Abstract
Informatics for Integrating Biology and the Bedside (i2b2) is one of seven projects sponsored by the NIH Roadmap National Centers for Biomedical Computing (http://www.ncbcs.org). Its mission is to provide clinical investigators with the tools necessary to integrate medical record and clinical research data in the genomics age, a software suite to construct and integrate the modern clinical research chart. i2b2 software may be used by an enterprise's research community to find sets of interesting patients from electronic patient medical record data, while preserving patient privacy through a query tool interface. Project-specific mini-databases (“data marts”) can be created from these sets to make highly detailed data available on these specific patients to the investigators on the i2b2 platform, as reviewed and restricted by the Institutional Review Board. The current version of this software has been released into the public domain and is available at the URL: http://www.i2b2.org/software.
Introduction
Many challenges exist when it comes to repurposing data from an electronic medical record system (EMRS) for research.1–4 Attempts to take the raw EMRS data and “clean it up” can be fruitless at a global, enterprise level, because one person's cleaning can be the destruction of another's important information. In i2b2, we have built software that may be used in a two-step process. First, data from the entire set of patients in the enterprise is exposed to the research community while preserving patient privacy, such that performing basic queries can yield a smaller set of patients interesting to the investigator. Second, the chosen set of patients is matched to controls from the enterprise database and a project-specific data mart is created. The data mart has the advantage of complying with The Health Insurance Portability and Accountability Act of 1996 (HIPAA) and the Institutional Review Board's (IRB's) requirements to limit the exposure of the enterprise patients, while making highly detailed data available on interesting patients to the investigator. Because the data is now a copy, separate from the enterprise data, the investigators are free to “clean” and manipulate it as much as desired for their own purposes with the i2b2 software. This allows patient medical record data, such as diagnoses, medications, and laboratory values, to be combined with clinical research data, such as from a case report form or data from a genome-wide association study, into a single cohesive unit such that it can be queried in an integrated manner.
Background
The repurposing of medical record data for clinical research holds high promise.1–5 Potentially such data are highly useful for research, representing some of the most important everyday clinical events of patients' lives as recorded by trained observers. If the adoption of EMRSs is to increase as anticipated,6 it is incumbent and opportune to develop methods for providing ways to look at this data across patients. However, this task is much more difficult than would first appear. EMRSs are typically built to look at data on single patients, not data across combinations of many patients. Attempts to overlay this functionality on existing EMRSs demonstrate that the functional and technical requirements of the transactional and analytical systems are in opposition.7
Unlike transaction systems that are optimized to show data regarding single patients, a system that supports queries that cut across multiple patients is more dependent on standard descriptors and annotations, queries can be challenging to specify, and these queries have complex implications for the privacy of the patients. Furthermore, attempting to “fit together” medical record data and clinical trial data at a person-level so that diseases, genes, and outcomes can be related to each other requires the existence of a very robust data model.
The i2b2 data model of the clinical research chart (CRC) is based on the “star schema”, a design proposed by Ralph Kimball.8 The star schema has a central “fact” table where each row represents a single fact. In i2b2, the facts are made up of observations about a patient. Observations about a patient are recorded by a specific observer within a specific time range (defined by start and end dates), regarding a specific concept, such as a lab test or disease, in the context of an encounter, such as a patient outpatient or inpatient visit. The concept can be any coded attribute about the patient, such as a code for diabetes, a medication the patient is on, or a specific test result. This manner of expressing a concept as an attribute in a row rather than designating it in a column is based on prior work known as the entity-attribute-value (EAV) model, first seen in early EMR implementations9 10 and later discussed by Nadkarni and Brandt11 and Johnson et al.12 It is extremely efficient to query data arranged in a star schema represented in an EAV format. This is because the concepts recorded in EAV format allow one very large index to be built which cuts across all patients' data in the fact table.7
The entire i2b2 software architecture is built on web services,13 and new i2b2 cells can be developed and added by teams outside the i2b2 core implementation by connecting them using these services. In this way the architecture is similar to that of the cancer Biomedical Informatics Grid (caBIG).14 However, unlike caBIG, the core data in i2b2 is instantiated according to a single relational model, not a compendium of object models, obviating the need for a data model standards repository like the caDSR. This greatly simplifies the query strategies in i2b2 and is designed to provide much improved performance as relational database technologies are used to interrogate the many billions of fact representations in i2b2 implementations such as at Partners HealthCare.
Design objectives
The design of the clinical research chart (CRC) and accompanying i2b2 software was focused around several goals. First and foremost was the goal to provide a secure presentation of patient information for research purposes. This presentation needed to be made in a software framework that could be easily extended, because much of the value of i2b2 was anticipated to occur through contributions from groups outside the core i2b2 team. Keeping this in mind, communication between modules in i2b2 was designed around web services that provide securable remote access to its various parts through well defined messages.13 Beside security and communication, the data model of the CRC was tuned to the needs of patient-specific information such that it had superior and scalable query performance while still being adaptable to new and unanticipated representations of healthcare information.
The i2b2 software was designed to support at least two general use cases within the mission of repurposing electronic health records and other patient data for research. One was to browse through all the enterprise data to find sets of patients that would be of interest for further research. A second was to use the data provided by the medical record to do a “deep dive” into the phenotype of the set of patients identified (possibly from the first use case) in support of genomic, outcome, and environmental studies.
System description
The i2b2 project allows the data to move back and forth between these two use cases. Data about a set of patients (metadata), is copied from the i2b2 enterprise database and placed into an i2b2 project database with the same data format and with the same data descriptors. Data that was collected independently of the electronic medical record can also be imported into the project database with its own data descriptors. In an i2b2 project database, new data can be derived from the raw EMR data in a multitude of different ways, such as through natural language processing, resulting in new, derived observations being created within the database.15 Often the new observations are more accurate or precise than the original raw observations, and can become the terms used in new queries and further data manipulations. The nature of the data schema and controlled terminologies allows the derived data and metadata tables to potentially (data ownership issues aside) be appended from a project database back into the enterprise tables.
An i2b2 “hive” is a set of server-side software modules (“cells”) that either store data or contain analysis methods that facilitate the repurposing of medical record data for research.13 The i2b2 hive manages both the enterprise-wide data and the distribution and access of data associated with the various projects that may have originated from cuts of the enterprise data. The data is generally loaded into the enterprise system using methods that take raw data from a hospital electronic medical record transaction system and place it into the CRC of the i2b2 hive. The feeding transaction systems may be the hospitals' registration and laboratory systems, provider-based medical record systems, or specially built electronic data capture systems. The implementation of the CRC currently supports Oracle and Microsoft SQL Server databases. These were chosen not only because of their industrial strength scalability, but also because many extract, transform, and load (ETL) tools exist to support these platforms. These tools are used to extract data from the clinical systems and place copies of it into the CRC. The core team has not developed connections to load data from specific EMR implementations, but teams outside i2b2 have been active on this front (M Kamerick, personal communication, 2009).
The infrastructure created by the i2b2 software allows investigators to perform queries on an enterprise data repository. This infrastructure consists of several cells of an i2b2 hive. The project management cell contains a set of data structures that associate users with passwords, IRB approvals, preferences, and (as created) new projects. When a user “logs on” to the i2b2 hive, they are using web services of the project management cell to authenticate themselves. Every time another part of the hive tries to perform an action on behalf of the user, it goes to the project management cell to gather the proper authorizations. Once authenticated, the user performs queries against a second cell of the hive, known as the data repository cell which contains the CRC of the enterprise. Each row of the main fact table in the CRC star schema database is associated with a term in another cell, called the “ontology” cell. This allows every term that is used to describe a patient in the enterprise CRC database to be used in a query.
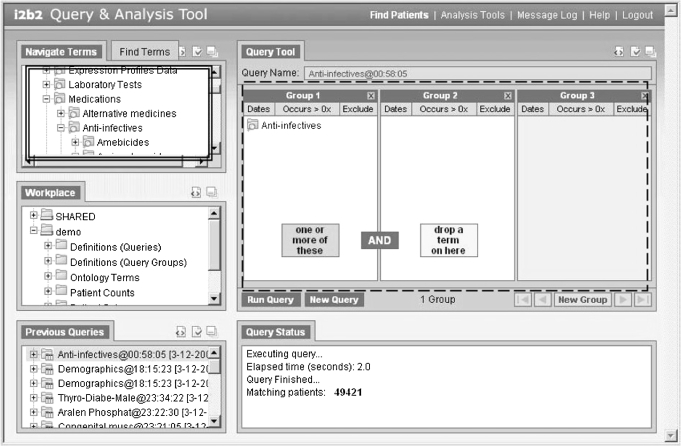
Using the i2b2 web client shown in figure 1 allows ad-hoc queries to be created by research clinicians throughout the enterprise and return aggregate numbers of patients that satisfy the queries. The terms used to create the queries are those in the data repository that are associated with patient observations. As shown in figure 1, metadata items are dragged from the “Navigate Terms” and “Find Terms” panels on the left (shown in the double lined box) and a query is created in the Venn-diagram-like panels (in the broken line box) on the right. If items are dragged into the same panel, they are logically OR'd together, and if they are dragged into different panels they are logically AND'd together. A similar interface has been described previously in detail.16 Other types of query tools are being developed by various groups to plug in to the i2b2 data repository cell. Of particular interest is a temporal alignment query tool that directs temporal queries of the clinical data.17
Figure 1.
The i2b2 web client is used as the interface for the enterprise users to construct queries. Patient attributes are dragged from the “Terms” panels into the “Query Tool” panels, and the patient sets that result after running the query which can be accessed and reused from the “Previous Queries” panels.
Using the i2b2 web client, patient sets may be created and aggregate counts on the demographics of these patients can be obtained. However, it only shows anonymous patient data, obfuscating the true results by adding or subtracting a small random number to the aggregate totals using a previously published method.18 This allows aggregate counts of the patient data to be displayed without the risk of disclosing the identities of the patients. The lists of patients that make up the patient sets are saved in the background, and line-item patient detail at the limited data set (LDS) level is available once data is extracted from the enterprise repository and placed into a specific project's data mart.
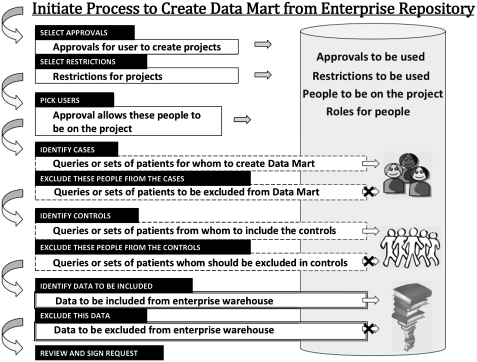
The process of creating a project's data mart is shown in figure 2. The investigator begins by selecting an approval (such as a recorded IRB approval) that allows them to create a project and include various kinds of patient detail. Restrictions may also be identified, such as restricting data to that of a single hospital. People who should be granted access to the data mart are then selected. Queries that were created in the broken line box of the i2b2 web client were saved as “previous queries”, and are now used to designate the patients for the new project (the “cases”). Previous queries can also designate patients to exclude from the project. Generally, a set of “cases” will be desired to have a matched set of “controls” in the data mart. A pool of patients that could be selected as matched controls can be selected from previous queries as well. Patient are usually matched by age, gender, race/ethnicity, and the number of observations in the medical record, or as closely as possible. Matching by the number of observations (facts) gives a rough approximation to hospital activity. Attempting to obtain an assessment of the match, we compare ranking of concepts in the two groups. The top ten diagnoses (not related to original selection), consistently give Spearman rank-order correlations of the two sets greater than 0.9.
Figure 2.
Sequential process for creating a project's data mart. The patient sets that were created in the query tool can be used in many places where the request is formulated, seen in the broken line boxes. The patient attribute terms from the i2b2 web client are used in the double line boxes to define the data that will be in the data mart.
Once the patients have been identified, the data to be included (and perhaps some to be specifically excluded) in the project data mart is selected from the available terms in the double lined box of the i2b2 web client. Proper authorizations to create the project are checked, and data can now be copied from the Enterprise data repository into the project data mart. This creates a persistent set of data for the project.
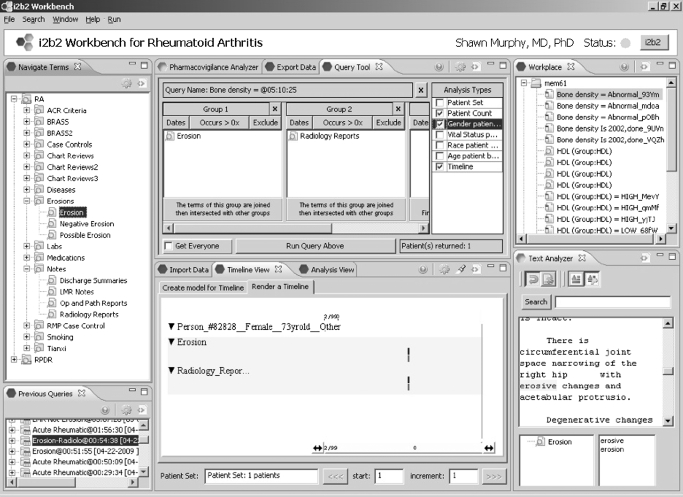
The new project data mart represents the next step in the data curation process. i2b2 software supports the data curation workflow by allowing the creation of new “observation-facts” in the fact table of the data mart, such as concepts identified through natural language processing (NLP). This allows a project to build up a catalog of new, more refined observations which represent progressive improvements in the medical record data. Illustrated in figure 3 is the i2b2 Java “Workbench”, which is based on the open-source Eclipse platform.19 This platform was chosen due to its popularity among developers and its well defined extensibility through plug-ins that are based on an OSGi-standard compliant implementation (http://www.osgi.org/). New terms generated from derived data (such as that resulting from chart reviews) are made available in the “Navigate Terms” window. This client extends the functionality available through the i2b2 web client, especially because it is known that the investigator has explicit IRB approval to view line-item patient detail. Even so, public health information (PHI) that is part of the 16 identifiers prohibited by limited data sets is either maintained in an unencrypted form in a separate repository, or encrypted in the data mart. To view this data, a decryption key is issued to those members of the project that are authorized to view PHI. Data export and import, multiple data visualizations, and mechanisms to validate newly derived NLP data are featured in the i2b2 Workbench. Various analysis types are available, and shown is a timeline view which plots the observations as bars in categories and according to times when they were observed. Clicking on the elements of the timeline can result in further detail, such as the Text Analyzer panel that shows how the “Erosion” term was derived form the Radiology Report through NLP.
Figure 3.
The i2b2 Eclipse Workbench is generally used as the interface for working with specific project databases. It offers views for exporting and importing data to the project database, as well as views focused on specialized analysis. The i2b2 software architecture is built with multiple “plug-in” points. The i2b2 web client, the Java i2b2 Workbench, and the (server-side) data repository cell all have open architectures for additional software plug-in functionality.
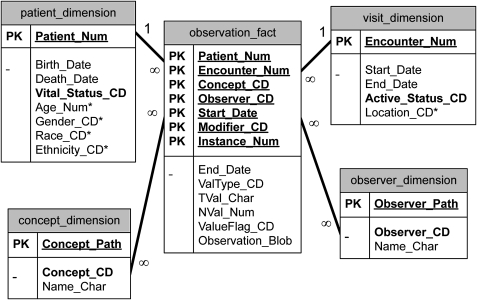
All of the operations that have been described rely on the highly flexible i2b2 patient data model, which at its core has a very simple design as seen in figure 4. The central fact table contains rows of observations about patients, with key attributes of patient ids, event ids, concept codes (possibly with associated modifier codes), observer codes, and dates associated with the observations. The fact table also contains value objects associated with the observations. The value objects are defined so they may be queried rapidly using relational database technology. This means they should obey at least two constraints. First, they should be expressed in the basic data types available in the database, such as numbers and simple strings. A complex value, such as blood pressure, for example “120/80-standing”, is reduced to three rows with concept “blood pressure” and modifiers “systolic”, “diastolic”, and “position” (preferably expressed as LOINC or other known standard, but this is not required). Second, the value type is specified in the ValType column, such as N=number, T=text, D=date, and so forth. This is a way of specifying the format for a value object in relational database technology. Numbers and dates are placed in the NVal_Num column which is a numeric data type. However, the TVal_Char column, a character data type, is also used in the specification of numeric values, and contains a representation of the operator, such as “equals”, “greater than”, or “less than”. ValueFlag is used to hold the source systems assessment of high/low/normal or normal/abnormal, if available.
Figure 4.
All patient data is held in the Star Schema of the data repository. The tables are represented by each box, and the columns are represented by the rows of the box. Columns that are underlined are part of the primary key of the table. Columns that are in bold must always be filled. Columns with an asterisk next to them enhance the usability of the data repository but are not required. Further information on the data model and its use is available from the i2b2 website.
Besides the fact table, called the Observation_Fact table in the i2b2 data repository, there are four other tables that help express the patient data. The Patient_Dimension table has one row for every patient in the database. This table contains the patient's Birth_Date, Death_Date, and a column Vital_Status_CD that describes if these dates are known, and if so, how accurately these dates are known. Date values are accompanied by an accuracy code to increase the efficiency of performing computations in relational database languages. Other columns in this table are specific to various i2b2 implementations and are not required. They may represent concepts that exist in the fact table, but for operational reasons may need be consolidated into one value per patient, such as current zip code. Business rules are applied to perform this consolidation.
The Visit_Dimension table allows periods to be represented that correspond roughly to patient encounters where observations were recorded. An “encounter” can involve a patient directly, such as a visit to a doctor's office, or it can involve the patient indirectly, such as running several tests tied together by the same tube of the patient's blood.
The Concept_Dimension table has vocabulary terms that map to the original codes used to specify observations made on the patient. The observation_fact table contains the original codes from the source systems with a three character prefix to avoid collisions of codes from various systems. Various standard concepts preloaded into i2b2 example terminologies include International Classification of Diseases (ICD), National Drug Code (NDC), and Logical Observation Identifiers Names and Codes (LOINC). These example terminologies were chosen due to their frequency of use in many source systems and practical accessibility. However, the i2b2 terminology systems do not recognize a difference between standard and local terminologies. Terms may be grouped into hierarchies. The hierarchical representation used in the concept table is similar to that of a hierarchical file system. The parent term is positioned in the “folder” position of the path, and the child term in the “file” position. For example, in the concept_dimension table (table 1), the parent “anti-infectives” can have the three children “penicillin”, “ampicillin”, and “Bactrim”. The children map to the NDC codes used in the fact table, but the path shows they are types of anti-infectives, and most importantly, the path allows group queries to be performed using Structured Query Language (SQL) in the data repository.
Table 1.
Layout of concept data representations in the concept dimension
| Concept_path | Concept_CD |
| Med-V2\ | |
| Med-V2\anti-infectives\ | |
| Med-V2\anti-infectives\penicillin\ | NDC:00002032902 |
| Med-V2\anti-infectives\ampicillin\ | NDC:60429002340 |
| Med-V2\anti-infectives\Bactrim | NDC:00003013850 |
The organization of concept strings described above allows the choice of hierarchies to dictate groups of concepts used in a query. This allows a simple SQL statement to be created that is easily optimized (and therefore very fast) in relational database systems. The paths allow us to use the general concept of anti-infectives in a query. The query below shows the use of the concept_path column to give all the codes for penicillin, ampicillin, and Bactrim. Therefore, if we wanted to get all patients seen on anti-infectives, we would run the following query:
Select distinct(patient_num)
From observation_fact
-
Where concept_cd in
(select concept_cd
from concept_dimension
where concept_path like
‘Med-V2\anti-infectives\%’)
The path of the concept is used to find and use all concept_cds that fall into the anti-infectives group. If we only wanted to find patients specifically on Bactrim, we would use the same query with the following concept_path:‘Med-V2\anti-infectives\Bactrim\%’
These patterns in SQL queries make it easy to implement automated query builders. Similar functionality using linked lists would require a much more complex implementation. These same patterns are used to create queries from the observer_dimension table. Rows in this table usually represent a clinician, but occasionally may point to a mechanical device, such as a continuous blood pressure monitor.
The database design described both allows new blocks of data to be added without disturbing the integrity of the old data, as well as allowing blocks of data to be copied out of a larger original database to a smaller one. The data about a set of patients can be copied from the i2b2 enterprise database and placed into an i2b2 project database with the same data format and with the same data descriptors while preserving powerful methods for querying the data.
The representations of the coding terminologies from two different systems can be combined, or kept separate depending on the degree of integration (and thus usability) that is required by the site. i2b2 does not provide an automated method to perform this function. If terminology systems are kept apart, the user will need to go to each terminology to perhaps choose very similar concepts to use within a query.
Status report
We have utilized the i2b2 tools to support projects across the Partners HealthCare system, and approximately 17 sites outside of Partners HealthCare are engaged in setting up their own i2b2 software systems to support enterprise discovery based on medical record data. A network of i2b2 sites is being created, such that multiple i2b2 sites can be queried at once, greatly increasing the power of the results.20
At Partners HealthCare, an enterprise level i2b2 data repository was created. This data repository contains data from inpatient clinical systems, billing systems, laboratory systems, and outpatient electronic medical record systems at Partners HealthCare. Data exists in the enterprise i2b2 data repository on 4.6 million patients. It consists of a total of 1.2 billion observations on these patients in the observation_fact table, including 440 million ICD9, DRG, COSTAR, and locally coded diagnoses, 70 million NDC, COSTAR and locally coded medications, 110 million CPT4, ICD9, COSTAR, and locally coded procedures, 540 million LOINC and locally coded laboratory test results, 50 million locally coded outcome variables, and 280 thousand genomic test results, all coupled to demographic, visit, and provider information.
Most queries constructed using the Query Tool and only requesting patient counts would complete within 10 s, many within several milliseconds. A data mart consisting of 550 million facts on 2.6 million patients completed building in a little over 1 h and 15 min on an 8×3 GHz processor machine with 32 GB RAM connected to a storage area network (SAN).
Four “driving biology projects” (DBPs) were established at Partners HealthCare in the context of the greater i2b2 federally funded grant, to provide use cases for the development of software and methods as are being described. These DBPs included asthma, major depressive disorder (MDD), obesity, and rheumatoid arthritis (RA). Each of these projects had slightly different goals, but generally was focused on extracting phenotypes from the medical record that could be used for genome-wide association studies. For example, the asthma project was looking for genotypes matched to people unresponsive to conventional asthma medication treatments, possibly because these patients have a different genotype than conventional responders. These could help subdivide asthma into several different recognized diseases and focus different treatments on each of the asthma variants. Other aspects of asthma have been studied within the i2b2 project framework using similar approaches.21 The MDD project was looking for genotypes matched to people unresponsive to serotonin reuptake blockers (eg, Prozac) for similar reasons.
When the RA project wished to use a definition of RA that required a specialized test result often not performed at Partners HealthCare, test results were extracted from narrative notes that recorded that result and placed it as a row in the fact table using previously described methods.15 The combination of more than three recorded observations of rheumatoid arthritis, visits one or more times to an arthritis clinic, and a highly positive anti-cyclic citrullinated peptide antibody test was then used as the proxy for the diagnosis of RA after it was determined to have a very high specificity when it was spot checked against 450 full reviews of patient medical records.
Discussion
Providing a data schema in i2b2 that allows very diverse patient data from the enterprise to be integrated into a few central tables allows developers to make i2b2 query tools that search and analyze the data that the enterprise makes available on its patients. Data can be provided from the enterprise data repository for individual projects. i2b2 tools can then be used to work with the project-level data that allow significant value to be added to the original data, as well as add data collected and discovered on the patients from systems outside the repository scope (such as clinical trials).
Two considerations limit the use of detailed patient data at the enterprise level. One consideration is patient privacy. When the entire patient population is exposed to the enterprise, many precautions must be taken so that single patients cannot be identified, and therefore the enterprise i2b2 web client can be used in the context of strictly anonymous patient data. A second consideration is that deep analysis of the data usually involves transforming data from the rough forms available in the medical record to clean and refined forms that are more appropriate for scientific study. Therefore, once a set of patients is identified in the enterprise i2b2 web client for further study, these patients and matching controls are extracted from the enterprise data repository into project-level data repositories. Project data repositories are created under specific IRB protocols that allow the exposure of PHI to the investigator. Therefore very detailed analysis is possible. Often the new, derived observations are more accurate or precise than the original raw observations, and can become the terms used in new queries and further data manipulations. This overall strategy can also be used to spawn sub-projects and personal data sets such that the risk of deleting or changing project data is minimized.
The i2b2 data architecture, where the structures of terminology and data representation are preserved in their entirety both at the enterprise and the project level, will allow the information in both repositories to flow seamlessly between the two. Only new data, not new data structures, need be copied between instances. This will be valuable both to update project databases with fresh enterprise data, as well as to move project data back into the enterprise repository. Although these capabilities are not implemented in the current i2b2 software, and many policies would need to be considered in their implementation, this is a very powerful aspect of the i2b2 data schemas. Overall, this allows a workflow where teams can build on each others' refinements and manipulations to the data and ultimately upon each other's ideas.
The reuse of data from the medical record for research has become increasingly strategic in performing cost-effective clinical studies.1–5 22 The i2b2 team has set up a Shared Research Informatics Network (SHRINE) which will be able to distribute i2b2 queries, increasing the ways to leverage this data for research with other Harvard hospitals, including the Beth Israel Deaconess Medical Center, the Dana Farber Cancer Institute, and Children's Hospital Boston.20 Setting up i2b2 instances at these institutions was accomplished as outlined in Weber et al. Among the issues encountered, placing the data into the i2b2 CRC was the most challenging, as local knowledge and resources were inevitably needed for the necessary data transformations. A similar network has been set up outside the core i2b2 team named the “i2b2 CICTR project”, a collaboration between the University of California at San Francisco (UCSF), University of California at Davis (UCD), University of Washington, Seattle (UW), Ohio State University, Rochester University, University of Pennsylvania, Mayo Clinic, and Washington University. The i2b2 CICTR project not only allows very large clinical studies to be performed across the i2b2 sites, but also works toward placing i2b2 Hives on the caGRID network (N Anderson, personal communication, 2009). Thus the expanding network of i2b2 sites not only allows i2b2 to serve the enterprise and individual investigator initiated projects, but also to tie together national initiatives and allow the projects to go beyond local boundaries.
Acknowledgments
The contributions of Rajesh Kuttan, Lori Phillips, Wensong Pan, and Janice Donahue have been invaluable for the construction and distribution of this software, and by John P. Glaser and Diane Keogh for contributions and support.
Footnotes
Funding: This work was funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54LM008748. Other funders: NIH.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Prokosch HU, Ganslandt T. Perspectives for medical informatics. Reusing the electronic medical record for clinical research. Methods Inf Med 2009;48:38–44 [PubMed] [Google Scholar]
- 2.Burgun A, Bodenreider O. Accessing and integrating data and knowledge for biomedical research. Yearb Med Inform 2008:91–101 [PMC free article] [PubMed] [Google Scholar]
- 3.Ohmann C, Kuchinke W. Future developments of medical informatics from the viewpoint of networked clinical research. Interoperability and integration. Methods Inf Med 2009;48:45–54 [PubMed] [Google Scholar]
- 4.Haux R, Knaup P, Leiner F. On educating about medical data management - the other side of the electronic health record. Methods Inf Med 2007;46:74–9 [PubMed] [Google Scholar]
- 5.Tannen RL, Weiner MG, Xie D. Use of primary care electronic medical record database in drug efficacy research on cardiovascular outcomes: comparison of database and randomized controlled trial findings. BMJ 2009;338:b81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.America, Congress of USA. Title VIII–Health information technology 2009:241–277 [Google Scholar]
- 7.Murphy SN. Data warehousing for clinical research. In: Liu L, Tamer OM, eds. Encyclopedia of database systems. Springer, 2009, ISBN: 978-0-387-49616-0 [Google Scholar]
- 8.Kimball R. The data warehousing toolkit. New York: John Wiley, 1997 [Google Scholar]
- 9.Stead WW, Hammond WE, Straube MJ. A chartless record–is it adequate? J Med Syst 1983;7:103–9 [DOI] [PubMed] [Google Scholar]
- 10.Friedman C, Hripcsak G, Johnson SB, et al. A generalized relational schema for an integrated clinical patient database. In: Miller RA, ed. Proceedings of the fourteenth annual symposium on computer applications in medical care. Los Alamitos, CA: IEEE Computer Society Press, 1990:335–9 [Google Scholar]
- 11.Nadkarni PM, Brandt C. Data extraction and Ad Hoc query of an entity-attribute-value database. J Am Med Inform Assoc 1998;5:511–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson SB, Paut T, Khenima A. Generic database design for patient management information. Proc AMIA Annu Fall Symp 1997:22–6 [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy SN, Mendis M, Hackett K, et al. Architecture of the open-source clinical research chart from informatics for integrating biology and the bedside. AMIA Annu Symp Proc 2007;548–52 [PMC free article] [PubMed] [Google Scholar]
- 14.Saltz J, Oster S, Hastings S, et al. CaGrid: design and implementation of the core architecture of the cancer biomedial informatics grid. Bioinformatics 2006;22:1910–6 [DOI] [PubMed] [Google Scholar]
- 15.Zeng QT, Goryachev S, Weiss S, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Medical Informatics and Decision Making 2006:6–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy SN, Gainer VS, Chueh H. A visual interface designed for novice users to find research patient cohorts in a large biomedical database. AMIA Annu Symp Proc 2003:489–93 [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Plaisant C, Quinn A, et al. Aligning temporal data by sentinel events: discovering patterns in electronic health records. ACM Computer-Human Interaction 2008:457–66 [Google Scholar]
- 18.Murphy SN, Chueh H. A security architecture for query tools used to access large biomedical databases. Proc AMIA Symp 2002:552–6 [PMC free article] [PubMed] [Google Scholar]
- 19.Eclipse Platform Technical Overview The eclipse foundation (February 2003), http://www.eclipse.org/whitepapers/eclipse-overview.pdf
- 20.Weber GM, Murphy SN, McMurry AJ, et al. The shared health research information network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc 2009;16:624–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Himes BE, Dai Y, Kohane IS, et al. Prediction of chronic obstructive pulmonary disease (COPD) in asthma patients using electronic medical records. J Am Med Inform Assoc 2009;16:371–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy S, Churchill S, Bry L, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Research 2009;19:1675–81 [DOI] [PMC free article] [PubMed] [Google Scholar]