Abstract
Among the hallmarks of aged organisms are an accumulation of misfolded proteins and a reduction in skeletal muscle mass (“sarcopenia”). We have examined the effects of aging and dietary restriction (which retards many age-related changes) on components of the ubiquitin proteasome system (UPS) in muscle. The hindlimb muscles of aged (30 months old) rats showed a marked loss of muscle mass and contained 2–3-fold higher levels of 26S proteasomes than those of adult (4 months old) controls. 26S proteasomes purified from muscles of aged and adult rats showed a similar capacity to degrade peptides, proteins, and an ubiquitylated substrate, but differed in levels of proteasome-associated proteins (e.g. the ubiquitin ligase E6AP and deubiquitylating enzyme USP14). Also, the activities of many other deubiquitylating enzymes were greatly enhanced in the aged muscles. Nevertheless, their content of polyubiquitylated proteins was higher than in adult animals. The aged muscles contained higher levels of the ubiquitin ligase CHIP, involved in eliminating misfolded proteins, and MuRF1, which ubiquitylates myofibrillar proteins. These muscles differed from ones rapidly atrophying due to disease, fasting, or disuse in that Atrogin-1/MAFbx expression was low and not inducible by glucocorticoids. Thus, the muscles of aged rats showed many adaptations indicating enhanced proteolysis by the UPS, which may enhance their capacity to eliminate misfolded proteins and seems to contribute to the sarcopenia. Accordingly, dietary restriction decreased or prevented the aging-associated increases in proteasomes and other UPS components and reduced muscle wasting.
Keywords: Aging, Deubiquitination, Proteasome, Protein Degradation, Skeletal Muscle, Ubiquitin, Sarcopenia
Introduction
A characteristic feature of aged organisms is the gradual loss of muscle mass (often termed “sarcopenia”), which is a major cause of frailty and morbidity among the elderly (1). This reduction in muscle mass with aging may result from a general decline in protein synthesis or alternatively from an enhancement of overall protein degradation rates, as occurs during the rapid atrophy of muscles induced by disuse, fasting, or disease (e.g. cancer or sepsis) in young or adult animals (2). Mammalian cells have two main pathways for protein degradation, the lysosomes, which function in autophagy and the breakdown of endocytosed proteins, and the ubiquitin proteasome system (UPS),4 which rapidly eliminates misfolded proteins and short-lived regulatory proteins, but also catalyzes the breakdown of the bulk of cell proteins (3). The proteolytic component in this pathway is the 26S proteasome, a large, ATP-dependent complex composed of the proteolytic 20S core particle and capped by one or two 19S regulatory particles (4). In this pathway, proteins destined for degradation are linked to a chain of ubiquitin molecules by a cascade of ubiquitin ligases (E1, E2, and E3) (5). The 19S regulatory particle contains subunits that bind the ubiquitin chain, six ATPases that unfold the protein substrate and translocate it into the 20S particle for hydrolysis, and three deubiquitylating enzymes or DUBs (Rpn11, Usp14/Upb6, and Uch37), which disassemble the ubiquitin chain, releasing the ubiquitin moieties. This sparing of the ubiquitin molecules enables the cell to maintain a high pool of free ubiquitin (6), which is crucial for ongoing proteolysis. In addition, 26S proteasomes have a set of loosely associated proteins that probably serve as cofactors or regulators (7) and may vary under different physiological states.
A number of studies have suggested that in aged rodents proteasomal proteolysis decreases in several tissues/organs (8–13). It is hard to reconcile such a reduced rate of proteolysis with the general loss of skeletal muscle mass characteristic of aging (sarcopenia) (1). During muscle atrophy in young adult animals induced by fasting, disuse, or disease (e.g. cancer cachexia, sepsis, renal or cardiac failure), there is a marked increase in protein ubiquitylation and proteolysis, especially of myofibrillar components (14, 15).
Data on the functioning of the UPS during the much slower loss of muscle in the elderly is controversial. There are conflicting reports about the levels of proteasome expression or activity (16–22). However, most of these conclusions about the effect of aging on proteasomal degradation in muscle or other tissues are based solely on measurements of the 20S core particle. Although some degradation of unfolded or denatured proteins may occur by the free 20S (23), the bulk of proteasome-mediated degradation, even of oxidatively damaged proteins (24), seems to require the 26S proteasome and ubiquitylation of the substrate. To understand the effects of aging it is therefore important to measure the capacity of 26S proteasomes to degrade their physiological substrates, ubiquitylated proteins, especially because the accumulation of ubiquitylated proteins is a hallmark of aging (25, 26) and protein ubiquitylation and content of ubiquitin conjugates are increased in muscle during various types of rapid atrophy (27, 28).
We therefore examined in 30-month-old Sprague-Dawley rats the effects of aging on the content and activity of 26S proteasomes, proteasome-associated regulatory proteins, and various other components of the UPS, including multiple deubiquitylating enzyme (DUB) and factors that bind ubiquitylated proteins and facilitate their degradation by the proteasome (e.g. p97). This experimental model was chosen because muscles of these aged animals undergo marked atrophy compared with muscles of adult (4-month-old) animals. In various organisms, dietary restriction is known to extend health span and lifespan (29). Therefore, we also studied the effects of dietary restriction (DR) on these aging-associated changes because DR can inhibit many of the deleterious effects of aging (29) including muscle wasting (30). Our studies indicate that in muscles of these aged animals with sarcopenia, multiple components of the ubiquitin proteasome system are markedly induced and that many of these aging-related changes can be prevented or reduced by dietary restriction.
MATERIALS AND METHODS
Chemicals
Creatine phosphate, creatine phosphate kinase, and casein were purchased from Sigma. Proteasome substrates Suc-LLVY-AMC, Boc-LRR-AMC, and Suc-YVAD-AMC were obtained from Bachem (Weil am Rhein, Germany). Ubiquitylated DHFR was a gift from Millennium, Inc. (Cambridge, MA). Antibodies used are listed in supplemental Table S2.
Experimental Animals
Thirty-nine male rats (colony originated from Harlan Sprague-Dawley, Houston, TX), 15 adult (4 months) and 24 aged (30 months) rats, were studied. Nine of the aged animals were maintained from 2 months of age on DR corresponding to 70% of ad libitum (AL) food intake of age-matched animals. All animals were kept under standardized barrier conditions (with 12/12 light/dark cycle) in a climate-controlled facility. In addition, five adults and five aged AL rats (from the same cohorts) were treated with dexamethasone (4 mg/kg intraperitoneal for 3 days). Based on lifespan expectancy of male SD rats (30), the 30-month-old rats were considered as aged (senescent). All animal experiments were approved by the Local Ethical Committee (N122/03 and N400/03).
Preparation of Rat Muscle Extracts
The triceps surae muscles were removed from anesthetized (chloral hydrate, 300 mg/kg, intraperitoneally) rats, weighed, and frozen in liquid nitrogen and stored at −80 °C until processed.
Frozen tissue from gastrocnemius muscle was homogenized in lysis buffer (50 mm Tris base, pH 7.4, 5 mm MgCl2, 250 mm sucrose, and fresh 2 mm ATP, and 1 mm DTT). Membrane fractions, nuclei, organelles, and cell debris were removed by centrifugation at 12,000 × g for 15 min and then ultracentrifugated at 100,000 × g for 1 h at 4 °C.
Immunoblotting
Equal amounts of protein were separated by SDS-PAGE, transferred to a PVDF or nitrocellulose membrane (Amersham Biosciences), and blotted with the primary antibody (see supplemental Table S1) containing 5% milk and 0.5% Tween 20. Immunodetection was performed using enhanced chemiluminescence detection (ECL Plus) method according to the manufacturer's protocol (Amersham Biosciences), and the membranes were exposed to X-OMAT AR film (Kodak) for visualization. Vinculin was identified as a useful loading control from a previous analysis of the proteome and transcriptome from the aged muscle (31).
Proteasome Measurement Using Active Site Labeling
Labeling of the proteasomal β-subunits using active site-directed probes was performed as reported previously (32, 33). In brief, the radiolabeled active site-directed probe AdaY[125I]Ahx3L3VS (0.5 × 106 cpm) or dansylated probe dansyl-Ahx3L3VS (1 mm) was added to 25 μg of tissue lysate and incubated for 2 h at 37 °C. Proteins were then separated by SDS-PAGE and visualized by autoradiography or immunoblotting using anti-dansyl antibodies. Two-dimensional nonequilibrium pH gradient and SDS-PAGE was performed as described previously (33). Autoradiograms were scanned as 16-bit tiff images and quantified by densitometry using the ImageJ software (rsb.info.nih.gov/ij/).
Proteasome Activity Assays Using Fluorogenic Substrates
One μg of enriched proteasome fraction (5 h pellet) or 10–30 ng of purified 26S proteasomes were assayed for chymotrypsin-like (Suc-LLVY-AMC), trypsin-like (Boc-LRR-AMC), and caspase-like activity (Suc-YVAD-AMC) as described previously (34). To measure 20S proteasome activity, lysates were prepared in buffer without ATP and with 0.02% SDS (34). The samples from individual animals were analyzed in a fluorescent plate reader (SPECTRAmax® Gemini's, Molecular Devices) at zero and every 5 min for 2 h using an excitation/emission ratio of 380/460 nm at 37 °C. All experiments were repeated three times.
Purification of Proteasomes
For each purification 60 mg of gastrocnemius muscle from 7 different animals were pooled for each age group. The affinity purification of proteasomes using the UBL domain as an affinity ligand was performed in the absence of NaCl as described previously (7).
Degradation of Ub5DHFR
50 nm Ub5DHFR were incubated with 4 nm of affinity purified 26S proteasome in reaction buffer (50 mm Tris, pH 8, 60 mm KCl, 10 mm MgCl2, 1 mm ATP, 1 mm DTT, 0.1 units/μl of creatine phosphate kinase, 10 mm creatine phosphate) at 37 °C for 0–2 h. The reaction was stopped with SDS sample buffer and the content of Ub5DHFR assessed by immunoblotting (anti-biotin antibody; Ub5DHFR is labeled with biotin at the first Ub moiety).
Profiling of DUBs
Labeling of DUBs using ubiquitin-based probes HA-Ub-VME and HA-Ub-Br2 was performed as described (35). In brief, 0.5 μg of HA-Ub-VME or HA-Ub-Br2 were added to 25 μg of tissue lysates and incubated for 1 h at 37 °C. Samples were then separated on an 8% SDS-PAGE and individual active DUBs were visualized by anti-HA immunoblotting.
RNA Isolation
Total RNA was isolated from muscles according to the TRIzol protocol (Invitrogen). RNA concentration, purity, and integrity were measured in a spectrophotometer, by running a denaturing agarose gel and by analysis using a 2100 Bioanalyzer (Agilent Technologies, Kista, Sweden). All samples used showed a high degree of integrity and in the qPCR runs, β-actin was run in parallel to assure equal loading. RNA was DNase treated (DNA-free, Ambion Inc.), according to the manufacturer's protocol, resulting in OD 260/280 values above 1.95.
qPCR
Analysis of relative mRNA levels in gastrocnemius muscle was performed using reverse transcription (standard reagents, Applied Biosystems) and real time PCR, with QuantiTect SYBR Green Mastermix (Qiagen) and the appropriate primer pairs (supplemental Table S2) in an ABI Prism 7000 instrument (Applied Biosystems). Real time analysis of SYBR Green chemistry values was performed as previously described (36).
Statistics
All statistical analysis were performed using Statistica 6.1 (Statsoft, Tulsa, OK). Comparisons of experimental groups were carried out with analysis of variance, and when significant differences were found, Bonferroni's post hoc test was used for pairwise comparisons. Statistical significance levels were set to: *, p < 0.05; **, p < 0.01; ***, p < 0.001. Test of correlation was accomplished with least-square regression using untransformed data and calculation of the coefficient of correlation, r.
RESULTS
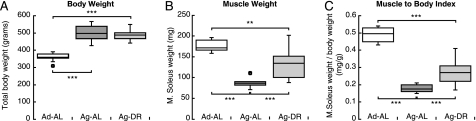
A colony of male Sprague-Dawley rats was maintained under standardized conditions and fed either AL or a restricted diet (DR), in which the rats consumed 70% of the caloric intake of age-matched AL controls. Aged rats showed reduced locomotor activity and disturbances of coordination and balance that was reduced by DR (30). DR rats lived on the average 6 months longer than the AL rats (36 versus 30 months).5 The animals were studied at 30 months, which is the average expected lifespan for AL maintained Sprague-Dawley rats (30), and were compared with 4-month-old adult rats, the age where muscle weight relative to body weight reaches its mature values (37). The average body weight of the AL rats peaked at 24 months and then declined. At 30 months, their mean body weight was 20% lower than peak weight, in accordance with earlier observations (30). At this time, the soleus muscles of the AL rats had lost 50% of their mass, but this marked atrophy was significantly smaller in 30-month-old aged rats maintained on DR (Fig. 1B). The body weights of the DR rats had not yet decreased significantly from their peak values at this age (Fig. 1A) (30). Due to this marked muscle atrophy and the concomitant accumulation of adipose tissue in the aged AL rats, the soleus/body weight ratio was 60% lower than in the adult rats (p < 0.001) and 40–50% lower than in the DR group (p < 0.001) (Fig. 1C).
FIGURE 1.
Aged rats (30 months old) have much less muscle mass, but greater body weights than adult (4 months old), and dietary restriction decreases this loss of muscle. A, boxplots illustrate the body weights of 10 adults (Ad-AL) and 10 aged rats fed AL (Ag-AL) or 9 aged rats maintained on dietary restriction (Ag-DR), as described under “Materials and Methods.” B, soleus muscle weight (mg) at 4 and 30 (AL and DR) months. C, ratio between soleus weights (mg) and body weights (g). Box limits represent upper and lower quartile values and are separated by the median (crossbar within box). Maximum and minimum values, which are not defined as outliers (circles), are illustrated using error bars. Outliers are defined as values deviating from the quartile borders by more than 1.5 times the interquartile distance. ***, p < 0.001.
Proteasome Levels Increase in Muscles of Aged Rats but Not in Animals on DR
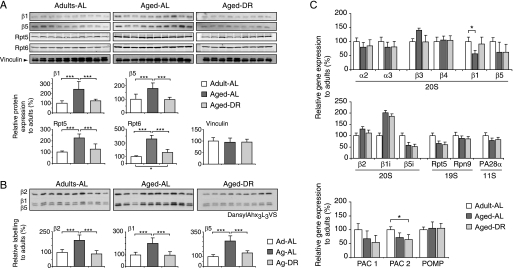
We analyzed the proteasome content of the triceps surae (the hind limb muscle composed of musculus gastrocnemius and musculus soleus). Immunoblot analysis showed a 2–3-fold increase in levels of the β1 and β5 subunits of the 20S and also subunits Rpt5 and Rpt6 of the 19S regulatory complex (Fig. 2A). These subunits are not found outside of these complexes (Fig. 4A). To clarify whether the aged proteasomes are indeed catalytically active, cell extracts were then treated with the specific active site probe, dansyl-Ahx3L3VS (32), which modifies covalently the active site threonine residues of the catalytic β1, β2, and β5 subunits (Fig. 2B). This approach also revealed a 2–3-fold increase in proteasomes in the aged muscle (Fig. 2B).
FIGURE 2.
Proteasome content increases in muscles of aged but not in rats on dietary restriction. A, levels of the 20S subunits, β1 and β5, and the 19S components, Rpt5 and Rpt6, were measured by immunoblotting with Vinculin as loading control. B, 20S proteasomes were covalently modified in crude extracts with the active site-directed probe dansyl-Ahx3L3VS followed by immunoblotting with anti-dansyl antibody. Each lane in panels A or B represents 25 μg of crude extract from gastrocnemius muscles of 4-month-old (n = 10) and 30-month-old AL (n = 10) and DR (n = 9) rats. Densitometry data and statistical analysis of protein levels (A) and proteasome activity (B) are plotted relative to the 4-month-old AL group (set to 100%). Error bars indicate standard deviation. C, changes in mRNA levels of several proteasome subunits as well as factors involved in proteasome assembly were assayed by qPCR. ***, p < 0.001.
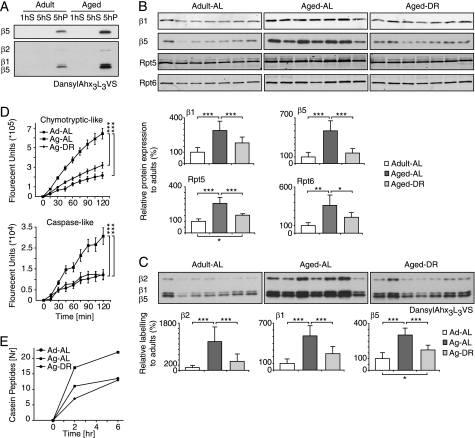
FIGURE 4.
Proteasome activity increases with age and this change is prevented by dietary restriction. A, to enrich for 26S proteasomes, muscle extracts were ultracentrifuged for 1 h at 100,000 × g to remove the microsomal fraction, and the supernatant (1hS) spun for 5 h at 100,000 × g. Proteasomes were detected only in the 5-h pellet (5hP) by immunoblotting (top) and using the active site probe dansyl-Ahx3L3VS (bottom) as described in the legend to Fig. 2; a representative result is shown. B and C, proteasome-enriched pellets were prepared from seven animals of each condition and proteasome content (B) and reactivity with the active site probe (C) were quantified. D, fluorogenic substrates were used to measure caspase-like (Suc-YVAD-AMC) and chymotrypsin-like (Suc-LLVY-AMC) activities. Error bars represent standard deviation. E, degradation of casein assessed by the appearance of casein fragments using tandem mass spectrometry. Proteasome-enriched fractions from 4-month (Ad-AL), 30-month (Ag-AL), or 30-month-old rat on DR (Ag-DR) were incubated with casein for different times at 37 °C, followed by LC-MS/MS analysis (see “Materials and Methods”). One of two independent experiments is shown. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Remarkably, the increase in 26S proteasomes was suppressed completely in aged animals on DR (Fig. 2, A and B). Because muscle wasting was reduced in these animals, these findings suggest that the buildup of proteasomes contributes to the loss of muscle mass in aged animals. Accordingly, the levels of these proteasome subunits in the aged AL group were inversely correlated with muscle weight (β1, r = −0.71, p < 0.05; β5, r = −0.69, p < 0.05), and no such inverse relationship was found in muscles of adult rats (β1, r = 0.42, ns; β5, r = 0.31, ns) or aged animals maintained on DR (β5, r = 0.27, ns; β1, r = 0.11, ns).
To determine whether this increase in proteasome content results from increased rates of subunit synthesis, we measured mRNA levels for several proteasomal subunits by qPCR. No significant increases in expression were observed except for the immunoproteasome subunit, β1i (Fig. 2C and below). To learn if proteasome assembly might increase with aging, we also measured the content of mRNA for 20S assembly factors PAC1, PAC2, and POMP1. Their expression did not change or even decrease with aging (Fig. 2D). These observations suggest that the large increase in proteasome content in aged muscles might result from enhanced translation or decreased rates of proteasome degradation in the aged muscles.
Immunoproteasomes Increase in Aged Muscle, but Are a Minor Fraction of the Proteasomes
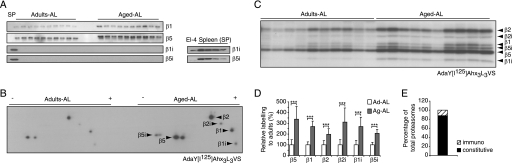
During infections, lymphokines, especially γ-interferon, induce alternative forms of the 20S proteasome called immunoproteasomes, which contain distinct catalytic subunits, termed β1i, β2i, and β5i. To learn if the increase in proteasomes with aging was due to the induction of immunoproteasomes, as was reported by some investigators (18) and suggested by the elevated transcription of β1i (Fig. 2C), we assayed expression of these alternative subunits by immunoblotting with antibodies against β1i, β2i, or β5i. However, they could not be detected in these muscles, although they were readily detected in spleen samples from these same animals (Fig. 3A). We therefore used a more sensitive assay of subunit activity involving a radioactive derivative of the affinity probe used in Fig. 2, B and C, the AdaY[125I]Ahx3L3VS (33). Two-dimensional gel electrophoresis revealed some induction of immunoproteasome subunits in muscles with age (Fig. 3B). Using standard SDS gels, we then determined if all of the rat muscles showed a similar induction, or if this result may be due to inflammation or disease in any individual rat. Similar levels of immunoproteasome subunits were in fact detected in all the aged animals, and like conventional 20S subunits, they were induced 2–3-fold in the aged muscles compared with adult rats. However, the degree of reaction with this probe suggested that immunoproteasomes constituted only about 10% of the total proteasomes in the muscles of these aged animals (Fig. 3E). Therefore, the age-related loss of muscle mass is associated primarily with a 2–3-fold increase in standard proteasomes.
FIGURE 3.
In aged muscles, immunoproteasomes are increased, but immunosubunits (β1i, β2i, and β5i) are still much less abundant than the standard catalytic subunits. A, representative immunoblot of the standard catalytic subunits of the proteasome, β1 (caspase-like activity), β5 (chrymotrypsin-like activity), and their homologs β1i and β5i in gastrocnemius muscle extracts prepared from rats. As a control, equal amounts of rat splenocytes (SP) were used to detect β1i and β5i adult and aged. B and C, gastrocnemius muscle extracts prepared from 4-month- and 30-month-old rats were incubated with the active site-directed probe AdaY[I125]Ahx3L3VS and analyzed by two-dimensional (B) and one-dimensional (C) gel electrophoresis. Each assay was performed in triplicates and a representative result is shown. D, densitometric quantification. E, distribution of 20S species as percent of total 20S proteasomes. ***, p < 0.001.
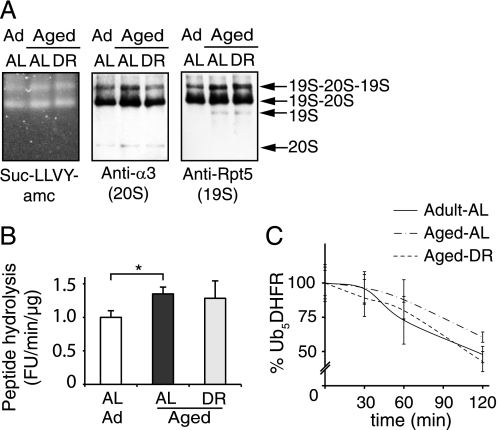
Muscle 26S Proteasomes from Aged and Adult Rats Have a Similar Ability to Degrade Peptides, Proteins, and Ubiquitylated Proteins
To determine whether the increase in muscle proteasome content with aging actually reflects greater proteolytic capacity, we prepared proteasome-rich fractions from the lysates by ultracentrifugation (Fig. 4A). Quantification of proteasomes by both immunoblot and active site probes confirmed the greater content in aged AL fed rats but not in the DR group (Fig. 4, B and C). Using these proteasome-enriched pellets, we were able to measure rates of hydrolysis of fluorogenic peptides that are specific substrates of its chymotrypsin-like and caspase-like sites (Fig. 4D) and of casein (Fig. 4E), a loosely folded protein that can be degraded by the proteasome without ubiquitylation. All these substrates were degraded 2–3-fold faster in aged AL animals, but not in muscles of the aged DR rats. Thus, the proteasomal particles that accumulate with aging appear fully active toward peptides and proteins that do not require ubiquitylation for degradation.
To assess their ability to degrade ubiquitylated proteins, we used a recently developed affinity purification method to isolate 26S proteasomes from any kind of tissue (7). Equal amounts of the triceps surae from seven animals in each group were pooled and homogenized. 26S proteasomes together with associated proteins were isolated in a single step using the ubiquitin-like (Ubl) domain from HHR23B as the affinity ligand (7). To assess their structural integrity, the isolated proteasomes were separated by native gel electrophoresis and analyzed by immunoblotting and by peptidase activity using a substrate overlay assay with Suc-LLVY-AMC (Fig. 5A). 26S proteasomes from adult and aged muscle were indistinguishable on native gels, and no change in the ratio of singly to doubly capped proteasomes was observed. The proteasomes purified from aged animals showed a small but consistent (∼35%) increase in the specific activity of the chymotrypsin-like site (p < 0.05) (Fig. 5B). To evaluate the ability of the particle to degrade ubiquitin conjugates, we used penta-ubiquitylated dihydrofolate reductase Ub5-DHFR (38) as a defined homogeneous globular substrate whose degradation requires ATP hydrolysis. No significant differences in its rate of degradation were found with proteasomes from adult and aged muscles (either AL or DR) (Fig. 5C). Together, these results indicate no functional impairment of the 26S proteasomes of the muscle with aging, in contrast to a prior report (18).
FIGURE 5.
26S proteasomes isolated from aged and adult muscles show similar size distributions and similar abilities to hydrolyze peptide substrates and ubiquitin conjugates. A, gastrocnemius muscles from individual animals were pooled (60 mg each; n = 7) and 26S proteasomes were isolated using the UBL affinity technique (7). Purified proteasomes were analyzed on native PAGE by peptidase activity (Suc-LLVY-AMC overlay assay) and immunoblot. B, the specific activity of purified 26S proteasomes was determined using Suc-LLVY-AMC and expressed relative to the activity of adult proteasomes. The averaged results from three different purifications are shown. C, to assay the degradation of a ubiquitylated protein, 50 nm Ub5DHFR was incubated with 2 nm 26S for 0, 30, 60, and 120 min. The amount of Ub5DHFR was determined by immunoblot and quantified by densitometry. The panel presents averages of two independent measurements, each analyzed by two sets of immunoblots. Error bars are SD. *, p < 0.05.
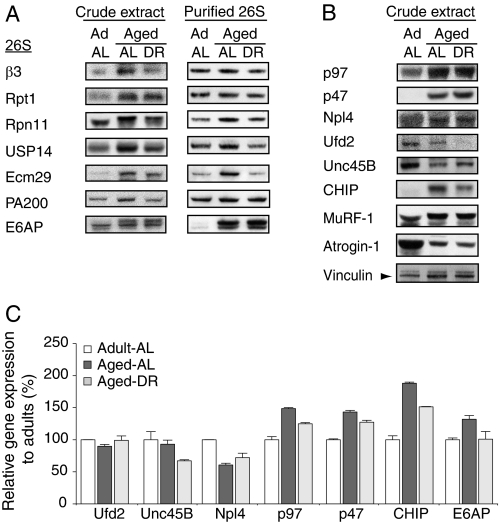
Effects of Aging on Proteasome-associated Proteins
Recently, we identified about 60 proteins associated with the 26S proteasomes that are affinity purified from rat muscle, including several ubiquitin ligases and DUBs (7). To determine if the proteins associated with the proteasome changed with aging, we measured the levels of four important proteasome-associated proteins, USP14, Ecm26, PA200, and E6AP, in muscle extracts and isolated proteasomes. USP14 is a DUB, which also regulates the degradative capacity of the 26S (6, 39). In yeast, Ecm29 was found to stabilize the 19S-20S association (40), and in mammalian cells, Ecm29 is largely found on vesicular structures of the endosomal pathway (41, 42). PA200 (Blm10 in yeast) can form hybrid 26S proteasomes, 19S-20S-PA200, and has been implicated in DNA repair (43). E6AP is an abundant ubiquitin ligase that was recently shown to also ubiquitylate selectively misfolded proteins bound to Hsp70 (44). In aged animals, levels of all four proteins increased in muscle, and with the exception of E6AP, these increases were prevented or reduced by DR (Fig. 6A). Accordingly, proteasomes of aged AL rats were enriched in USP14 and Ecm29 relative to 26S subunits, β3 and Rpt1, but not in muscle proteasomes from the DR group. Remarkably, the content of E6AP, although barely detectable by immunoblot on proteasomes from adult animals, was dramatically increased on the 26S proteasomes from aged rats in both the AL fed and DR groups (Fig. 6A). Thus, proteasomes in adult and aged muscles differ in the levels of associated proteins, and DR can block or reduce most of these changes.
FIGURE 6.
Aged and adult muscles differ in the proteins associated with the 26S proteasome and their contents of ubiquitin ligases and the p97/VCP complex samples. 60 mg of gastrocnemius muscles from 7 animals in each group were pooled and homogenized. Equal amounts of crude extracts (40 μg/lane) and isolated proteasomes (0.4 μg/lane) were analyzed by immunoblot. A, β3 is a 20S subunit, Rpt1 is found in the 19S base, and Rpn11 in the 19S lid. USP14, Ecm29, and PA200 are proteasome-associated proteins and E6AP is a ubiquitin ligase. B, levels of p97 and its cofactor p47 were increased in the aged muscle, whereas its associated ubiquitin ligase Ufd2 and the myofibril assembly factor Unc45B were decreased. CHIP, an E3 that ubiquitylates unfolded proteins, also increased in muscle with aging but less in animals on DR. Atrogin-1/MAFbx levels decreased in aged muscle, whereas the MuRF1 content doubled. Vinculin served as loading control for panels A and B. C, mRNA levels of several proteins assayed in panels A and B determined by qPCR.
The Ubiquitin Ligases CHIP, E6AP, and MuRF1, Increase in Muscle with Aging
A characteristic feature of aged animals is the accumulation in cells of damaged or misfolded proteins (45). Therefore, the induction of E6AP on the proteasome with aging is intriguing in light of its reported ability to selectively ubiquitylate such unfolded proteins (44). We therefore measured the content of the ubiquitin ligase, CHIP, which promotes the selective degradation of unfolded proteins bound to Hsp70 or Hsp90 (46, 47). CHIP was also induced in muscles of aged rats, although, unlike E6AP, it does not associate with the proteasome (Fig. 6, A and B). In extracts of aged muscles, CHIP levels were much higher than in adult muscles, and this age-associated increase was reduced in the DR group (Fig. 6B). The increased content of CHIP appeared to result from changes in gene expression, because the levels of CHIP mRNA were also increased in the aged muscles (Fig. 6C).
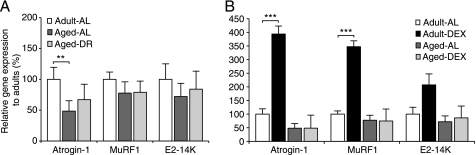
In muscles atrophying due to disuse, fasting, or disease, the expression of the ubiquitin ligases, MuRF1 and Atrogin-1/MAFbx, but not CHIP or E6AP (Fig. S2), is markedly induced and is essential for the rapid loss of muscle weight (2). However, unlike CHIP and E6AP mRNAs, Atrogin-1/MAFbx and MuRF1 expression was not higher in the aged sarcopenic rats, as measured by qPCR (Fig. 7A). At the protein level, Atrogin-1/MAFbx decreased in accord with the fall in mRNA, whereas MuRF1 protein increased in aged rats from both AL fed and DR groups (Fig. 6B). Thus, our findings distinguish the rapid atrophy in adults from the age-associated sarcopenia in contrast to a previous study, which reported an increase in Atrogin-1/MAFbx and MuRF1 mRNA in the tibialis anterior of aged rats (48).
FIGURE 7.
Expression of the ubiquitin ligases, Atrogin-1/MAFbx and MuRF1, and the E2, E2–14K, does not increase in muscles of aged animals and cannot be induced by dexamethasone. Real time PCR analysis of mRNAs encoding Atrogin-1/MAFbx and MuRF1, and the E2 ubiquitin conjugating enzyme E2-14K. A, muscle expression in 4-month and 30-month-old rats fed AL (AL) or DR. B, expression in muscle in 4-month and 30-month-old AL rats treated with dexamethasone and age-matched controls. Individual samples were run in triplicates and expressed relative to adult control samples. Error bars are S.E. **, p < 0.01; ***, p < 0.001.
To further document this difference, the two groups of rats were treated with a high dose of the synthetic glucocorticoid, dexamethasone, which is highly catabolic to muscle. In adult animals both Atrogin-1/MAFbx and MuRF1 were induced. However, no such change occurred in the aged rats after dexamethasone administration (Fig. 7B). These findings are in accord with a prior finding that glucocorticoids fail to stimulate muscle protein breakdown in aged rats as they do in younger animals (16).
The Content of Ubiquitylated Proteins Increases in Aged Muscle Despite Their Increased Content of DUBs and Proteasomes
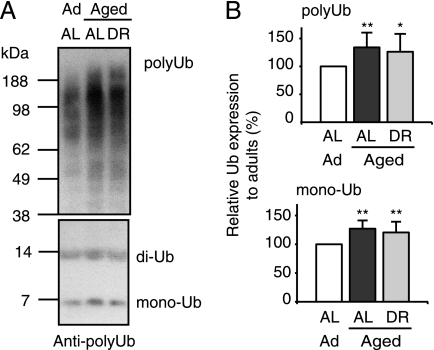
The increased levels of 26S proteasomes and several ubiquitin ligases in the aged muscles suggest an enhanced rate of proteolysis by the UPS. Furthermore, immunoblotting of the muscle extracts with an anti-ubiquitin antibody revealed that the muscles from aged rats contained greater amounts of ubiquitylated proteins as well as slightly more free ubiquitin than those from adult rats (Fig. 8). An increase in free ubiquitin and ubiquitin conjugates with aging was also reported by others in rat muscles (48, 49). Because the 26S proteasomes isolated from aged muscle are fully active in degrading ubiquitylated proteins, the elevated levels of ubiquitylated proteins strongly suggests higher overall rates of protein ubiquitylation that exceed the rates of conjugate degradation.
FIGURE 8.
In aged (sarcopenic) muscle levels of free ubiquitin and ubiquitylated proteins are increased. Equal amounts of crude extracts derived from pooled muscles from 7 animals per group (40 μg/lane; see Fig. 6) were analyzed by immunoblot for poly- and monoubiquitin content. Densitometric analysis of seven is presented to the right, *, p < 0.05; **, p < 0.01.
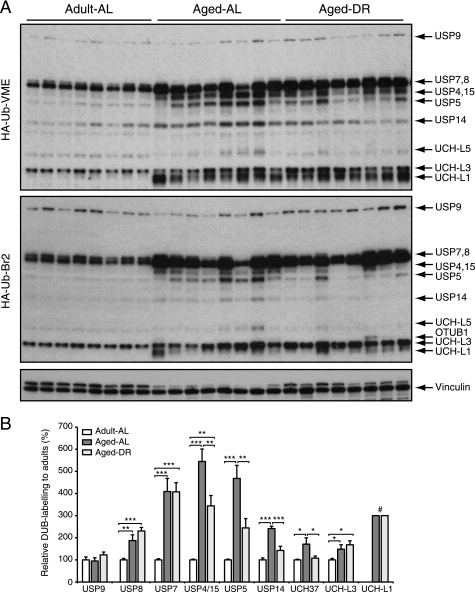
However, increased conjugate levels could also result from decreased rates of protein deubiquitylation. To test if deubiquitylation may be less efficient in muscles of aged animals, we compared the levels of various DUBs in the extracts by activity profiling using ubiquitin-based covalent probes (35). Surprisingly, large increases in the activities of 11 different DUBs were evident in the muscles of aged rats (Fig. 9). The individual DUBs were identified by comparison of the molecular weights of the labeled bands with those in an extract of EL-4 cells, where the bands of derivatized DUBs were identified by mass spectrometry (supplemental Fig. S1) (35). The enzymes that showed the most dramatic increases in activity in the aged muscles were USP7, USP4, and USP15 (Fig. 9). Like 26S proteasomes, the levels of the proteasome-associated DUBs USP14 and UCH37 (7, 35, 50) were elevated in aged animals but suppressed after DR. Accordingly, the affinity-purified proteasomes from aged rats contained more USP14 than those from adults (Fig. 5A).
FIGURE 9.
In aged (sarcopenic) muscle, deubiquitylating activities are increased. Deubiquitylating enzyme activity profiling using two ubiquitin-based probes (HA-Ub-VME and HA-Ub-Br2) that target complementary DUB subsets. The identity of the DUBs was determined by comparison with labeled DUBs identified in EL-4 cell extract; for details see supplemental Fig. S1 (35). Error bars are SD. *, p < 0.05; **, p < 0.01; ***, p < 0.001; # indicates that levels in adults were too low to be quantified.
This surprisingly large increase in the activity of many DUBs with aging, together with the 2–3-fold elevation in 26S proteasomes, should accelerate the breakdown of ubiquitin conjugates. Therefore, the increased content of ubiquitylated proteins in the aged muscles results most likely from higher rates of ubiquitylation (Fig. 8B), perhaps as a consequence of the presence of more damaged proteins and the increases in CHIP, E6AP, and MuRF1. In muscles undergoing rapid atrophy due to fasting or denervation, the total levels of ubiquitin conjugates also increased despite accelerated proteolysis (28). Their accumulation resulted from enhanced rates of ubiquitylation (27), which exceeded their rates of degradation.
The p97/VCP ATPase Complex and Its p47 Cofactor Increase in Aged Muscles
Another critical factor in degradation by the UPS of both damaged cytosolic proteins (24) and misfolded membrane or secretory proteins via the ERAD pathway (51) is the p97/VCP ATPase complex (the homolog of yeast CDC48). This complex also plays a critical anabolic role in myofibril assembly in Caenorhabditis elegans (52). It is noteworthy that levels of p97 were markedly increased in the aged muscle (Fig. 6B) together with a small increase in p97 mRNA (Fig. 6C), and these increases with aging were attenuated by dietary restriction (Fig. 6C). Normally, p97 functions together with a variety of protein cofactors (53, 54). To assess the possible significance of this increase in p97 content with aging, we analyzed the protein and mRNA levels of several of its cofactors reported to function in protein degradation and/or myofibril assembly (Fig. 6C). In C. elegans, the chaperone unc45B catalyzes myosin filament formation and is then degraded in a process involving p97 and the ubiquitin ligases CHIP and Ufd2 (52). The levels of Unc45 were lower in the aged muscles, suggesting a decline in assembly of new myofibrils. Interestingly, DR reduced this decline in Unc45 expression with aging.
The p97 complex functions together with Npl4 in the ERAD pathway (51) and in the breakdown of damaged cytosolic proteins (24). Surprisingly, levels of both Npl4 protein and its mRNA decreased in the aged muscle. On the other hand, another p97 cofactor, p47, was markedly induced in the muscles with aging in both AL fed and DR rats. p47 binds monoubiquitylated proteins and functions in membrane fusion events in dividing cells (54). The marked induction of p97 together with p47 in the sarcopenic but not in DR rats suggests that this particular complex might contribute also to accelerated proteolysis.
DISCUSSION
Although sarcopenia is common in elderly humans and is a well documented phenomenon in aged animals, its underlying mechanisms have not been defined. Such studies are complicated because of changes in the elderly in the pattern of activity, food intake, hormonal levels, and susceptibility to disease, all of which can influence muscle protein content. The rat model studied here showed dramatic muscle wasting, which dietary restriction counteracts (Fig. 1). Although DR alters many age-related metabolic changes, its affects on protein turnover have not been previously investigated. A number of studies of aged humans and rodents have shown decreased rates of protein synthesis in muscle, which was associated with insulin resistance (16) (Fig. 1). Our studies demonstrate a number of unexpected biochemical features of the muscles of sarcopenic rats, especially their increased content of proteasomes and other components of the UPS strongly suggesting enhanced rates of protein degradation.
Accumulation of Proteasomes in Skeletal Muscle with Aging
In the aged rats (Fig. 1) (30, 37), the atrophied hindlimb muscles contained at least 2–3-fold higher levels of 26S proteasomes than those of adult animals (Figs. 2–4). Moreover, after purification, they showed normal activity toward peptides, proteins, and ubiquitin conjugates (Figs. 4 and 5). Elevated 20S proteasome levels were previously reported in muscles from aged F344/BN and LOU rats (17–19, 55), but Ferrington and co-workers (18, 55) found the expression of the 19S (PA700) unchanged. However, in those studies proteasomes were measured after ultracentrifugation in a buffer lacking ATP, which is necessary to maintain the structural integrity of the 26S. Thus, the lack of 19S components is probably due to the method used.
The accumulation of proteasomes in aged muscle is surprising because multiple reports show a decline in expression of proteasome subunits during aging in other tissues (8–13, 56). On the other hand, Dasuri et al. (57) recently reported an increase in 26S subunits in liver but a decline in brain from aged rats. These observations suggest that proteasome levels and presumably rates of proteolysis change in a distinct manner with aging in muscle, which is differentially lost in sarcopenic animals.
Interestingly, elevated 20S levels were also reported in muscles of aged, sarcopenic (22 month) mice. Overexpression of the transcription factor PGC1-α in these mice prevented muscle wasting and suppressed proteasome levels in adult and aged animals (58). In our study, the enhanced capacity of the UPS in muscle appears to be a regulated response, because the increase in proteasome content with age was attenuated by dietary restriction (Fig. 2). However, the underlying mechanisms are quite unclear. For example, the accumulation of proteasomes occurred without any increase in mRNA (Fig. 2), in accord with prior studies in aged muscle (59, 60). By contrast, in muscles atrophying due to disuse, fasting, or various systemic diseases, there is a characteristic set of transcriptional changes including induction of the mRNA for many 26S subunits (61, 62).
Consequently, the accumulation of proteasomes in aged muscle must be due to enhanced subunit translation, more efficient assembly of the 26S particles (although no change in expression of assembly factors was seen), or slower degradation of the 26S particles. Many particles and organelles are eliminated by autophagy, and proteasomal proteins have been detected in lysosomal vacuoles of rat liver (63). Several studies report that the capacity for lysosomal proteolysis is reduced in postmitotic cells with aging (64). We found an accumulation of lipofuscin in aged skeletal muscle fibers,6 which strongly suggests some functional impairment of the lysosomal degradative pathway. In addition, treatment of a rat muscle cell line and adult animals with chloroquine, which impairs lysosomal proteolysis, results in increased proteasome content. In addition, Cuervo and colleagues (63, 64) reported that autophagy declines in liver with aging but this effect is reduced by DR. Activation of autophagy in muscle by DR could explain our findings that DR prevented the increase in proteasomes with age without any change in proteasome mRNA levels.
Effect of Aging on Proteasome Activity in Muscle
There are multiple reports that some of the activities of the proteasome peptidase are reduced in aged tissues through oxidative covalent modifications (8, 11, 67–69) or oxidation of cysteines (18). However, no such defect in degradative capacity with aging was seen in the present study. Instead, our data indicate that proteasome content increases during sarcopenia, and that these particles retain their full ability to function in protein breakdown.
Accumulation of Damaged Proteins with Aging
The accumulation of oxidized or otherwise damaged proteins is a hallmark of aging that is believed to reflect increased generation of oxygen radicals, decreased defenses against reactive oxygen species, or reduced capacity to degrade such proteins (70). Several groups have reported an increased content of oxidatively damaged proteins in aged rodent muscle (48, 49) as well as of the chaperones, Hsp70 (48) and Hsp90 (18). These heat shock proteins selectively bind unfolded proteins and are induced as part of a cellular stress response that is triggered by the buildup of misfolded proteins (71). Together these findings suggest that the enhanced capacity of the UPS may be a response to the increased generation of damaged polypeptides. Strong support for this conclusion was the finding of elevated levels of the ubiquitin ligases, CHIP and E6AP, in muscle of aged animals (Fig. 6). CHIP ubiquitinates misfolded or mutated proteins in the cytosol that are bound to Hsp70 or Hsp90 (46), and recently a similar function in cellular quality control has been reported for E6AP (44). Interestingly, E6AP, unlike CHIP, accumulates on the 26S proteasome in the aged muscle (Fig. 6B). The function of ubiquitin ligases on the proteasome is not known, but in yeast, the HECT E3 ligase, Hul5, on the proteasome appears to promote substrate degradation by extending ubiquitin chains (72). Mutations of E6AP cause severe mental retardation (73), and the present observations suggest that E6AP may be an important regulator of proteasomal degradation of abnormal proteins. In this context, the robust increase in p97 in aged muscle (Fig. 6B) is also of special interest because p97 is essential for both the extraction of misfolded ubiquitylated proteins from the endoplasmic reticulum in the “ERAD” pathway (51) and the elimination of oxidatively damaged proteins in the cytosol (24). Recently, we have found that p97 activity is critical in muscle atrophy and is induced following denervation.7 The p97/VCP complex has also been implicated in defense against neurodegenerative diseases (74), and a lack of this ATPase leads to cytosolic inclusions and dysfunction of multiple organs (75).
Levels of DUBs Increase in Muscles of Aged Rats
In yeast, decreases in the pool of free ubiquitin from the pool can limit the rate of proteolysis by the proteasome (76, 77). Free ubiquitin is in turn released during degradation of ubiquitylated proteins by the DUBs associated with the 26S proteasome. Thus, to achieve high degradation rates, the capacity for deubiquitylation probably needs to be increased also. Surprisingly, 11 DUB enzymes were strongly up-regulated in the muscles of aged rats (Figs. 8 and 9), including USP14 and Uch37, which are known to be associated with the 26S proteasome. USP14 and UCH37 trim off ubiquitin from the polyubiquitin chain and release ubiquitin monomers for re-use. In addition, USP14 controls gate opening into the 20S proteasome and facilitates substrate degradation (39). On the other hand, the yeast homolog of USP14, Ubp6, regulates the overall rate of proteolysis and appears critical in replenishing the free ubiquitin pool in yeast (6, 76). Similarly, USP5 hydrolyzes anchorless ubiquitin chains (78, 79) and appears to work downstream of Rpn11, an intrinsic DUB on the proteasome, to prevent the binding of free ubiquitin chains to the 19S, which would inhibit proteolysis. Together these data illustrate important roles for deubiquitylation in the aged muscle, probably ensuring a supply of ubiquitin for enhanced proteolysis but perhaps also serving additional regulatory functions.
Differences between Sarcopenia and Rapid Atrophy Due to Inactivity or Disease
Loss of muscle mass can occur through increased protein degradation but also decreased protein synthesis or through some combination of these responses. With aging, sarcopenia develops over months to years depending on the species, unlike the rapid loss of muscle weight induced by fasting, disuse, and in various catabolic diseases, where marked atrophy (20–50% loss) can occur in rodents in several days. In these types of rapid atrophy there is a common program of changes in the transcription of a set of atrophy-related genes (“atrogenes”) (2, 61, 62). The various biochemical changes observed here clearly distinguish the muscles of aged rats from those undergoing rapid atrophy in adult animals. Upon denervation or fasting, the atrophy-specific ubiquitin ligases, Atrogin-1/MAFbx and MuRF1, are induced by members of the FOXO family of transcription factors, and this induction is essential for the rapid weight loss (2, 80, 81). Inhibition of FOXOs prevents their induction and the loss of muscle mass upon denervation, fasting, or glucocorticoid treatment (65, 66, 81). In contrast, in the aged muscles, mRNAs for Atrogin-1/MAFbx, MuRF1, and the Ub-conjugating enzyme, E2-14K, were unchanged or lower than adult levels (Fig. 7A) (36). Also, treatment with the glucocorticoid, dexamethasone (Fig. 7B), failed to induce Atrogin-1/MAFbx or MuRF1 or to cause muscle wasting, as it does in adult animals (80, 82). However, MuRF1 protein increased in aged muscle from both AL fed and DR groups (Fig. 6B), whereas Atrogin-1/MAFbx protein decreased (in accord with the mRNA data). Unlike these atrogenes, the ubiquitin ligases, CHIP and E6AP, increased markedly in the aged muscle, although they do not rise in rapidly atrophying muscles (supplemental Fig. S2). Their induction may reflect adaptations in the aged muscle to eliminate more efficiently misfolded proteins.
Transcriptional profiling (59, 60) and mass spectrometry (31) of muscles from aged rodents showed a reduced expression of myofibrillar components, which presumably reflects decreased protein synthesis in aged muscle (83–85). The finding of increased content of proteasomes and other UPS components (e.g. MuRF1) argues that proteolysis also increases in these muscles and may contribute to muscle wasting in the aged rats. DR decreased levels of proteasomes and several other UPS components toward the levels observed in adult animal and partially inhibited the development of sarcopenia (Fig. 1B). The modest dietary restriction (70% of AL) administered here slows but does not prevent the development of sarcopenia (Fig. 1). In related work, we found that 36-month-old rats maintained on DR eventually showed the same extent of muscle wasting as 30-month-old rats fed AL.5
In summary, the present findings do not support the widespread view that with aging, the capacity of the organism for protein degradation declines irreversibly. At least for skeletal muscle, when sarcopenia is dramatic, the muscles showed multiple indications of enhanced activity of the ubiquitin proteasome system. The generality of these changes to other tissues of aged animals, especially the central nervous system, will be important to explore to distinguish general features of aged tissues from muscle-specific responses that are part of the pathogenesis of sarcopenia. Because muscle appears to decrease in mass by distinct mechanisms in aging and during atrophy induced by disuse, fasting, or systemic disease, if the latter conditions develop in an aged sarcopenic organism, these distinct mechanisms could have additive effects in causing further loss of muscle mass and functional impairment.
Supplementary Material
Acknowledgments
We thank Dr. Huib Ovaa for a generous supply of the proteasome-specific probe dansyl-Ahx3L3VS and the DUB-specific probe HA-Ub-VME. We are also grateful to Dr. M. Ingelman-Sundberg for use of the ultracentrifuge and M. Dethavong for valuable assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM51923 from the NIGMS and AR055255 from the NIA (to H. C. B. and A. L. G.), grants from the Ellison Foundation (to H. C. B. and A. L. G.), the Swedish MRC Project 10800 and the M & M Wallenbergs Foundation (to B. U.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
M. Grönholdt-Klein, M. Altun, A. Fahlström, and B. Ulfhake, manuscript in preparation.
M. Altun and B. Ulfhake, unpublished observations.
R. Piccirillo and A. L. Goldberg, unpublished data.
- UPS
- ubiquitin proteasome system
- DUB
- deubiquitylating enzyme
- DR
- dietary restriction
- DHFR
- dihydrofolate reductase
- dansyl
- 5-(dimethylamino)naphthalene-1-sulfonyl.
REFERENCES
- 1.Evans W. J. (1995) J. Gerontol. A Biol. Sci. Med. Sci. 50, 5–8 [DOI] [PubMed] [Google Scholar]
- 2.Lecker S. H., Goldberg A. L., Mitch W. E. (2006) J. Am. Soc. Nephrol. 17, 1807–1819 [DOI] [PubMed] [Google Scholar]
- 3.Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. (1994) Cell 78, 761–771 [DOI] [PubMed] [Google Scholar]
- 4.Voges D., Zwickl P., Baumeister W. (1999) Annu. Rev. Biochem. 68, 1015–1068 [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover A. (2005) Nat. Rev. Mol. Cell Biol. 6, 79–87 [DOI] [PubMed] [Google Scholar]
- 6.Hanna J., Meides A., Zhang D. P., Finley D. (2007) Cell 129, 747–759 [DOI] [PubMed] [Google Scholar]
- 7.Besche H., Haas W., Gygi S., Goldberg A. (2009) Biochemistry 48, 2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulteau A. L., Petropoulos I., Friguet B. (2000) Exp. Gerontol. 35, 767–777 [DOI] [PubMed] [Google Scholar]
- 9.Bulteau A. L., Szweda L. I., Friguet B. (2002) Arch. Biochem. Biophys. 397, 298–304 [DOI] [PubMed] [Google Scholar]
- 10.Keller J. N., Hanni K. B., Markesbery W. R. (2000) Mech. Ageing Dev. 113, 61–70 [DOI] [PubMed] [Google Scholar]
- 11.Keller J. N., Huang F. F., Markesbery W. R. (2000) Neuroscience 98, 149–156 [DOI] [PubMed] [Google Scholar]
- 12.Petropoulos I., Conconi M., Wang X., Hoenel B., Brégégère F., Milner Y., Friguet B. (2000) J. Gerontol. A Biol. Sci. Med. Sci. 55, B220–227 [DOI] [PubMed] [Google Scholar]
- 13.Zhang L., Li F., Dimayuga E., Craddock J., Keller J. N. (2007) FEBS Lett. 581, 5543–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecker S. H., Solomon V., Mitch W. E., Goldberg A. L. (1999) J. Nutr. 129, 227S–237S [DOI] [PubMed] [Google Scholar]
- 15.Mitch W. E., Goldberg A. L. (1996) N. Engl. J. Med. 335, 1897–1905 [DOI] [PubMed] [Google Scholar]
- 16.Attaix D., Mosoni L., Dardevet D., Combaret L., Mirand P. P., Grizard J. (2005) Int. J. Biochem. Cell Biol. 37, 1962–1973 [DOI] [PubMed] [Google Scholar]
- 17.Bardag-Gorce F., Farout L., Veyrat-Durebex C., Briand Y., Briand M. (1999) Mol. Biol. Rep. 26, 89–93 [DOI] [PubMed] [Google Scholar]
- 18.Ferrington D. A., Husom A. D., Thompson L. V. (2005) FASEB J. 19, 644–646 [DOI] [PubMed] [Google Scholar]
- 19.Hepple R. T., Qin M., Nakamoto H., Goto S. (2008) Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R1231–1237 [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Vicente M., Sovak G., Cuervo A. M. (2005) Exp. Gerontol. 40, 622–633 [DOI] [PubMed] [Google Scholar]
- 21.Selsby J. T., Judge A. R., Yimlamai T., Leeuwenburgh C., Dodd S. L. (2005) Exp. Gerontol. 40, 37–42 [DOI] [PubMed] [Google Scholar]
- 22.Whitman S. A., Wacker M. J., Richmond S. R., Godard M. P. (2005) Pflugers Arch. 450, 437–446 [DOI] [PubMed] [Google Scholar]
- 23.Jariel-Encontre I., Bossis G., Piechaczyk M. (2008) Biochim. Biophys. Acta 1786, 153–177 [DOI] [PubMed] [Google Scholar]
- 24.Medicherla B., Goldberg A. L. (2008) J. Cell Biol. 182, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goto S., Takahashi R., Kumiyama A. A., Radák Z., Hayashi T., Takenouchi M., Abe R. (2001) Ann. N.Y. Acad. Sci. 928, 54–64 [DOI] [PubMed] [Google Scholar]
- 26.Goldberg A. L. (2003) Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 27.Solomon V., Baracos V., Sarraf P., Goldberg A. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12602–12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wing S. S., Haas A. L., Goldberg A. L. (1995) Biochem. J. 307, 639–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piper M. D., Bartke A. (2008) Cell Metab. 8, 99–104 [DOI] [PubMed] [Google Scholar]
- 30.Altun M., Bergman E., Edström E., Johnson H., Ulfhake B. (2007) Physiol. Behav. 92, 911–923 [DOI] [PubMed] [Google Scholar]
- 31.Altun M., Edström E., Spooner E., Flores-Moralez A., Bergman E., Tollet-Egnell P., Norstedt G., Kessler B. M., Ulfhake B. (2007) Muscle Nerve 36, 223–233 [DOI] [PubMed] [Google Scholar]
- 32.Berkers C. R., Verdoes M., Lichtman E., Fiebiger E., Kessler B. M., Anderson K. C., Ploegh H. L., Ovaa H., Galardy P. J. (2005) Nat. Methods 2, 357–362 [DOI] [PubMed] [Google Scholar]
- 33.Kessler B. M., Tortorella D., Altun M., Kisselev A. F., Fiebiger E., Hekking B. G., Ploegh H. L., Overkleeft H. S. (2001) Chem. Biol. 8, 913–929 [DOI] [PubMed] [Google Scholar]
- 34.Kisselev A. F., Akopian T. N., Woo K. M., Goldberg A. L. (1999) J. Biol. Chem. 274, 3363–3371 [DOI] [PubMed] [Google Scholar]
- 35.Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K. D., Ploegh H. L., Kessler B. M. (2002) Chem. Biol. 9, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 36.Edström E., Altun M., Hägglund M., Ulfhake B. (2006) J. Gerontol. A Biol. Sci. Med. Sci. 61, 663–674 [DOI] [PubMed] [Google Scholar]
- 37.Edström E., Ulfhake B. (2005) Aging Cell 4, 65–77 [DOI] [PubMed] [Google Scholar]
- 38.Lam Y. A., Huang J. W., Showole O. (2005) Methods Enzymol. 398, 379–390 [DOI] [PubMed] [Google Scholar]
- 39.Peth A., Besche H. C., Goldberg A. L. (2009) Mol. Cell 36, 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleijnen M. F., Roelofs J., Park S., Hathaway N. A., Glickman M., King R. W., Finley D. (2007) Nat. Struct. Mol. Biol. 14, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 41.Gorbea C., Goellner G. M., Teter K., Holmes R. K., Rechsteiner M. (2004) J. Biol. Chem. 279, 54849–54861 [DOI] [PubMed] [Google Scholar]
- 42.Gorbea C., Pratt G., Ustrell V., Bell R., Sahasrabudhe S., Hughes R. E., Rechsteiner M. (2010) J. Biol. Chem. 285, 31616–31633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt M., Haas W., Crosas B., Santamaria P. G., Gygi S. P., Walz T., Finley D. (2005) Nat. Struct. Mol. Biol. 12, 294–303 [DOI] [PubMed] [Google Scholar]
- 44.Mishra A., Godavarthi S. K., Maheshwari M., Goswami A., Jana N. R. (2009) J. Biol. Chem. 284, 10537–10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guarente L. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 483–488 [DOI] [PubMed] [Google Scholar]
- 46.Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) Nat. Cell Biol. 3, 93–96 [DOI] [PubMed] [Google Scholar]
- 47.Murata S., Chiba T., Tanaka K. (2003) Int. J. Biochem. Cell Biol. 35, 572–578 [DOI] [PubMed] [Google Scholar]
- 48.Clavel S., Coldefy A. S., Kurkdjian E., Salles J., Margaritis I., Derijard B. (2006) Mech. Ageing Dev 127, 794–801 [DOI] [PubMed] [Google Scholar]
- 49.Cai D., Lee K. K., Li M., Tang M. K., Chan K. M. (2004) Arch. Biochem. Biophys. 425, 42–50 [DOI] [PubMed] [Google Scholar]
- 50.Stone M., Hartmann-Petersen R., Seeger M., Bech-Otschir D., Wallace M., Gordon C. (2004) J. Mol. Biol. 344, 697–706 [DOI] [PubMed] [Google Scholar]
- 51.Raasi S., Wolf D. H. (2007) Semin. Cell Dev. Biol. 18, 780–791 [DOI] [PubMed] [Google Scholar]
- 52.Kim J., Löwe T., Hoppe T. (2008) Trends Cell Biol. 18, 264–272 [DOI] [PubMed] [Google Scholar]
- 53.Alexandru G., Graumann J., Smith G. T., Kolawa N. J., Fang R., Deshaies R. J. (2008) Cell 134, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuberth C., Buchberger A. (2008) Cell Mol. Life Sci. 65, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Husom A. D., Peters E. A., Kolling E. A., Fugere N. A., Thompson L. V., Ferrington D. A. (2004) Arch. Biochem. Biophys. 421, 67–76 [DOI] [PubMed] [Google Scholar]
- 56.Zeng B. Y., Medhurst A. D., Jackson M., Rose S., Jenner P. (2005) Mech. Ageing Dev. 126, 760–766 [DOI] [PubMed] [Google Scholar]
- 57.Dasuri K., Zhang L., Ebenezer P., Liu Y., Fernandez-Kim S. O., Keller J. N. (2009) Mech Ageing Dev. 130, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wenz T., Rossi S. G., Rotundo R. L., Spiegelman B. M., Moraes C. T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20405–20410 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Pattison J. S., Folk L. C., Madsen R. W., Childs T. E., Booth F. W. (2003) Physiol. Genomics 15, 34–43 [DOI] [PubMed] [Google Scholar]
- 60.Welle S., Brooks A. I., Delehanty J. M., Needler N., Bhatt K., Shah B., Thornton C. A. (2004) Exp. Gerontol. 39, 369–377 [DOI] [PubMed] [Google Scholar]
- 61.Lecker S. H., Jagoe R. T., Gilbert A., Gomes M., Baracos V., Bailey J., Price S. R., Mitch W. E., Goldberg A. L. (2004) FASEB J. 18, 39–51 [DOI] [PubMed] [Google Scholar]
- 62.Sacheck J. M., Hyatt J. P., Raffaello A., Jagoe R. T., Roy R. R., Edgerton V. R., Lecker S. H., Goldberg A. L. (2007) FASEB J. 21, 140–155 [DOI] [PubMed] [Google Scholar]
- 63.Cuervo A. M., Palmer A., Rivett A. J., Knecht E. (1995) Eur. J. Biochem. 227, 792–800 [DOI] [PubMed] [Google Scholar]
- 64.Cuervo A. M. (2008) Trends Genet. 24, 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S. J., Di Lisi R., Sandri C., Zhao J., Goldberg A. L., Schiaffino S., Sandri M. (2007) Cell Metab. 6, 458–471 [DOI] [PubMed] [Google Scholar]
- 66.Zhao J., Brault J. J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S. H., Goldberg A. L. (2007) Cell Metab. 6, 472–483 [DOI] [PubMed] [Google Scholar]
- 67.Conconi M., Szweda L. I., Levine R. L., Stadtman E. R., Friguet B. (1996) Arch. Biochem. Biophys. 331, 232–240 [DOI] [PubMed] [Google Scholar]
- 68.Grune T., Shringarpure R., Sitte N., Davies K. (2001) J. Gerontol. A Biol. Sci. Med. Sci. 56, B459–467 [DOI] [PubMed] [Google Scholar]
- 69.Hayashi T., Goto S. (1998) Mech. Ageing Dev. 102, 55–66 [DOI] [PubMed] [Google Scholar]
- 70.Friguet B. (2006) FEBS Lett. 580, 2910–2916 [DOI] [PubMed] [Google Scholar]
- 71.Goff S. A., Goldberg A. L. (1985) Cell 41, 587–595 [DOI] [PubMed] [Google Scholar]
- 72.Crosas B., Hanna J., Kirkpatrick D. S., Zhang D. P., Tone Y., Hathaway N. A., Buecker C., Leggett D. S., Schmidt M., King R. W., Gygi S. P., Finley D. (2006) Cell 127, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 73.Matsuura T., Sutcliffe J. S., Fang P., Galjaard R. J., Jiang Y. H., Benton C. S., Rommens J. M., Beaudet A. L. (1997) Nat. Genet. 15, 74–77 [DOI] [PubMed] [Google Scholar]
- 74.Hirabayashi M., Inoue K., Tanaka K., Nakadate K., Ohsawa Y., Kamei Y., Popiel A. H., Sinohara A., Iwamatsu A., Kimura Y., Uchiyama Y., Hori S., Kakizuka A. (2001) Cell Death Differ. 8, 977–984 [DOI] [PubMed] [Google Scholar]
- 75.Watts G. D., Wymer J., Kovach M. J., Mehta S. G., Mumm S., Darvish D., Pestronk A., Whyte M. P., Kimonis V. E. (2004) Nat. Genet. 36, 377–381 [DOI] [PubMed] [Google Scholar]
- 76.Hanna J., Leggett D. S., Finley D. (2003) Mol. Cell. Biol. 23, 9251–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kimura Y., Yashiroda H., Kudo T., Koitabashi S., Murata S., Kakizuka A., Tanaka K. (2009) Cell 137, 549–559 [DOI] [PubMed] [Google Scholar]
- 78.Reyes-Turcu F. E., Horton J. R., Mullally J. E., Heroux A., Cheng X., Wilkinson K. D. (2006) Cell 124, 1197–1208 [DOI] [PubMed] [Google Scholar]
- 79.Reyes-Turcu F. E., Shanks J. R., Komander D., Wilkinson K. D. (2008) J. Biol. Chem. 283, 19581–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., Pan Z. Q., Valenzuela D. M., DeChiara T. M., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Science 294, 1704–1708 [DOI] [PubMed] [Google Scholar]
- 81.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004) Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gomes M. D., Lecker S. H., Jagoe R. T., Navon A., Goldberg A. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balagopal P., Rooyackers O. E., Adey D. B., Ades P. A., Nair K. S. (1997) Am. J. Physiol. 273, E790–800 [DOI] [PubMed] [Google Scholar]
- 84.Dardevet D., Sornet C., Taillandier D., Savary I., Attaix D., Grizard J. (1995) J. Clin. Invest. 96, 2113–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Welle S., Thornton C., Jozefowicz R., Statt M. (1993) Am. J. Physiol. 264, E693–698 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.