Abstract
Mayo Clinic's Enterprise Data Trust is a collection of data from patient care, education, research, and administrative transactional systems, organized to support information retrieval, business intelligence, and high-level decision making. Structurally it is a top-down, subject-oriented, integrated, time-variant, and non-volatile collection of data in support of Mayo Clinic's analytic and decision-making processes. It is an interconnected piece of Mayo Clinic's Enterprise Information Management initiative, which also includes Data Governance, Enterprise Data Modeling, the Enterprise Vocabulary System, and Metadata Management. These resources enable unprecedented organization of enterprise information about patient, genomic, and research data. While facile access for cohort definition or aggregate retrieval is supported, a high level of security, retrieval audit, and user authentication ensures privacy, confidentiality, and respect for the trust imparted by our patients for the respectful use of information about their conditions.
Introduction
Most academic medical centers confront the challenge of collecting, organizing, and retrieving vast quantities of heterogeneous data for research, quality improvement, outcomes analyses, or best practice discovery. Typically, such information resides within scores if not hundreds of disparate databases, registries, data collections, and departmental systems. With luck, a large fraction of clinical data will be aggregated into an electronic medical record, although such transactional systems make for poor cross-patient aggregation and retrieval environments.
Data warehouses have emerged in many industries during the past 15 years as a standard method for managing enterprise data.1 However, their adoption by healthcare organizations has been less pervasive, no doubt due to the complexity and heterogeneity of biomedical, operational, and clinical data.
Nevertheless, substantial progress has been made at organizations such as Intermountain Healthcare,2 who are building on their 30-year legacy of clinical decision support systems. Indeed, current development efforts at Intermountain include sophisticated modeling of data elements, composite concepts, and ‘detailed clinical models.’3 4
Similarly, the long tradition of COSTAR5 at the Massachusetts General Hospital has spawned a new generation of enterprise data warehousing optimized for translational research across the Harvard clinical communities.6 7
Most recently, a large multi-site effort has begun to integrate genomic data with high-throughput patient enrollment in an anonymized ‘non-human subject’ data model at Vanderbilt.8 This database, together with Marshfield Clinic, Northwestern, Group Health of Puget Sound, and Mayo Clinic, are engaged in an ongoing project to algorithmically define clinical phenotype from electronic records and clinical warehouses as part of the eMERGE9 (electronic Medical Records and Genomics) consortium, funded by NHGRI.
Mayo patient record history
The legacy of the Mayo patient record as an organized resource to support research and quality improvement dates back well over a century.10 Working with structured paper documents that organized where information should be put, such as laboratory results or physical examination findings, Mayo has supported the notion of explicitly ‘missing information’ since 1907. Augmenting this have been comprehensive indices of patient diagnoses or surgeries, initially on manually curated 5×7 index cards, incrementally invoking machine automating from IBM tabulating card technology through fully electronic databases by the 1970s. These data collections have proven to be an invaluable resource for disease natural history, outcomes analyses, and evidence discovery.11
Early efforts to integrate information organization into a semantically well-formed structure began by the early 1990s,12 coupled with a coordinated infrastructure for registry creation and information retrieval.13 14 This was enhanced by sustained efforts in the domain of clinical vocabulary and ontology,15 particularly as it pertained to patient information retrieval.16
Inevitably, these efforts evolved toward data warehousing, as Mayo along with other healthcare organizations began to adopt data warehousing principles. Our first effort was a joint development with IBM to create a comprehensive, single site clinical data repository, derived from electronic medical record and billing systems resources.17 This proved successful in many respects, not the least of which was the formal integration of NLP18 (natural language processing) based on the open-source UIMA19 (unstructured information management architecture) platform from IBM. The core NLP pipeline has been released into open source as part of the OHNLP (Open Health Natural Language Process) Consortium.20
However, a systematic normalization of this data, including consistent information models, shared vocabulary, and systematic ETL (extraction, transform, and load) was recognized as a critical need for a semantically integrated data store for all enterprise data. Whereas our initial, more targeted benefits were fairly quickly realized utilizing copies of transactional data models and a novel query tool, the architecture and infrastructure were neither flexible nor scalable enough for handling larger and more complex enterprise-wide queries. Data modeling was handled in a cursory fashion and metadata were not systemically captured. The focus was on querying existing data, the results of which reinforced the need for, and challenges associated with, Data Governance. Hence, Mayo commenced work on a data repository built on industry standard data warehousing principles starting in 2005, called the Enterprise Data Trust.
Semantically integrated warehousing
The Mayo Enterprise Data Trust (EDT) is a collection of data from internal and external transactional systems and data repositories, optimized for business intelligence and ad hoc data delivery. The EDT is the source of truth for data that are aggregated into it. Data for the EDT are integrated from all facets of the Mayo enterprise: practice, research, education, and administration across all Mayo sites. It is built using data warehousing practices that evolved from the banking and manufacturing communities, which have become industry-standard methods within the data warehousing community. Consistent with those practices, new sources of data and analytical capabilities are added as business needs and priorities dictate.
In the course of developing and building the EDT, we asserted principles about the data that define our approach to modeling, data transformation, and maintenance. These principles are that the EDT contents will be:
Subject oriented: data that give information about a particular subject area instead of about a company's ongoing operations
Integrated: data that are gathered into the data warehouse from a variety of sources and merged into a coherent whole
Time-variant: all data in the data warehouse are identified with a particular time period; this permits exact re-execution of a query made at a point in time, regardless the amount of subsequent data that have been added.
Non-volatile: data are stable in a data warehouse; more data are added but data are never removed.
The EDT is one component of Mayo's Enterprise Information Management (EIM) initiative that includes Enterprise Data Governance, Enterprise Data Modeling, Enterprise Vocabulary, and Enterprise Managed Metadata Management. Each component of Mayo's EIM initiative progresses as a whole to deliver trustworthy data necessary for the enterprise to engage in reporting and analytical activities. Examples of analytical activities supported by EIM include research, quality management, practice improvement, outcomes improvement, cost reduction, and individualized evidence-based medicine.
Enterprise Data Governance
Mayo's Enterprise Data Governance (EDG) oversees all of Mayo's data as an enterprise asset. EDG establishes and enforces policies, principles, and standards to optimize Mayo's enterprise data assets through a Data Governance Committee, comprising 15 members from across Mayo's three-campus enterprise. Members include the Executive Dean for Clinical Practice, Executive Dean for Education, CIO, CMIO, Chief Planning Officer, and others. One of us (CGC) serves as Vice-Chair for Data Standards. Operational activities are carried out through a data stewardship program, which is comprised of approximately 10 individuals responsible solely for activities that improve the modeling, standardization, and quality of Mayo's enterprise data and metadata. EDG is responsible for the definition of vocabularies and reference information used in Mayo's data. Industry-standard vocabularies and reference information are linked whenever possible, enhancing the ability to collaborate with other organizations. EDG is closely supported by Mayo IT, which provides and manages the infrastructure and applications supporting both vocabulary and metadata activities. The importance of EDG cannot be overstated as it is charged with fundamentally improving the standardization and quality of data captured by the source systems, which all downstream systems must rely upon for subsequent processing and interpretation.
Enterprise Data Modeling
Mayo's Enterprise Data Modeling (EDM) provides a context for Mayo enterprise activities. It provides a tiered model, based on current and future state, for the organization of Mayo's data assets into:
Subjects—the highest level areas that define the activities of the enterprise (eg, Individual)
Concepts—the collections of data that are contained in one or more subject areas (eg, Patient, Provider, Employee, Referrer, Volunteer, etc)
Business Information Models—the organization of the data that support the processes and workflows of the enterprise's defined Concepts.
EDM provides a consistent roadmap for the collection of data by Mayo's IT systems, enhancing the integration and availability of data for the entire range of enterprise activities.
Where practical, EDM embraces external data standards relevant to the topics at hand, for example HL721 Reference Information Model (RIM) artifacts. It is by design, however, necessarily reflective of Mayo and its integrated practice, education, and research environment. In the research domain, we have diligently worked to be compliant with caBIG22 and BRIDG23 24 models; indeed the first author (CGC) chairs the BRIDG board of directors which coordinates a consensus clinical trials data model across HL7, NCI, FDA, and CDISC. Similarly, our partnership with Intermountain Healthcare has forged similarity between elements of Mayo's EDM process and Intermountain Healthcare's detailed clinical models.
Enterprise Vocabulary System
A symbiotic requirement with modeling for the EDT is a common vocabulary infrastructure to ensure the comparability and consistency of Mayo's data at the data element level. Mayo has worked diligently in the terminology services arena at HL7 and caBIG, creating the open-source family of terminology management resources built around LexGrid.25 Mayo's Enterprise Vocabulary, in partnership with Intermountain Healthcare and GE Healthcare, leverages the LexGrid platform to create a comprehensive terminology management solution which includes:
Coding systems, vocabularies, and ontologies collected and compiled into a common terminology model
Backbone thesauri within the Mayo environment that normalize external and internal coding systems into domain-specific master lists
Value sets which address the use-case and data-element specific needs for ‘small’ vocabulary lists
Mapping resources which track the provenance of thesauri to coding systems, value sets to thesauri, and coding system to coding system equivalencies
Terminology services APIs based on the HL7 Common Terminology Services and the caBIG LexEVS specifications.
Enterprise Managed Metadata Environment
Mayo's Enterprise Managed Metadata Environment (EMME) provides Mayo with context for the systems, applications, and data that comprise the enterprise data asset. These information resources are the means for obtaining knowledge from Mayo's ever-growing data pool. EMME is designed to help facilitate technical and business end-users' understanding of the data, allowing for more accurate and rapid retrieval of the data necessary to make informed, insightful decisions in a well-timed manner. The primary software environment for EMME is the ASG-Rochade system.
Enterprise Data Trust
Each of the preceding initiatives are strategic and necessary components to bring value to Mayo's Enterprise Data Trust. Without them, the EDT would be a collection of diverse and unintegrated data which would rely exclusively on the user to determine relationships and meaning of the data in order to provide analytical value.
Given the framework of EIM, the EDT integrates data from across the Mayo enterprise into a time variant Atomic Data Store (ADS). The time variant nature of the ADS allows for the historical collection and lineage of data and metadata, giving the ability to create datasets for a given point in time. The ADS is the core of the EDT environment and contains integrated data from source systems across the Mayo enterprise in a third-normal form database structure. An ad hoc query environment is created from the aggregation of dimensionally modeled concepts, in turn facilitating end-user ‘views’ of the data they require. Data integration (extraction, transformation, and loading) for the EDT environment is done within the framework of IBM's InfoSphere Information Server.
Data security within the EDT environment provides enterprise-level authentication, row-based authorization based on policy and roles, and complete auditing of all data access. Data security is consistent across the EDT environment through Teleran, a proxy layer that monitors and logs all database activity and enforces role-based security.
Data delivery and analysis is tool independent and multiple tools can be supported based on specific user needs. The focus of typical end-user analysis and reporting has been SAS and SAP's BusinessObjects.
The EDT may be usefully contrasted with the enterprise EHR (Electronic Health Records) at Mayo. Simplistically, the EHR is a transactional system invoked during real-time care of one patient to document clinical findings, events, interventions, and document reports. The EDT, in contrast, is a non-transactional system which is intended to aggregate information about many patients to inform outcomes research, best evidence generation, clinical quality improvement, and genomic association analyses among other analytical activities.
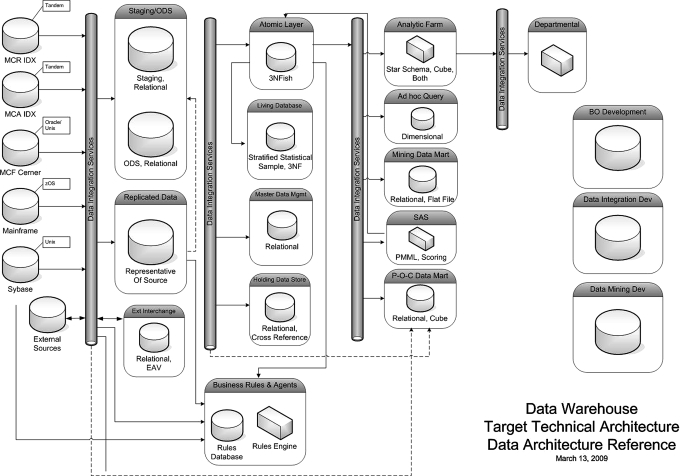
Foundational technologies used in the EDT environment include IBM p-series servers, AIX, IBM DB/2 UDB (Data Warehouse Edition), IBM InfoSphere Information Server, Teleran iSight & iGuard, SAP BusinessObjects, Sybase PowerDesigner, and ASG-Rochade. Figure 1 shows the high level architecture of the overall data warehouse. Mayo is incrementally instantiating each component of the architecture as the needs of the projects dictate.
Figure 1.
Data integration proceeds from left to right. Leftmost are the primary data sources, including the EMR environments for each major campus (not shown are the multitude of departmental data system feeds such as laboratories). Moving right, the data are integrated into staging and replication services, with further refinement (and rightward movement) into normalized versions of the information (‘Atomic Layer’, ‘Living Database’) which are dependent upon Master Data (standards). The right-most full column are the various presentation of data derivatives (subsets) to users, applications, and systems. The free-standing objects on the extreme right are support and development technical environments that support the maintenance and refinement of the overarching Enterprise Data Trust. Dotted lines indicate ‘short cuts’ in the data curation process, where some information is transformed directly to project-specific data marts. BO, business objects; EAV, entity, attribute, value; ODS, operational data store.
Incremental assembly
The resources required to analyze, design, extract, transform, and load data resources into a warehouse with the semantic rigor of the EDT are large. It is not feasible to complete this in totality for all data sources across the enterprise at once. Thus, a strategic sequence of data loads was needed.
Mayo addressed the sequencing problem by prioritizing projects and allowing them in turn to dictate the modeling and data integration sequencing. The first project was, reasonably enough, the core load of critical elements from the Patient Concept: patient identifiers, demographics, and basic institutional data. Two subsequent projects (cancer-center patient profiles and referral pattern analyses) re-used this core concept, and added elements from additional Enterprise Data Model Concepts. Specifically these included: Appointment, Biological Material, Diagnosis, Disease, Location, Medical Specialty, Order, Organization, Patient, Pharmaceuticals and Biologics, Referral, and Referrer.
Using this incremental process, each subsequent project finds itself with more data already ‘there’, and therefore needs to add incrementally smaller amounts of information to achieve that project's goal. Bootstrapping over many projects, the EDT asymptotically is completed, although at an affordable pace and demonstrating immediate benefits and return on investment for the projects that sponsored the incorporation of this data. To provide some order of magnitude for our as-yet incomplete EDT population, table 1 outlines the approximate content by information category.
Table 1.
Approximate counts of key data elements contained in the Enterprise Data Trust
| Information category | Count |
| Unique patients | 7015420 |
| Diagnoses statements (since 1995) | 64 million |
| Laboratory (since 1995) | |
| Unique laboratory tests | 6557 |
| Total test results | 268 million |
| Clinical documents (clinical notes and path reports) | 60 million |
A high-profile project currently underway is a special case, since its goal is to migrate the NLP resources from the first generation repository to a more robust warehouse design in the EDT. These NLP resources will enrich the diagnostic data and related detailed information such as drug utilization, as documented in clinical and pathology notes.
Discussion
Clinical and biomedical research information is, by its nature, complex. Bringing order, normalization, and semantic consistency to information on the scale of an enterprise such as Mayo Clinic, which has 50 000 employees, is a formidable task; it requires a thoughtful plan, significant investment, and strong executive sponsorship. For all academic medical centers to undertake redundant work would be hugely inefficient. As we are in the midst of a fundamental investment by government and the stimulus package on health data standardization, enhanced interoperability, and patient-centered care models, it seems self-evident that cooperation around an intellectual commons for developing shared, standards-based infrastructure for clinical and research data organization must emerge. Mayo is committed to sharing common vocabulary and terminology infrastructures, and is finalizing internal evaluations that would support the open-specification of data models and related infrastructure. Our work on the EDT modeling is not finished, and in some measure the dynamic modeling of a vibrant enterprise will never be completed. However, it seems likely that at least the high-level organization of healthcare enterprise data would benefit from a shared model to support interoperability.
What differentiates the Mayo EDT from similar clinical data repositories at peer academic health centers is our focus on Data Governance. This focus includes substantial resource investment in consensus information models (informed by prevailing health information technology standards) and prescribed terminology ‘value sets’ for applications and messages throughout the enterprise. The ADS is normalized into semantically consistent data elements that can sustain comparable and consistent query for information drawn from across the enterprise. The extensions of Data Governance that operationalize these data normalizations are primarily the Enterprise Data Modeling and Enterprise Terminology Services activities. Thus, the single most important lesson applicable to organizations that seek to emulate Mayo's success with the EDT is to begin with a well-established Data Governance organization and process that authoritatively represents the institution's interests.
Computer science fashion has introduced many semantic web technologies into informatics infrastructures with variable success. A common question is whether modern triple-store technology and its corresponding manifestations such as RDF representation and SPARQL-based query offer material advantages over our technology choice of conventional SQL databases. A proper analysis is beyond the scope of this report, however prevailing opinion is that RDF and SQL data storage strategies are at some level isomorphic. That being said, it is widely acknowledged that today SQL-database environments have significant maturity advantages manifest by superior performance, security, design artifacts, and integration into analytic resources such as statistical packages and report generators.
The greatest limitation confronting the EDT is attributable to the elegance and comprehensiveness of its data normalization. The Data Governance processes such as modeling and shared vocabulary management, are resource intensive and time consuming. Institutions that prioritize a rapid start and immediate results will not find our approach to data integration and retrieval a satisfactory path. The investment Mayo is making in the EDT and its concomitant Enterprise Information Management infrastructure is strategic, consistent with the heritage of our organization. It is improbable that any organization could, from a standing start, complete the scope of organizational change, model and vocabulary content development, integration of existing data sources, and deployment of database and query infrastructure for their complete institutional data in under a decade; most organizations may not have the patience for such timelines and corresponding costs. However, the advantages of information transparency within the enterprise more than justify these time and resource investments.
Functionally, the impact of well-structured, easily queriable information about clinical events, risks, outcomes, and resource utilization fundamentally transforms a healthcare organization's capacity for quality improvement, research productivity, and best-practice monitoring. The transformation can be so profound that understanding the data-intensive opportunities for improvement requires a cultural transition away from exclusively process-oriented improvements to a more holistic systems orientation. Internalizing the new opportunities for systems improvement will require education, engagement, and most importantly, visible success. Mayo is already realizing palpable success from our cancer center projects, which is enabling analysis of clinical trial capture and accrual patterns, patient volumes, and clinical trial patient filtering, and will eventually optimize patient options for clinical trials. An infection analytics project has standardized the data definition and capture of infection and infection-related case data across the enterprise. The project has enabled a single standardized, enterprise-based reporting and analysis environment for infection data, and is currently seeking to further automate the identification of healthcare acquired infections. Other deliverables to data include referral analysis, balanced scorecard reporting, quality dashboards, and ad hoc reports across a wide spectrum of clinical practice, research, and administrative requests.
Mayo's century-long tradition of fostering the curation of patient data has fully migrated into the current information age through top-down data governance and bottom-up project mandates, and manifested into a cohesive Enterprise Data Trust.
Footnotes
Funding: Dr Chute was supported in part by NHGRI eMERGE project cooperative agreement (U01-HG04599) and the National Center for Biomedical Ontology (N01-HG04028), one of the NIH National Centers for Biomedical Computing. Other funders: NIH; NHGRI eMERGE; AT&T Foundation.
Competing interests: None.
Contributors: All authors made substantial contributions to conception, design, analysis and interpretation of data. CGC drafted the manuscript, and all authors were involved in revising it critically for important intellectual content and final approval. CGC is guarantor.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Inmon W. Building the data warehouse. Hoboken, NJ: John Wiley & Sons, 1993 [Google Scholar]
- 2.Clayton PD, Narus SP, Huff SM, et al. Building a comprehensive clinical information system from components. The approach at Intermountain Health Care. Methods Inf Med 2003;42:1–7 [PubMed] [Google Scholar]
- 3.Huff SM, Rocha RA, Coyle JF, et al. Integrating detailed clinical models into application development tools. Stud Health Technol Inform 2004;107(Pt 2):1058–62 [PubMed] [Google Scholar]
- 4.Parker CG, Rocha RA, Campbell JR, et al. Detailed clinical models for sharable, executable guidelines. Stud Health Technol Inform 2004;107(Pt 1):145–8 [PubMed] [Google Scholar]
- 5.Murphy SN, Morgan MM, Barnett GO, et al. Optimizing healthcare research data warehouse design through past COSTAR query analysis. Proc AMIA Symp 1999:892–6 [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy SN, Mendis M, Hackett K, et al. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annu Symp Proc 2007:548–52 [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy SN, Mendis ME, Berkowitz DA, et al. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc. 2006:1040. [PMC free article] [PubMed] [Google Scholar]
- 8.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008;84:362–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Main_Page (accessed 23 Dec 2009).
- 10.Kurland LT, Molgaard CA. The patient record in epidemiology. Sci Am 1981;245:54–63 [DOI] [PubMed] [Google Scholar]
- 11.Melton LJ., 3rd History of the Rochester Epidemiology Project. Mayo Clin Proc 1996;71:266–74 [DOI] [PubMed] [Google Scholar]
- 12.Beeler GW, Jr, Gibbons PS, Chute CG. Development of a clinical data architecture. Proc Annu Symp Comput Appl Med Care 1992:244–8 [PMC free article] [PubMed] [Google Scholar]
- 13.Van Grevenhof P, Chute CG, Ballard DJ. A common and clonable environment to support research using patient data. Proc Annu Symp Comput Appl Med Care 1991:358–62 [PMC free article] [PubMed] [Google Scholar]
- 14.Chute CG. Clinical data retrieval and analysis. I've seen a case like that before. Ann N Y Acad Sci 1992;670:133–40 [DOI] [PubMed] [Google Scholar]
- 15.Chute CG, Yang Y, Evans DA. Latent Semantic Indexing of medical diagnoses using UMLS semantic structures. Proc Annu Symp Comput Appl Med Care 1991:185–9 [PMC free article] [PubMed] [Google Scholar]
- 16.Chute CG, Yang Y. An overview of statistical methods for the classification and retrieval of patient events. Methods Inf Med 1995;34:104–10 [PubMed] [Google Scholar]
- 17.Rhodes RA. Healthy Approach to Data: IBM and Mayo Clinic team up to create massive patient database. IBM Systems Magazine 2002 [Google Scholar]
- 18.Savova GK, Ogren PV, Duffy PH, et al. Mayo clinic NLP system for patient smoking status identification. J Am Med Inform Assoc 2008;15:25–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.http://domino.research.ibm.com/comm/research_projects.nsf/pages/uima.index.html (accessed 23 Dec 2009).
- 20.http://ohnlp.org (accessed 23 Dec 2009).
- 21.http://www.hl7.org (accessed 23 Dec 2009).
- 22.https://cabig.nci.nih.gov/ (accessed 23 Dec 2009).
- 23.Fridsma DB, Evans J, Hastak S, et al. The BRIDG project: a technical report. J Am Med Inform Assoc 2008;15:130–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.http://www.bridgmodel.org/ (accessed 23 Dec 2009).
- 25.Pathak J, Solbrig HR, Buntrock JD, et al. LexGrid: a framework for representing, storing, and querying biomedical terminologies from simple to sublime. J Am Med Inform Assoc 2009;16:305–15 [DOI] [PMC free article] [PubMed] [Google Scholar]