Polymer-based gene carriers have been increasingly proposed as a safer alternative to viral vectors, due to their ease of synthesis, flexibility in the size of the transgene to be delivered, and minimal host immune responses. A major challenge to apply polycation/DNA nanoparticles in vivo, however, is the poor colloidal stability and serum stability, which lead to rapid aggregation followed by macrophage uptake, and premature dissociation of nanoparticles and release of DNA payload, respectively. Together they conspire to yield extremely low gene delivery efficiency through intravenous injection. To improve efficiency, an ideal polycation/DNA delivery system should satisfy the conflicting requirements of high stability in extracellular environment and endolysosomal compartments, so that the nanoparticles maintain their small size and integrity in circulation and endosomal sequestration, followed by low stability in cytosol and nucleus so as to allow for DNA release and transcription.
Recently, we have synthesized a series of polyethylene glycol-block-polyphosphoramidate (PEG-b-PPA)/DNA micellar nanoparticles for liver-targeted gene delivery.[1] The PEG corona of the micelles around the PPA/DNA core effectively reduced protein adsorption in physiological media and led to improved serum stability and enhanced gene transfection efficiency in the rat liver after infusion through the tail vein or bile duct, as compared with PPA/DNA nanoparticles and PEI/DNA complexes. In a series of PEG-b-PPA carriers prepared with 12 KDa PEG, we observed increasing DNA binding affinity as the molecular weight of PPA block increased from 3 KDa to 80 KDa. Interestingly however, micelles formed with PEG12K-b-PPA128K (the molecular weight of PPA block is 128 KDa) exhibited instability in solution with physiological salt concentration and released most of encapsulated plasmid DNA 4 h after transfection in HEK293 cells.[2] The quick DNA unpacking ability of these micelles as a result of salt instability could be advantageous for intracellular unpacking of DNA cargo upon delivery to the cytosol and nucleus. To avoid the premature release of encapsulated plasmid DNA in extracellular milieu or in endocytic compartments, we have introduced reversible reduction-sensitive cross-links to the micelles. The resulting micellar nanoparticles showed dual sensitivity that affords stability of the nanoparticles in extracellular and endocytic environments and DNA unpacking ability in cytosols and nuclei (Fig. 1a). As reported previously,[3,4] disulfide crosslinking is readily reduced in the cytosol and nucleus where the L-glutathione concentration is two orders of magnitude higher than that in the more oxidizing endocytic compartments and in extracellular environment. We have demonstrated that these disulfide-crosslinked PEG12K-b-PPA128K/DNA micelles have significantly improved stability in serum and salt solutions. Moreover, different from other reported disulfide-crosslinked complex or nanoparticle systems,[5–7] upon reduction of disulfide crosslinks in cytosol and nucleus, the micelles became unstable and hence the release of the DNA cargo can be regulated more effectively, due to the low DNA compaction ability of PEG12K-b-PPA128K in physiological ionic strength. More importantly, we showed that the dual-sensitive micellar nanoparticles mediated enhanced and prolonged gene transfection efficiency in vitro.
Figure 1.
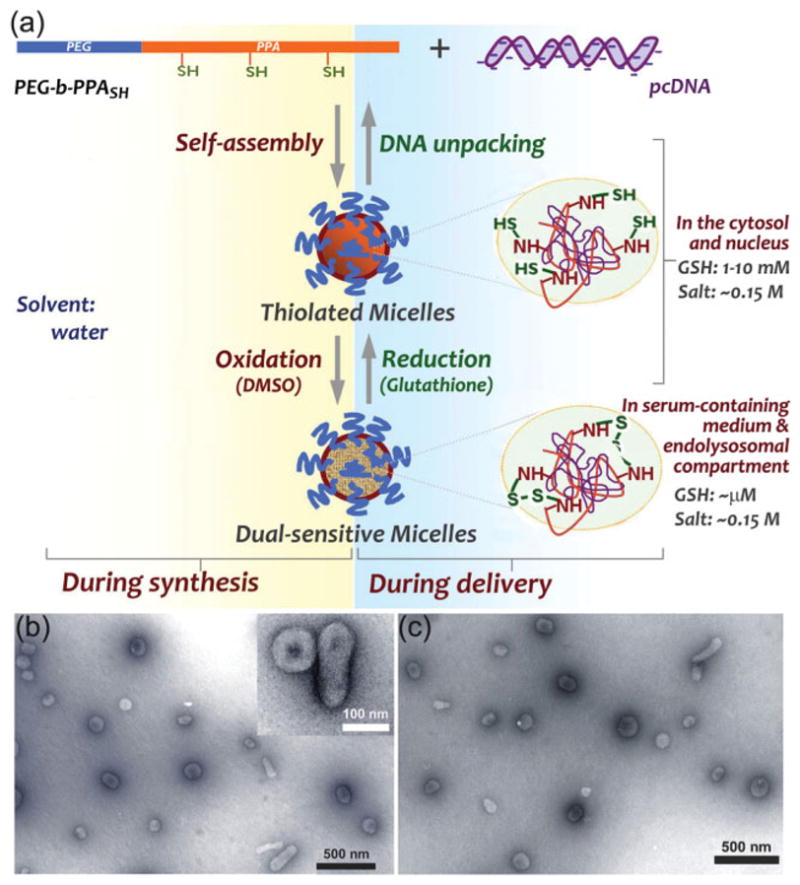
a) Preparation of dual-sensitive micelles. Micelles are prepared in distilled water to yield compact nanoparticles, then oxided in the presence of DMSO. The disulfide-crosslinked micelles are stable in blood, extracellular melieu, and the endolysosomal compartment where the glutathione (GSH) concentration is in the micromolar range. These crosslinks can be reduced when they reach the cytosol and nucleus where GSH concentration is in the range of 1–10 mM; the reduced micelles become unstable due to the salt-sensitive nature of the PEG12K-b-PPA128K carrier, thus releasing unpacked DNA. b,c) TEM images of dual-sensitive micelles prepared with 18.8% thiolated copolymer in deionized water (b) and after incubation with 0.15 M NaCl for 30 min (c).
To introduce the disulfide crosslinking, the PPA block of PEG12K-b-PPA128K polymer was modified with Traut’s reagent at different thiolation degrees (see the Supporting Information, Fig. S1a).[8] Traut’s reagent was chosen because the thiolation of PPA block does not change the positive charge density on the PPA block.[9] The crosslinked micellar nanoparticles exhibited similar particle size as the noncrosslinked micelles. The difference in particle sizes between micelles in water and in 0.15 M NaCl solution was used as an indicator of complex stability of the micelles (Fig. S1b). The complex stability of the crosslinked micelles increased with the thiolation degree until it reached 18.8%, at which point there was no difference in particles size between the two media. The PEG12K-b-PPA128K with a thiolation degree of 18.8% was used for all the following experiments and is hereby referred to as the crosslinked micellar nanoparticles.
Transmission electron microscopy (TEM) images showed that these crosslinked micellar nanoparticles were mostly spherical with diameters ranging from 100 to 150 nm (Fig. 1b), corroborated well with the size (123.4 ± 8.7 nm) measured by dynamic light scattering (DLS) method. There were less than 10% of micelles assuming elongated morphology with a diameter of 60–80 nm and length of about 200 nm. Consistent with the size measurement by DLS, we did not observe any change in size for the crosslinked nanoparticles after incubation in 0.15 M NaCl solution under TEM (Fig. 1c), suggesting the robust complex stability in salt containing medium. In contrast, the size of the noncrosslinked micelles increased from 90 nm to around 220 nm rapidly after addition to the 0.15 M NaCl solution.
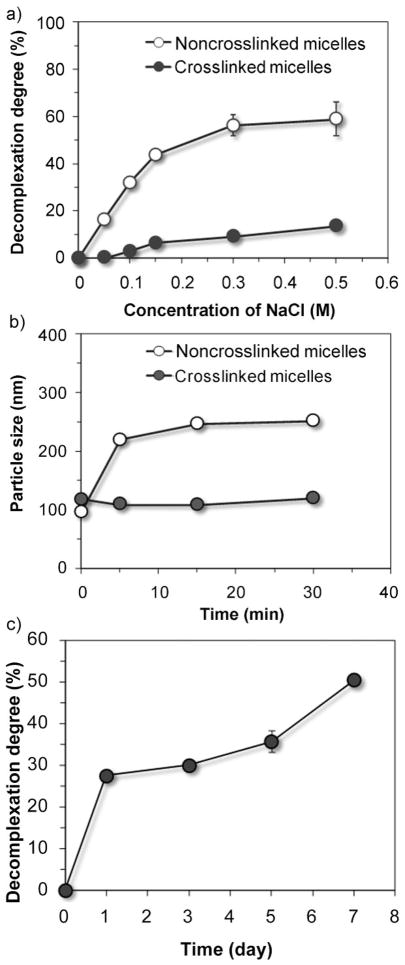
The salt stability was also confirmed by using decomplexation assay with ethidium bromide. The fluorescence efficiency of ethidium bromide increased dramatically upon binding to double stranded DNA by intercalation.[10] If the complexation between plasmid DNA and polymers is disrupted or relaxed in the presence of salt, ethidium bromide intercalation with uncomplexed DNA can result in an increase in fluorescence intensity; and the decomplexation degree of micelles can be calculated using Equation (1). As the concentration of salt went up gradually from 0 to 0.5 M, increasing amount of DNA was decomplexed from the noncrosslinked micelles, as indicated by an increase in decomplexation degree to 44% (Fig. 2a). In contrast, the decomplexation degree of the crosslinked micelles increased slightly to 7%. In serum stability test, the noncrosslinked and crosslinked micelles were challenged by fetal bovine serum (FBS). After incubation with 10% (v/v) FBS for 30 min, the particle size of noncrosslinked micelles increased to around 250 nm (Fig. 2b), whereas the average size of the crosslinked micelles remained unchanged. These results collectively indicate that the shielding effect of PEG corona and disulfide crosslinks effectively enhanced the complex stability in salt solution and colloidal stability in serum containing medium.
Figure 2.
Stability of noncrosslinked and dual-sensitive micelles in the presence of NaCl solution of different concentrations (a) or 10% fetal bovine serum (b). c) Degree of de-complexation of DNA after incubating the dual-sensitive micelles with 15 mM L-glutathione for different intervals at 37 °C.
To validate the hypothesis that the crosslinked micelles can recover its salt sensitivity and unpack plasmid DNA in the reducing environment, the crosslinked micelles were incubated with 15 mM L-glutathione (a concentration similar to cytosolic level) at 37 °C.[3] At different time intervals after the addition of L-glutathione, the micelles were challenged with 0.15 M NaCl solution and the decomplexation degree was measured. Interestingly, crosslinked micelles regained the salt sensitivity much more slowly than we had expected. One hour after incubation with L-glutathione, the decomplexation degree of crosslinked micelles increased slightly by only 3% (data not shown), suggesting that most of disulfide crosslinks were still intact. A more significant increase in salt sensitivity was observed at 24 h after incubation, when the decomplexation degree increased to 28% (Fig. 2c). The decomplexation degree increased gradually and reached a comparable level to that of noncrosslinked micelles by day 7. This observation suggests that the cleavage of disulfide crosslinks was a relatively slow process in a reduction microenvironment similar to that found in the cytosol and nucleus.[3]
The transfection efficiency of noncrosslinked micelles was first screened by transfecting HEK293 cells with micelles prepared at different N/P ratios (the molar ratios of nitrogen atoms in PPAs to phosphorus atoms in DNA) from 1 to 15. Micelles prepared with an N/P ratio of 8 showed the highest transfection efficiency at two days after transfection (see the Supporting Information, Fig. S2). Therefore, both noncrosslinked and crosslinked micelles at the N/P ratio of 8 were used to evaluate the effect of dual sensitivity on transfection efficiency. As shown in Figure 3, the PEI/DNA complexes mediated the highest transfection efficiency on day 1. The transgene expression decreased slightly on day 3, and dropped by 90% on day 5 and then to the background level on day 7. Transfection efficiency of the noncrosslinked micelles (e.g., salt-sensitive) was much lower than that of PEI/DNA nanoparticles. The transgene expression level mediated by noncrosslinked micelles peaked on day 3 and decreased to the background level on day 7. In contrast, the transfection efficiency of the crosslinked micelles (e.g., dual sensitive) was 2- and 4-fold higher than noncrosslinked micelles on days 1 and 3, respectively, likely due to the protective and stabilization effect of the crosslinking. Transgene expression maintained for four days at the peak level, which was only 6-fold lower than the peak expression level by PEI/DNA nanoparticles. By day 10, 35% of the peak expression level was still observed in cells transfected by the dual-sensitive micelles.
Figure 3.
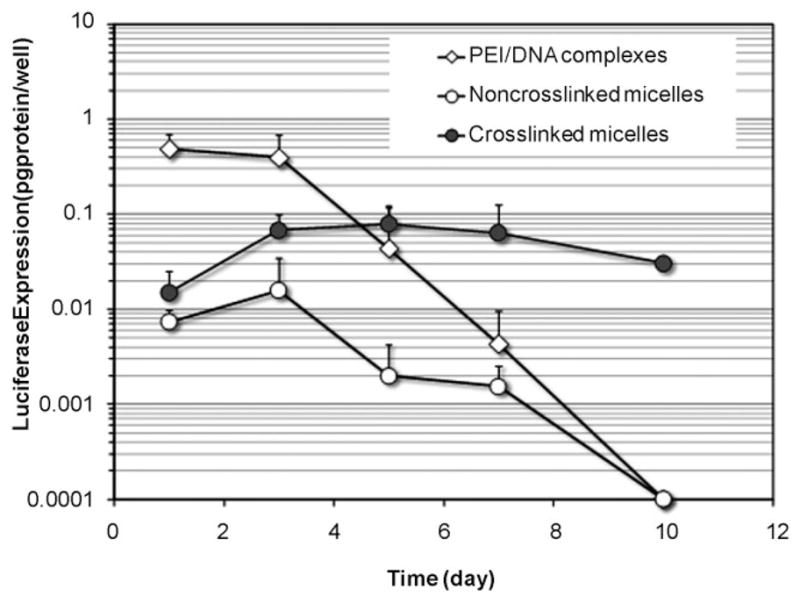
Transgene expression level mediated by dual-sensitive micelles and noncrosslinked micelles in HEK293 cells over time, in comparison with PEI/DNA nanoparticles. Each data point represents average ± standard division (n = 4).
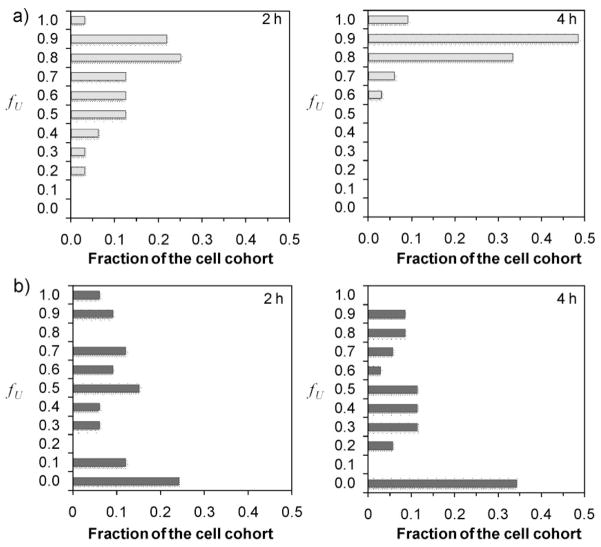
The fact that the crosslinked dual-sensitive micelles enhanced and significantly prolonged transgene expression compared with noncrosslinked micelles can be related to the different DNA unpacking mechanism from reversibly crosslinked micelles. As demonstrated in Figure 2c, the restoration of salt sensitivity for the crosslinked micelles was a slow process. Therefore, the unpacking of crosslinked micelles may be significantly delayed compared with that of noncrosslinked micelles. Based on this hypothesis, we used a recently developed quantum dot (QD)-FRET technique to study the intracellular DNA unpacking kinetics of both noncrosslinked and crosslinked micelles. PEG12K-b-PPA128K or thiolated PEG12K-b-PPA128K block polymers and plasmid DNA were labeled with Cy5 and QD605, respectively. After cell uptake of Cy5 and QD-labeled micelles, the high signal-to-noise ratio in QD-mediated FRET enabled sensitive detection of discrete changes in micelle stability. From confocal image stacks obtained at each time point, the distributions of intact and unpacked micelles in the cytosol, endolysosomal compartment, and nucleus were quantified with a particle-specific voxel-based method adapted from a previous report using a pixel-based algorithm.[11] In this experiment, thiolated PEG12K-b-PPA128K polymer with a thiolation degree of 8.8% was used because crosslinked micelles formed with block copolymers with higher thiolation degrees induced strong fluorescence at 505 nm upon excitation of QDs, which interfered seriously with the signal from Lysotracker Green (for the localization of endo/lysosomes). In Figure 4, histograms were generated to compare the fractions of cohorts of cells having specific volume fractions of unpacked DNA ( fU) at 2 and 4 h post-transfection. There is a marked difference in the overall unpacking rate and unpacked plasmid DNA fraction between noncrosslinked and crosslinked micelles from 2 to 4 h time points. At 2 h after transfection, the majority (~88%) of cells transfected with noncrosslinked micelles had an fU > 0.5, which indicated that more than 50% of the internalized DNA was unpacked among these cells. In contrast, nearly 50% of cells transfected with dual-sensitive micelles showed an fU > 0.5 at 2 h time point. Among all the cells, an average of 37% of internalized DNA was unpacked as compared to 73% from noncrosslinked micelles. At 4 h, the histogram for noncrosslinked micelles was significantly shifted toward higher fU values. All cells had an fU value of higher than 0.6 and, on average, 80% of all internalized DNA was unpacked, whereas dual-sensitive micelles showed a much delayed unpacking of plasmid DNA with nearly the same fU histogram as that at 2 h.
Figure 4.
Overall distribution of unpacked DNA inside transfected cells. The overall intracellularly unpacked DNA from noncrosslinked micelles (a) and dual-sensitive micelles (b) at 2 and 4 h post-transfection. At each time point, histograms show the fraction of an analyzed cohort of cells (30–34 cells) with a particular binned volume fraction of unpacked DNA (fU).
In addition to the overall distribution of unpacked plasmid DNA in the cells transfected with micelles, the unpacking of micelles in three subcellular compartments (endo/lysosomes, cytosol, and nucleus) was also evaluated. Among cells transfected with noncrosslinked and crosslinked micelles, low volume fractions of unpacked DNA were localized in endolysosomal compartments at 2 and 4 h time points (Fig. S3), indicating that most of the noncrosslinked and crosslinked micelles found in the endo/lysosomes remained unpacked at these time points. In addition, the majority of cells had an f′U (endo/lyso) value < 0.2 (Fig. S4), confirming that most of the unpacked DNA was not found in this compartment. The majority of unpacked DNA from noncrosslinked and crosslinked micelles was localized in the cytosolic compartment and nucleus (Fig. S4). Interestingly, when more noncrosslinked micelles unpacked at 4 h, most unpacking happened in cytosolic compartment, since there was a significant upshift of fU (cyto) (Fig. S3) and f′U (cyto) (Fig. S4), and downshift of fU (nuc) and f′U (nuc) values in the histogram of noncrosslinked micelles. As for the crosslinked micelles, the distribution pattern of unpacked plasmid DNA in cytosolic compartment and nucleus did not change significantly over these two hours. More importantly, there was no appreciable increase of DNA unpacking from the dual-sensitive micelles from 2 h to 4 h in the cytosol and nucleus compartments. Especially, at 4 h, an average of 83% of intracellular dual-sensitive micelles remained unpacked, as compared to 31% for the noncrosslinked micelles. This difference confirms our hypothesis that the dual-sensitive micelles yielded a significantly delayed unpacking of DNA compared with noncrosslinked micelles at early time points. Monitoring longer term DNA unpacking events has been challenging due to the fluorescence quenching of the Cy5 signal.
In this report, we have prepared a reversibly crosslinked micellar nanoparticles that are capable of releasing encapsulated DNA in response to the concentration of reduction agent (e.g., L-glutathione) and ionic strength (e.g., at physiological salt concentration). Such a property was used to prevent extracellular DNA release and regulate intracellular DNA unpacking. Compared with noncrosslinked micelles, the dual-sensitive micelles showed significantly enhanced complex stability and colloidal stability in salt and serum containing media, and more sustained DNA release at cytosolic salt and glutathione concentrations. Using a more quantitative QD-FRET method, we demonstrated that the dual-sensitive micelles unpacked DNA primarily in the cytosol and nucleus, and the DNA unpacking was significantly delayed compared with noncrosslinked micelles at 4 h after transfection. Due to the unique DNA unpacking behavior, the dual-sensitive micelles mediated enhanced and much prolonged gene expression for at least 10 days in HEK 293 cells.
Experimental
Preparation and Characterization of Dual-Sensitive Micelles
The synthesis of PEG12K-b-PPA128K/DNA micelles with VR1255 plasmid DNA, and characterization of size and surface charge of micelles by DLS and TEM imaging were conducted according to the methods we reported previously [1]. For crosslinked micelles, plasmid DNA was incubated with thiolated copolymer similarly; then the micelle solution was transferred to a dialysis bag with MWCO of 3,500 Da (Spectrapor, Rancho Dominguez, CA) and dialyzed against 0.5% (v/v) DMSO for 24 h to oxidize thiol groups. Complete oxidization and removal of residual DMSO was then achieved by dialyzing the micelle solution against DI water for additional 48 h.
Determination of the Decomplexation Degree of Micelles in the Presence of Salts
One hundred μL of micelle or DNA solution containing 1 μg plasmid DNA was mixed with an equal volume of ethidium bromide solution (1 μg mL−1). Concentrated NaCl solution (5 M) was added to the mixed solution to bring the salt concentration to a desired value. After incubation at room temperature for 10 min, the fluorescence intensity (I) of the micelle and DNA solution in the absence of salts (Im and IDNA, respectively) and in the presence of salts (Im,s and IDNA,s, respectively), were measured using a microplate reader (SpectraMax, Molecular Devices, Sunnyvale, CA, USA) with excitation and emission wavelengths of 310 and 590 nm, respectively. The decomplexation degree of the micelles was calculated according to
| (1) |
In Vitro Gene Transfection
Transfection of HEK293 cells (American Type Culture Collection, Manassas, VA) with nanoparticles and characterization of transfection efficiency were conducted according to the same method we described previously [1]. To evaluate the transfection efficiency on days 3, 5, 7 and 10, another set of cells transfected with nanoparticles were split at 1:5 ratio on day 3 to prevent over-confluence of cells. On day 5, the cells were split again 1:5 ratio and 80% of the cells were harvested for the measurement of luciferase activity. On day 10, all the cells were harvested for the measurement of luciferase activity. To compare the total transgene expression level on days 5, 7, and 10 with that at earlier time points, the total luciferase activity obtained on days 5, 7, and 10 were multiplied by factors of 5, 25, and 125, respectively (see the Supporting Information for details).
Live Cell Imaging and Quantifying Distribution of Complexed or Unpacked DNA
The preparation of QD605-labeled plasmid DNA and Cy5-labeled polymers and the analysis of intracellular trafficking study was conducted according to a method we reported previously [11]. Detailed procedures are provided in Supporting Information.
Supplementary Material
Acknowledgments
The authors thank Prof. Michael McCaffery at the Integrated Imaging Center at Johns Hopkins University for his assistance with TEM imaging. We also thank Dr. Yi-Ping Ho and Dr. Yong Ren for their technical assistance. This study was supported by grants from the National Institutes of Health, the National Institute of General Medical Sciences GM073937 (HQM), the National Institute of Biomedical Imaging and Bioengineering HL089764 (KWL), and the National Science Foundation ECCS-0725528 and CBET-0546012 (THW). Supporting Information is available online from Wiley InterScience or from the author.
Contributor Information
Xuan Jiang, Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, MD 21218 (USA).
Yiran Zheng, Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD 21218 (USA).
Dr. Hunter H. Chen, Department of Biomedical Engineering, Johns Hopkins University, School of Medicine, 720 Rutland Avenue, Baltimore, MD 21231 (USA)
Prof. Kam W. Leong, Department of Biomedical Engineering, Duke University, Durham, North Carolina 27708 (USA)
Prof. Tza-Huei Wang, Department of Biomedical Engineering, Johns Hopkins University, School of Medicine, 720 Rutland Avenue, Baltimore, MD 21231 (USA), Department of Mechanical Engineering, Johns Hopkins University, Baltimore, MD 21218 (USA)
Prof. Hai-Quan Mao, Email: hmao@jhu.edu, Department of Materials Science and Engineering and Whitaker Biomedical Engineering Institute, Johns Hopkins University, 206 Maryland Hall, Baltimore, MD 21218 (USA)
References
- 1.Jiang X, Dai H, Ke CY, Mo X, Torbenson MS, Li ZP, Mao HQ. J Controlled Release. 2007;122:297. doi: 10.1016/j.jconrel.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen HH, Ho YP, Jiang X, Mao HQ, Wang TH, Leong KW. Nano Today. 2009;4:125. doi: 10.1016/j.nantod.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauhuber S, Hozsa C, Breunig M, Gopferich A. Adv Mater. 2009;21:3286. doi: 10.1002/adma.200802453. [DOI] [PubMed] [Google Scholar]
- 4.Soundara Manickam D, Oupicky D. J Drug Target. 2006;14:519. doi: 10.1080/10611860600834409. [DOI] [PubMed] [Google Scholar]
- 5.Kakizawa Y, Harada A, Kataoka K. J Am Chem Soc. 1999;121:11247. [Google Scholar]
- 6.Oupicky D, Carlisle RC, Seymour LW. Gene Ther. 2001;8:713. doi: 10.1038/sj.gt.3301446. [DOI] [PubMed] [Google Scholar]
- 7.Neu M, Germershaus O, Mao S, Voigt KH, Behe M, Kissel T. J Controlled Release. 2007;118:370. doi: 10.1016/j.jconrel.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Mao HQ, Roy K, Troung-Le VL, Janes KA, Lin KY, Wang Y, August JT, Leong KW. J Controlled Release. 2001;70:399. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- 9.Miyata K, Kakizawa Y, Nishiyama N, Harada A, Yamasaki Y, Koyama H, Kataoka K. J Am Chem Soc. 2004;126:2355. doi: 10.1021/ja0379666. [DOI] [PubMed] [Google Scholar]
- 10.Lepecq JB, Paoletti C. J Mol Biol. 1967;27:87. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- 11.Chen HH, Ho YP, Jiang X, Mao HQ, Wang TH, Leong KW. Mol Ther. 2008;16:324. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.