] [Google Scholar]
- 3.Key Capabilities of an Electronic Health Record System - Letter Report: Committee on Data Standards for Patient Safety, Board on Health Care Services, Institute of Medicine, 2003. National Academies Press, Washington DC: Available at http://www.iom.edu/Reports.aspx [PubMed] [Google Scholar]
- 4.Hospital accreditation standards: accreditation policies, standards, elements of performance (HAS): Joint Commission on the Accreditation of Healthcare Organizations, 2009
- 5.ASTM Standard E2369-05e1 “Standard Specification for Continuity of Care Record (CCR)”. West Conshohocken, PA: ASTM International, 2005. DOI:10.1520/E2369-05E01 [Google Scholar]
- 6.Dickinson G, Fischetti L, Heard S, eds. HL7 EHR System Functional Model - Draft Standard for Trial Use: Health Level Seven, July 2004. Available at http://www.hl7.org/ [Google Scholar]
- 7.Medicare and Medicaid Programs; Electronic Health Record Incentive Program; Final Rule Centers for Medicare & Medicaid Services, Department of Health and Human Services. Federal Register Vol. 75, No. 144, July 2010. Available at http://www.gpoaccess.gov/fr/ [PubMed]
- 8.Jha AK, DesRoches CM, Campbell EG, et al. Use of electronic health records in U.S. hospitals. N Engl J Med 2009;360:1628–38 [DOI] [PubMed] [Google Scholar]
- 9.International Health Terminology Standards Development Organisation, owner of SNOMED CT. http://www.ihtsdo.org/
- 10.Consolidated Health Informatics (CHI) Initiative; Health Care and Vocabulary Standards for Use in Federal Health Information Technology Systems Department of Health and Human Services. Federal Register Vol. 70, No. 246, Dec 2005. Available at http://www.gpoaccess.gov/fr/
- 11.Giannangelo K, Fenton SH. SNOMED CT survey: an assessment of implementation in EMR/EHR applications. Perspect Health Inf Manag 2008;5:7. [PMC free article] [PubMed] [Google Scholar]
- 12.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med 1993;32:281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care 1994:235–9 [PMC free article] [PubMed] [Google Scholar]
- 14.Hole WT, Srinivasan S. Discovering missed synonymy in a large concept-oriented Metathesaurus. Proc AMIA Symp 2000:354–8 [PMC free article] [PubMed] [Google Scholar]
- 15.UMLS RRF Browser. http://www.nlm.nih.gov/research/umls/implementation_resources/metamorphosys/RRF_Browser.html
- 16.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Stud Health Technol Inform 2001;84:216–20 [PMC free article] [PubMed] [Google Scholar]
- 17.Chute CG, Elkin PL, Fenton SH, et al. A clinical terminology in the post modern era: pragmatic problem list development. Proc AMIA Symp 1998:795–9 [PMC free article] [PubMed] [Google Scholar]
- 18.Elkin PL, Mohr DN, Tuttle MS, et al. Standardized problem list generation, utilizing the Mayo canonical vocabulary embedded within the Unified Medical Language System. Proc AMIA Annu Fall Symp 1997:500–4 [PMC free article] [PubMed] [Google Scholar]
- 19.Brown SH, Miller RA, Camp HN, et al. Empirical derivation of an electronic clinically useful problem statement system. Ann Intern Med 1999;131:117–26 [DOI] [PubMed] [Google Scholar]
- 20.Warren JJ, Collins J, Sorrentino C, et al. Just-in-time coding of the problem list in a clinical environment. Proc AMIA Symp 1998:280–4 [PMC free article] [PubMed] [Google Scholar]
- 21.Wang SJ, Bates DW, Chueh HC, et al. Automated coded ambulatory problem lists: evaluation of a vocabulary and a data entry tool. Int J Med Inform 2003;72:17–28 [DOI] [PubMed] [Google Scholar]
- 22.The CORE Problem List Subset of SNOMED CT. http://www.nlm.nih.gov/research/umls/Snomed/core_subset.html
- 23.Humphreys BL, McCray AT, Cheh ML. Evaluating the coverage of controlled health data terminologies: report on the results of the NLM/AHCPR large scale vocabulary test. J Am Med Inform Assoc 1997;4:484–500 [DOI] [PMC free article] [PubMed] [Google Scholar]