Abstract
Replication fork protection complex Swi1-Swi3 and replication checkpoint mediator Mrc1 are required for maintenance of replication fork integrity during the course of DNA replication in the fission yeast Schizosaccharomyces pombe. These proteins play crucial roles in stabilizing stalled forks and activating replication checkpoint signaling pathways. Although they are conserved replication fork components, precise biochemical roles of these proteins are not known. Here we purified Mrc1 and Swi1-Swi3 proteins and show that these proteins bind to DNA independently but synergistically in vitro. Mrc1 binds preferentially to arrested fork or D-loop-like structures, although the affinity is relatively low, whereas the Swi1-Swi3 complex binds to double-stranded DNA with higher affinity. In the presence of a low concentration of Swi1-Swi3, Mrc1 generates a novel ternary complex and binds to various types of DNA with higher affinity. Moreover, purified Mrc1 and Swi1-Swi3 physically interact with each other, and this interaction is lost by mutations in the known DNA binding domain of Mrc1 (K235E,K236E). The interaction is also lost in a mutant form of Swi1 (E662K) that is specifically defective in polar fork arrest at a site called RTS1 and causes sensitivity to genotoxic agents, although the DNA binding affinity of Swi1-Swi3 is not affected by this mutation. As expected, the synergistic effect of the Swi1-Swi3 on DNA binding of Mrc1 is also lost by these mutations affecting the interaction between Mrc1 and Swi1-Swi3. Our results reveal an aspect of molecular interactions that may play an important role in replication pausing and fork stabilization.
Keywords: Cell Cycle, DNA-binding Protein, DNA-Protein Interaction, DNA Replication, Protein-Protein Interactions, Fork Protection Complex, Fork Stabilization, Replication Fork
Introduction
Living cells continually suffer damages from exogenous and endogenous genotoxic agents, and the progression of DNA replication forks can often be impeded by such obstacles. In addition, forks can also pause transiently during the normal process of chromosome replication, upon encountering sites where non-nucleosomal proteins are bound tightly (1, 2). Failure to stabilize such stalled forks can result in their collapse; therefore, strict monitoring of ongoing forks is crucial for genome integrity. To prevent fork collapse and to ultimately complete replication, stalled forks need to be dealt with swiftly by cellular surveillance mechanisms.
The DNA replication checkpoint (S-phase checkpoint) is one of the mechanisms that suppress replication initiation or elongation in response to fork stall, and many components of the DNA damage response are required either for the stabilization of or for restart of stalled forks (3–5). This slowing of S-phase in response to DNA damage is often called the intra-S-phase checkpoint to distinguish this from other checkpoints in S-phase, such as S-M checkpoint, which suppress entry into mitosis until DNA replication is completed (3, 6, 7). In the fission yeast Schizosaccharomyces pombe, the protein kinase Cds1 is a critical effector of the replication checkpoint, which is required both to stabilize stalled forks and to help prevent premature entry into mitosis (6). Cds1 activation is restricted to S-phase and specifically depends on the replication checkpoint mediator Mrc1 (8). The budding yeast Mrc1 protein and other orthologs of Mrc1 in Xenopus laevis and humans (known as Claspin) are also required for checkpoint activation at stalled forks, indicating that this aspect of Mrc1 function has been conserved throughout evolution (9–11). Interestingly, recent genetic studies suggest checkpoint-independent functions of Mrc1 (12, 13). However, the detailed mechanism remains unclear. Biochemically, Sar et al. (14) have reported that the purified human Claspin binds preferentially to branched DNA molecules in vitro. Similarly, in fission yeast, Zhao and Russell (15) have reported that the N-terminal half of the fission yeast Mrc1 binds preferentially to branched DNA in vitro, and the DNA binding domain has been identified. These results suggest that Claspin/Mrc1 directly interacts with various branched structures of DNA associated with fork arrest.
It was proposed that Swi1 and Swi3 form a “replication fork protection complex,” which is required for stabilization of stalled forks in fission yeast (16). The Swi1-Swi3 complex is also required for promotion of imprinting by a programmed fork-pausing event, which is necessary for switching at the mating-type locus mat1 (16, 17) and for prevention of fork collapse at natural fork barriers in the spacer regions of ribosomal DNA (rDNA)3 repeats (16, 18, 19). However, differential functions of Swi1 action at these pausing/arrest sites have been suggested because a swi1 mutant (E662K), which cannot cause fork arrest at the mat1-proximal polar replication terminator RTS1, still is able to cause fork pausing/arrest at the imprinting site MPS1 (mat1 pause site 1) and the natural fork barriers in rDNA sites Ter1–Ter3 (17, 19). Moreover, Swi1-Swi3 is required for efficient activation of Cds1 (16). However, because Cds1 is not required for mating-type switching, Swi1 has both Cds1-dependent and -independent activities (18). The Swi1-Swi3 complex is evolutionarily conserved and is homologous to Tof1-Csm3 complex in budding yeast and Tim (Timeless)-Tipin complex in humans (16, 20–23). Also in budding yeast, Tof1-Csm3 is required for coordinated arrest of DNA synthesis (24, 25) and contributes to Rad53 (Cds1 homolog) activation (26), and similar roles exist in humans (20–23). Interestingly, recent studies in yeasts have suggested that Mrc1 and Swi1-Swi3/Tof1-Csm3 function in promotion of DNA replication at hard-to-replicate segments or under some genetic backgrounds with initiation deficiency or even under normal growth conditions without involving their checkpoint function (13, 27, 28). Tim-Tipin and Swi1-Swi3 have also been suggested to be required for chromatin loading of Claspin in human cells (23), in Xenopus egg extracts (29), and in fission yeast cells (30), but their biochemical properties have not been characterized in detail.
Swi1-Swi3 and Mrc1 are likely to be parts of the DNA replication machinery. Indeed, in budding yeast, Xenopus egg extracts, and mammalian cells, the homologs of these proteins have been reported to interact with the replication factors, such as Cdc45-MCM2–7-GINS complex, which is thought to act as the DNA helicase responsible for unwinding the parental DNA duplex at DNA replication forks (20, 21, 24, 29, 31, 32). In budding yeast, Mrc1 and Tof1 are needed to prevent uncoupling of the replication machinery from DNA synthesis when replication is arrested (24), and this may be mediated by interaction of Mrc1 with MCM (minichromosome maintenance) or DNA polymerase ϵ (33, 34). Physical interaction of Tof1-Csm3 with Mrc1 was also reported (35). Also in fission yeast, Mrc1 has been suggested to negatively regulate Cdc45 and MCM helicase for recovery from replication block (12). However, little is known about the nature of the interaction between Mrc1 and Swi1-Swi3 (or these homologs) or about how this interaction could contribute to their actions at the fork. Although Mrc1 and Claspin have been shown to bind to DNA (14, 15), biochemical properties of the Swi1-Swi3 complex are not known. Therefore, analyses of Mrc1-Swi1-Swi3 interactions and their effects on the properties of these proteins would be important to unravel the molecular actions of these proteins.
In this study, we purified fission yeast Mrc1 and Swi1-Swi3 proteins and characterized their biochemical properties in vitro. Our results show that Swi1-Swi3 possesses double-stranded DNA binding activity and facilitates the DNA binding of Mrc1 to generate a ternary complex on double-stranded DNA. Direct interaction between Mrc1 and Swi1-Swi3 in vitro was shown by coimmunoprecipitation experiments. Previously identified DNA binding mutants of Mrc1 (K235E,K236E and Δ221–228) (15) do not form the ternary complexes with Swi1-Swi3. These mutants do not interact with Swi1-Swi3, suggesting that the Swi1-Swi3-interacting domain may overlap with the DNA binding domain. Also, the previously identified RTS1 arrest-defective mutant of Swi1 (E662K) (17) forms a complex with Swi3, but this mutant neither forms the ternary complex nor interacts with Mrc1, although its DNA binding affinity is not affected. We have also generated other mutants that are defective in checkpoint responses and characterized these proteins. On the basis of the results presented, we will discuss modes of interaction of Mrc1 and Swi1-Swi3 proteins with DNA.
EXPERIMENTAL PROCEDURES
Expression Vectors for Fission Yeast Mrc1 and Swi1-Swi3
For the expression of Mrc1 in Escherichia coli cells, full-length Mrc1-FLAG (tagged at the C terminus) was cloned at the BamHI sites of pT7–7/pQE30, resulting in generation of RGS-His6-Mrc1-FLAG. For the expression of Mrc1 Δ221–283 or Mrc1-K235E,K236E in E. coli, the NsiI-PvuII fragment of the wild-type Mrc1 was replaced by the same fragment from pKT753 (Δ221–283) or pKT754 (K235E,K236E) (kindly provided by Dr. Paul Russell at the Scripps Institute), respectively. For the coexpression of Swi1 and Swi3, full-length Swi1-FLAG (obtained from pREP41-Swi1-FLAG) and Swi3 were cloned at the NdeI/KpnI and the BamHI/PstI of pETDuet-1 vector (Novagen), respectively. Mutants of Swi proteins were constructed by the QuikChange multisite-directed mutagenesis kit (Stratagene) on pETDuet-1 vector harboring wild-type swi1 and swi3 as a template. Oligonucleotide primers used for amplification are listed in Table 1. The sequences of the cloned segments were confirmed by sequencing. For expression, BL21-CodonPlus-RP cells (Stratagene) transformed with each expression plasmid were grown at 37 °C, and the expression was induced overnight at 18 °C after the addition of isopropyl 1-thio-β-d-galactopyranoside (1 mm for Mrc1 or 0.1 mm for Swi1-Swi3).
TABLE 1.
Oligonucleotide primers used for construction of mutant proteins
a The sequence of this oligonucleotide is derived from anti-sense sequence; all others are derived from sense sequence.
Protein Purification
Two liters of E. coli cells expressing Mrc1 or Swi1-Swi3 were harvested, washed once with 60 ml of ice-cold phosphate-buffered saline, and then resuspended in 120 ml of buffer A (40 mm Hepes-KOH (pH 7.6), 100 mm potassium glutamate, 1 mm EDTA, 10% glycerol, 1 mm DTT, and protease inhibitor mixture (1 tablet for 50–100 ml of solution; Roche Applied Science). After incubation on ice for 5 min, the cells were lysed by sonication. The supernatant was collected by centrifugation at 15,000 rpm at 4 °C for 30 min and then mixed with 3 ml of Ni2+-NTA Superflow (Qiagen) pre-equilibrated with buffer A. The mixtures were incubated at 4 °C for 1.5 h on a rotator. The beads were collected, washed three times with 100 ml of buffer A and once with 30 ml of buffer A containing 30 mm imidazole, and then resuspended in buffer B (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.05% Triton X-100, and protease inhibitor). Bound protein was eluted with 15 ml of buffer B containing 150 mm imidazole. The eluted fraction was mixed with 0.5 ml of anti-FLAG M2 affinity gel (Sigma) equilibrated with buffer B and incubated at 4 °C for 1.5 h on a rotator. After washing three times with 15 ml of buffer B, bound protein was eluted four times by incubation at 4 °C for 20 min each with 0.5 ml of buffer B containing 0.4 mg/ml FLAG peptide. The eluted fraction was further purified by a MonoQ PC 1.6/5 column (GE Healthcare) using the SMART system (GE Healthcare). Prior to assays, a portion of the MonoQ fractions was dialyzed against 10 mm Tris-HCl (pH 7.0) for 3 h at 4 °C.
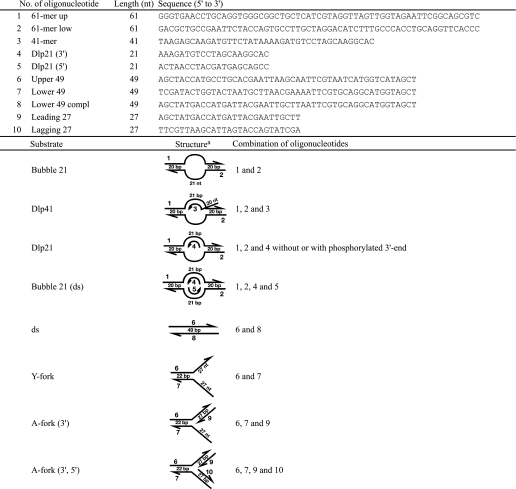
DNA Substrates
Forked, bubble, or D-loop substrates were constructed by annealing oligonucleotides as indicated in Table 2. The 5′-end of an oligonucleotide indicated was phosphorylated by T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (GE Healthcare) prior to annealing. Each substrate was separated by native polyacrylamide gel electrophoresis and isolated and purified as described previously (36, 37).
TABLE 2.
Oligonucleotides used for preparation of substrate DNAs and schematic drawing of their structures
a The numbers in the right column are those of the oligonucleotides used. The direction of the arrow is from 5′ to 3′. nt, nucleotide.
Antibodies
Anti-Mrc1 or anti-Swi3 antibody was developed in rabbits using the N-terminal 180-amino acid Mrc1 (30) or full-length (181-amino acid) Swi3 polypeptide as antigen, respectively. IgG was purified from antisera using a Protein A affinity column. Anti-FLAG M2 or anti-penta-His antibody was purchased from Sigma or Qiagen, respectively.
Gel Shift Assays
Reactions were conducted as described previously (37). After the run, gels were dried and autoradiographed. When two or more proteins were present in the reaction mixtures, they were mixed prior to the addition to the reaction. DNA substrate was added at a concentration of 4 nm. Quantification of the band intensities was performed by Multi Gauge software version 3.1 (Fuji Photofilm).
Nuclease Footprinting
Reactions were performed as described previously (38). 0.002 units of DNase I (TAKARA) or 0.04 units of P1 nuclease (Roche Applied Science) was added in each reaction. The samples were run on 10% denaturing polyacrylamide gel electrophoresis.
Immunoprecipitation Assay
Immunoprecipitation assays were conducted in 100 μl of reaction mixtures containing IP buffer (50 mm Tris-HCl (pH 7.0), 5 mm EDTA, 1% Triton X-100, 1 mm DTT, and 100 mm NaCl) or buffer A (40 mm Hepes/KOH (pH 7.6), 40 mm potassium glutamate, 1 mm EDTA, 1 mm DTT, 8 mm magnesium acetate, 1 mm CaCl2, and 10% glycerol) using IgG purified from anti-Mrc1 serum. Reactions normally contained 200 ng of Mrc1 and 100 ng of Swi1-Swi3, unless otherwise stated, and were incubated at 25 °C for 10 min. Fifty fmol of double-stranded DNA (49 bp in length) were added where indicated. In reactions without DNase I digestion, IP buffer was used. In reactions involving DNase I digestion, buffer A was used, and DNA was digested with 0.2 units of DNase I at 37 °C for 5 min, before the addition of the antibody. Following incubation with the antibody at 4 °C for 1 h, Protein A-Sepharose beads (GE Healthcare) was added, and incubation was continued at 4 °C for 1 h with rotation. Beads were washed six times (4 min each) with 1 ml of IP buffer at room temperature. Bound proteins were eluted from the beads with 20 μl of 2 × SDS sample buffer, and 50% of the eluted samples were run on 10% SDS-PAGE unless otherwise stated. The gel was visualized by silver staining or was transferred to nylon membrane for Western blotting analyses. Band intensity was quantified by LAS 3000 Multi Gauge software (Fuji Photofilm).
Native Gel Electrophoresis
In order to analyze protein-protein interactions, reaction mixtures were directly applied onto 6% native polyacrylamide gel (acrylamide/bisacrylamide = 29:1). Reactions were similar to those used in gel shift assays. Mrc1 and Swi1-Swi3 were present at 64 and 128 nm, respectively, and non-radiolabeled Y-fork substrate was used at a concentration of 40 nm. After electrophoresis, gel was silver-stained to visualize proteins.
Measurements of Sensitivities of Yeast Mutant Strains to Hydroxyurea
swi1Δ (EN3182) or swi3Δ (EN3366) cells harboring pREP41 plasmids expressing various swi1 or swi3 mutants, respectively, were cultured to exponential phase in EMM + uracil, and 5-fold dilution series of the cells were spotted on EMM + uracil with or without thiamine plates containing 0, 3, 5, 7, or 9 mm hydroxyurea (HU) and incubated at 25 and 30 °C. Fission yeast strains used in this study were YM71 (h− leu1-32 ura4-D18), EN3182 (h− leu1-32 ura4-D18 swi1::kan), EN3366 (h− leu1-32 ura4-D18 swi3::kan), Y393 (h90 leu1-32 ura4-D18), Y400 (h90 leu1-32 ura4-D18 swi1::kan), E111 (h90 his2 ade5-210 swi1-111), and JZ277 (h90 his2 ade6-M210 swi1-E662K).
RESULTS
Binding of Mrc1 Protein to D-loop or Bubble-like Structures
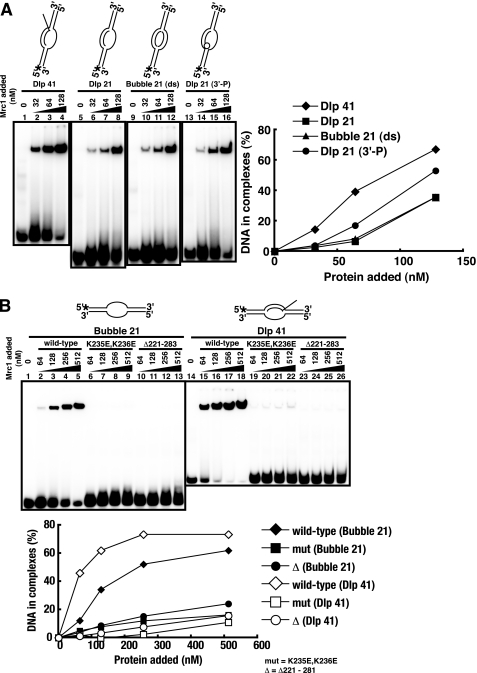
The full-length Mrc1 protein, tagged with His6 and FLAG at the N terminus and the C terminus, respectively, was purified from E. coli (supplemental Fig. S1A) and was examined in gel shift assays using D-loop/bubble-related structures as substrates (Fig. 1A) (14, 15). DNA substrates used are D-loop carrying a 41-mer invading strand with 5′-tail (Dlp 41; lanes 1–4), D-loop carrying a 21-mer invading strand with no tail (Dlp 21; lanes 5–8), bubble-like structure with duplex unwound segments (Bubble 21 (ds); lanes 9–12), and D-loop carrying a 21-mer invading strand with a 3′-phosphorylated terminus (Dlp 21 (3′-P); lanes 13–16). The phosphorylation of the 3′-end can interfere with binding of a structure-specific DNA-binding protein carrying a pocket structure (TT-pocket; three-prime terminus binding pocket) recognizing the 3′-end of DNA (39, 40). Mrc1 can bind to these structures (15), and affinity to the D-loop with a 41-mer invading strand is the highest, suggesting that structures involving the 5′-tail (double-branched structure) may play a role in high affinity binding. Phosphorylation of the 3′-end of the invading strand did not reduce the binding but slightly stimulated it, indicating that the 3′-end recognition is not involved in binding of Mrc1 protein.
FIGURE 1.
DNA binding activities of the wild-type and mutant Mrc1 on various substrates. Histidine- and FLAG-tagged Mrc1 protein (wild-type or mutant) was purified from E. coli. Gel shift assays of the purified fractions with various substrates were conducted as described previously (37). The structures of substrates are schematically drawn at the top of each panel. Bubble and D-loop are 61-mers containing a 21-nucleotide unpaired segment. The concentration of the substrate in each reaction was 4 nm, and the concentrations of the proteins added are indicated in the figure. A, D-loop or bubble binding activities of the wild-type Mrc1 protein. Substrates used in this assay are D-loop with a 41-mer invading strand carrying a 5′-tail (Dlp 41, lanes 1–4), D-loop with a 21-mer untailed invading strand (Dlp 21, lanes 5–8), bubble with melted duplex segments (Bubble 21 (ds), lanes 9–12), and D-loop with a 3′-phosphorylated untailed invading strand (Dlp 21 (3′-P), lanes 13–16). B, bubble (lanes 1–13) or D-loop (lanes 14–26) binding activities of the wild-type (lanes 1–5 and 14–18), K235E,K236E (lanes 6–9 and 19–22), and Δ221–283 (lanes 10–13 and 23–26) Mrc1 proteins. In A and B, the intensity of each band was quantified, and values representing the fractions of bound DNA were plotted against protein concentrations. Radioactive termini and a phosphorylated 3′-end of DNA are indicated by asterisks and an open circle, respectively. Mrc1 binds to D-loop-like structures, and mutations in the DNA binding domain abolish this binding.
A previous report identified the DNA binding domain of Mrc1 protein (amino acids 160–317) and point mutations (K235E,K236E) abrogating its DNA binding activity (15). The same amino acid substitutions or an internal deletion (Δ221–283) were introduced into the full-length protein, and the mutant proteins were purified. They were tested for DNA binding in gel shift assays (Fig. 1B). Consistent with previous report (15), these mutant proteins were defective in DNA binding (lanes 6–13 and 19–26), confirming that this segment is important for DNA binding of Mrc1 protein in vitro.
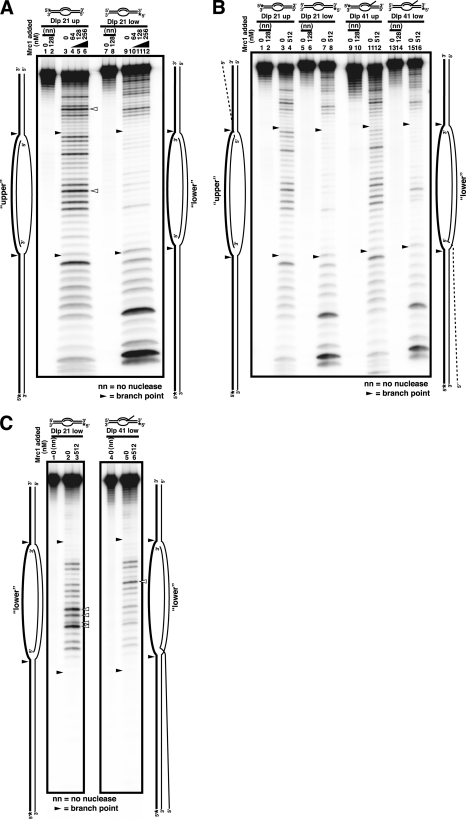
Nuclease Protection Assays of Mrc1 Protein with Various DNA Structures
To determine the mode of binding of Mrc1, nuclease footprinting analyses were conducted with various DNA structures (Fig. 2). Reactions were performed as reported previously with 3 nm substrates and 0.002 units of DNase I (Fig. 2, A and B) or 0.04 units of P1 nuclease (Fig. 2C) (38). Unexpectedly, specific protection was not detected on both template strands in DNase I footprinting on D-loop or bubble-related structures (Fig. 2A). Even at a higher concentration of Mrc1 where a specific protein-DNA complex is generated in gel shift assays (Fig. 1B), no significant protection was detected (Fig. 2B). In P1 nuclease footprinting, increased sensitivity in the middle of the unwound single-stranded segment was detected in the presence of Mrc1 (Fig. 2C, open triangles). This may indicate that Mrc1 interacts mainly with the duplex segments or branched structures, thus causing hypersensitivity in the unbound single-stranded segment.
FIGURE 2.
Nuclease protection analyses of interaction of Mrc1 with bubble or d-loop-related structures. A–C, nuclease footprinting assays were conducted as described previously (38). The D-loop substrates (Dlp) carry a 21-nucleotide unpaired segment surrounded by 20-bp duplex segments with a 21- or 41-mer invading strand (Dlp 21 or Dlp 41, respectively). A schematic drawing of each substrate is presented at the top and at the sides of each gel. The upper or lower strand of the template was radioactively labeled with 32P as indicated by asterisks (marked as upper or lower, respectively, and the labeled strands are indicated by thick lines in the drawings at the sides of the panels). The nuclease used was DNase I (A and B) or P1 nuclease (C). Low (A) or high (B) concentration ranges of Mrc1 were examined. Filled and open arrowheads indicate the junctions between the duplex and melted segments and the nuclease-hypersensitive sites generated in the presence of Mrc1, respectively. Mrc1 does not generate a strong nuclease-resistant footprint on DNA substrates.
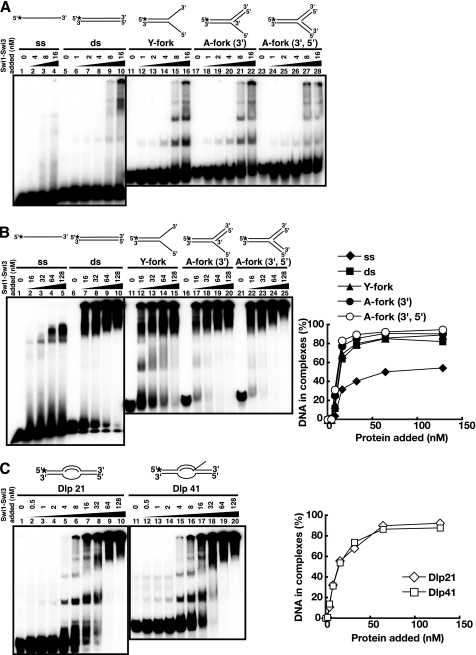
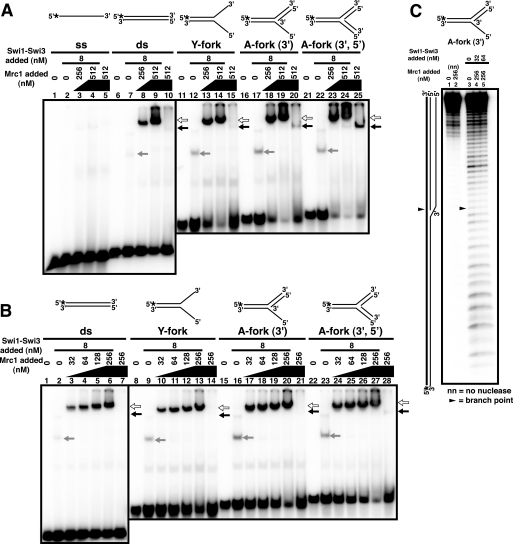
DNA Binding of the Swi1-Swi3 Protein Complex
The Swi1-Swi3 complex was purified from the E. coli cells coexpressing both proteins (see supplemental Fig. S1). Standard gel shift assays were performed with various DNA substrates (Fig. 3). The purified Swi1-Swi3 complex bound to both single-stranded and double-stranded DNA, but the affinity to double-stranded DNA was higher by more than 1 order of magnitude (Fig. 3A, lanes 5–10). It also bound to various forked DNA structures (lanes 11–28), but the affinity was similar to that to a simple double-stranded DNA. At low concentrations of protein (below 16 nm), distinct multiple shifts were observed, notably on arrested fork DNAs (lanes 15, 16, 21, 22, 27, and 28), indicative of stepwise association of increased numbers of protein molecules on the substrate DNA. However, at higher concentrations, complexes that got stuck at the well or migrate for only a short distance accumulated (Fig. 3B), indicative of generation of aggregates of proteins on substrate DNAs. On D-loop structures, specific binding was also detected at 4 nm (Fig. 3C, lanes 5 and 15), although aggregates were generated at higher concentrations. These results suggest that the Swi1-Swi3 complex may recognize branched or fork like structures with higher affinity, although the preference for specific structures may not be very significant. In DNase I footprinting studies, significant protection was not detected on arrested fork structures (A-fork (3′) representing the arrested fork like structure carrying a 3′-end of the putative nascent leading strand at the branch and A-fork (3′,5′) representing the arrested fork like structure carrying both 3′- and 5′-ends of the putative nascent leading and lagging strands at the branch), except for weak protection around the branch point on A-fork (3′,5′) (supplemental Fig. S2). Thus, we conclude that the Swi1-Swi3 complex binds generally to duplex DNA with relatively high affinity in vitro.
FIGURE 3.
DNA binding activities of the Swi1-Swi3 protein complex on various substrates. Swi1-Swi3 protein was purified from E. coli cells as a complex. Standard gel shift assays were conducted with various substrates as schematically drawn at the top of each panel. Low (A) or high (B) concentration ranges of Mrc1 were examined. Substrates used were single-stranded (ss; lanes 1–4 of A and lanes 1–5 of B), double-stranded (ds; lanes 5–10 of A and lanes 6–10 of B), Y-fork (lanes 11–16 of A and lanes 11–15 of B), A-fork (3′) (lanes 17–22 of A and lanes 16–20 of B), A-fork (3′,5′) (lanes 23–28 of A and lanes 21–25 of B), Dlp 21 (lanes 1–10 of C), and Dlp 41 (lanes 11–20 of C). In B and C, the intensity of each band was quantified, and values representing the fractions of bound DNA were plotted against protein concentrations. The Swi1-Swi3 complex binds to duplex DNA with high affinity.
The Swi1-Swi3 Complex Facilitates the Binding of Mrc1 to DNA
It has been reported that chromatin loading of Claspin depends on Tim1/Tipin in mammalian cells (23). In fission yeast, efficient chromatin binding of Mrc1 was shown to require the presence of Swi1-Swi3 (30). Therefore, we examined whether the addition of both proteins in the reaction mixtures affects the DNA binding properties of each protein (Fig. 4). The addition of Mrc1 protein in the presence of a low concentration of Swi1-Swi3 led to the appearance of a shifted band of a strong intensity at a novel position (Fig. 4, A and B, indicated by white arrows). Titration of Mrc1 protein indicated that this complex is generated at the concentration of Mrc1 that is lower than that required for the formation of Mrc1-DNA complex (Fig. 4, A and B, black arrows). The shifted band migrated more slowly than the Swi1-Swi3-DNA (gray arrows) or Mrc1-DNA complex did, suggesting that it may represent the ternary complex between the substrate DNA and the two proteins. In order to identify the proteins present in these complexes, we added antibodies to the complex. The addition of anti-FLAG antibody caused the hypershift of the putative ternary complex band (supplemental Fig. S3, lane 4, white arrow). On the other hand, the addition of anti-Mrc1 antibody caused the disappearance of the initial band (supplemental Fig. S3, lane 2, black arrow) and the appearance of a fast migrating band that comigrates with the Swi-DNA complex (supplemental Fig. S3, lane 3, gray arrow), suggesting that the anti-Mrc1 antibody can disrupt the Swi-Mrc1 interaction. Because the FLAG tag is at the C terminus and anti-Mrc1 antibody was developed against the N-terminal 180 amino acids, this result suggests that Swi1-Swi3 complex may interact with the N-terminal domain of Mrc1.
FIGURE 4.
Swi1-Swi3 facilitates binding of Mrc1 to DNA. Mrc1 and Swi1-Swi3 complex were mixed prior to the addition to each reaction. Mrc1 protein fraction was titrated as indicated in the figure. Substrate structures are schematically drawn at the top of each panel. A and B, gel shift assays on various DNA substrates. Mrc1 was titrated, as indicated, in the presence of 8 nm Swi1-Swi3. High (A) and low (B) concentration ranges of Mrc1 were examined. Substrates used were single-stranded (ss; lanes 1–5 of A), double-stranded (ds; lanes 6–10 of A and lanes 1–7 of B), Y-fork (lanes 11–15 of A and lanes 8–14 of B), A-fork (3′) (lanes 16–20 of A and lanes 15–21 of B), and A-fork (3′,5′) (lanes 21–25 of A and lanes 22–28 of B). Lanes 2, 7, 12, 17, and 22 in A and lanes 2, 9, 16, and 23 in B, Swi1-Swi3 alone; lanes 5, 10, 15, 20, and 25 in A and lanes 7, 14, 21, and 28 in B, Mrc1 alone (512 and 256 nm, respectively). The mobility-shifted bands containing Mrc1 alone, Swi1-Swi3 alone, or a combination of both proteins are indicated by black, gray, or white arrows, respectively. C, DNase I footprinting analyses of binding of Swi1-Swi3 and Mrc1 on A-fork (3′). Radioactive termini of DNA are indicated by asterisks in the schematic drawings. The position of the branch point is indicated by filled triangles. Note that the binding of Mrc1 alone to the arrested fork like structure is less efficient than that to the D-loop like structures (Fig. 1).
Generation of this band was observed even on double-stranded DNA, and slightly more efficient formation of the putative ternary complex was observed on other fork like substrates (Fig. 4, A (lanes 6–25) and B), but very little enhancement was observed with single-stranded DNA (Fig. 4A, lanes 1–5). These results indicate that the presence of both proteins facilitates their binding to DNA. However, DNase I footprinting analyses on A-fork (3′) indicated the absence of significant protection even in the presence of both proteins except for the weak protection near the branch point (Fig. 4C, lanes 4 and 5). These results suggest a highly mobile DNA binding or quick on-off rates. In order to address this issue, we measured the off-rate of the binding by challenging the complex with a cold competitor DNA (supplemental Fig. S4). As a control, we used PriA, the bacterial arrested fork-binding protein, that is known to generate a strong footprint on the A-fork (3′) substrate (38, 40). Mrc1 and Swi1-Swi3 generate a ternary complex that migrates slowly on a gel (supplemental Fig. S4, lane 2, black arrow). The addition of a 10-fold excess of cold competitor to a preformed complex of Mrc1-Swi resulted in disruption of nearly 80% of the complex within 1 min after the addition of the competitor (supplemental Fig. S4, lane 3). In contrast, the PriA-DNA complex was stable even after 10 min under the same condition (Fig. S4, lanes 6–9). These results suggest that the off-rate of the Mrc1-Swi-DNA complex is very rapid, providing an explanation for the lack of strong protection from nuclease attack (Fig. 4C).
Bindings of Swi1-Swi3 alone, Mrc1 alone, and the combination of Swi1-Swi3 and Mrc1 to double-stranded DNA and Y-fork structure were similarly competed by poly(dI/dC) (supplemental Fig. S5, lanes 1–14 and 15–28, respectively). These results show that the binding is largely nonspecific to DNA structures.
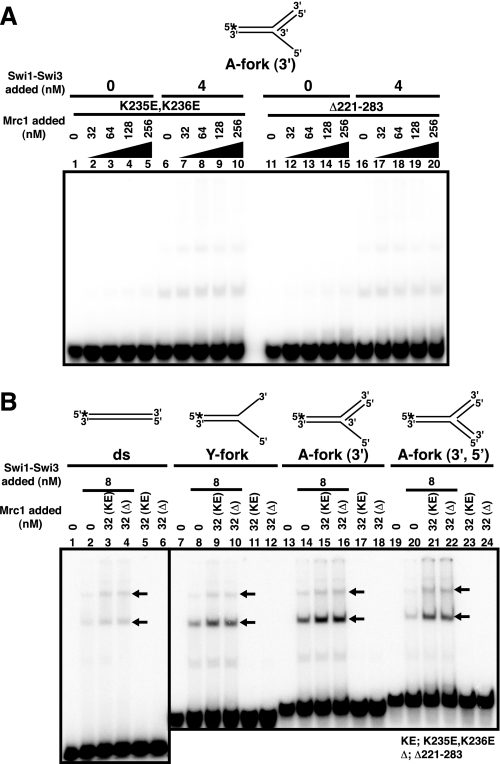
DNA Binding-deficient Mutants of Mrc1 Fail to Form the Ternary Complex with Swi1-Swi3
We have tested DNA binding mutants (K235E,K236E and Δ221–283) of Mrc1 in gel shift assays with A-fork (3′) in the presence of Swi1-Swi3 protein and have shown that they lack DNA binding activity almost completely, as was reported in an N-terminal fragment of Mrc1 (Fig. 5A) (15). This mutant was reported to cause mild sensitivity to HU in fission yeast cells (15). Although the intensities of the two shifted bands generated by the Swi1-Swi3 complex slightly increased (Fig. 5B, black arrows), the bands representing the ternary complex did not appear (compare with Fig. 4B). This was the case for other fork substrates as well (Fig. 5B, lanes 7–24). These results suggest that the DNA binding domain of Mrc1 may be important for ternary complex formation with Swi1-Swi3 and DNA. This indicates two possibilities. The first is that DNA binding activity of Mrc1 plays a direct role in efficient formation of the ternary complex. The second is that these mutants are defective in interaction with Swi1-Swi3 protein.
FIGURE 5.
DNA binding mutants of Mrc1 fail to generate the ternary complex on DNA. Substrate structures are schematically drawn at the top of each panel. A, gel shift assays with K235E,K236E (KE; lanes 1–10) and Δ221–283 (Δ; lanes 11–20) at various concentrations indicated in the absence (lanes 1–5 and 11–15) or presence (lanes 6–10 and 16–20) of the Swi1-Swi3 complex (4 nm) on the A-fork (3′) substrate. The Mrc1 mutants and Swi1-Swi3 complex were mixed prior to the addition to each reaction. B, gel shift assays with the mutant Mrc1 proteins (K235E,K236E and Δ221–283) on various substrates. Mutant Mrc1 and Swi1-Swi3 complex were added at 32 and 8 nm, respectively. Substrates used were double-stranded (ds; lanes 1–6), Y-fork (lanes 7–12), A-fork (3′) (lanes 13–18), and A-fork (3′,5′) (lanes 19–24). The positions of the two shifted bands generated by the Swi1-Swi3 complex are indicated by black arrows.
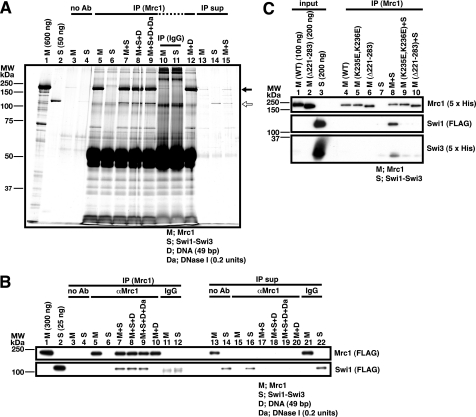
Physical Interaction between Mrc1 and Swi1-Swi3
In order to discriminate these possibilities, we first examined the physical interaction between Mrc1 and Swi1-Swi3 by pull-down assays with anti-Mrc1 antibody. The immunoprecipitate contained Swi1 protein, as shown by silver staining (Fig. 6A, lane 7, indicated by a white arrow). Western blotting with the antibodies against the tag (FLAG and penta-His for Mrc1 and Swi1-Swi3, respectively) also confirmed the coimmunoprecipitation of Mrc1 and Swi1-Swi3 (Fig. 6B, lane 7). The presence of DNase I did not affect the interaction, indicating that the two proteins can interact with each other in the absence of DNA (Fig. 6, A and B, lanes 8 and 9).
FIGURE 6.
Coimmunoprecipitation of Mrc1 and Swi1-Swi3 with anti-Mrc1 antibody. Proteins and DNA (M, Mrc1; S, Swi1-Swi3 complex; D, 49-bp duplex DNA) were mixed as indicated in the figure, and Mrc1 was immunoprecipitated by anti-Mrc1 antibody (IP). A, immunoprecipitation was conducted without antibody (lanes 3 and 4), with anti-Mrc1 antibody (lanes 5–9 and 12), or with normal rabbit IgG (lanes 10 and 11). Half of the immunoprecipitates were loaded onto 10% SDS-PAGE. 10% of the unbound fractions after centrifugation with Protein A-Sepharose beads were loaded in lanes 13–15 (IP sup). The gel was subjected to silver staining. Protein fractions used in this assay (one-half of the input) were run in lanes 1 and 2. Black and white arrows indicate Mrc1 and Swi1 proteins, respectively. Swi3 protein is not visible under this condition. B, immunoprecipitation was conducted without antibody (lanes 3 and 4), with anti-Mrc1 antibody (lanes 5–10), or with normal rabbit IgG (lanes 11 and 12). One-sixth of the immunoprecipitates were loaded onto the gel. 10% of the unbound fractions after centrifugation were loaded in lanes 13–22 (IP sup). Protein fractions used in this assay (one-half of the input) were run in lanes 1 and 2. C, immunoprecipitation was conducted in the presence of the wild-type or mutant (K235E,K236E or Δ221–283) Mrc1 proteins, as shown, with anti-Mrc1 antibody (lanes 4–10). The entire immunoprecipitates were applied on the gel (lanes 4–10). The input proteins of the indicated amount were loaded in lanes 1–3. In B and C, proteins were detected by Western blotting using the antibody indicated on the right of each panel. The positions of molecular weight markers are shown on the left of each panel. Complex formation between Mrc1 and Swi1-Swi3 can be detected by immunoprecipitation assays and is disrupted in Mrc1 K235E,K236E and Δ221–283 mutants. The positions of the two shifted bands generated by the Swi1-Swi3 complex are indicated by black arrows.
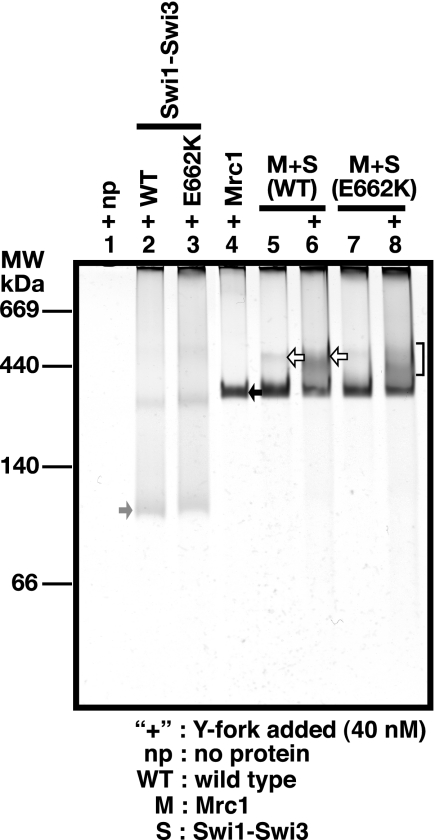
Native gel electrophoresis was employed to directly assess the conformation of Mrc1 and Swi proteins (Fig. 7). Proteins with different combinations were loaded onto a 6% native polyacrylamide gel and silver-stained. Combination of Swi1-Swi3 and Mrc1 (lane 5) migrated on the gel at a position (white arrow) distinct from that of Swi1-Swi3 or Mrc1 protein alone (lane 2 or 4; gray or black arrow, respectively), suggesting that these proteins can form a complex in solution without DNA substrate, consistent with the results of the immunoprecipitation assay (see Fig. 6, A and B), whereas the addition of 40 nm Y-fork substrate (+) enhanced the intensity of the complex band as indicated by a white arrow in lane 6. Comparison with molecular weight standards indicates that the Swi1-Swi3 complex is about 120 kDa (gray arrow), and Mrc1 migrates around 200 kDa (black arrow), suggesting that the Swi1-Swi3 complex is a 1:1 heterodimer and that Mrc1 forms a homodimer.
FIGURE 7.
Analyses of complex formation between Mrc1 and Swi1-Swi3 in native gel electrophoresis. Mrc1 and Swi1-Swi3 (wild-type or E662K) proteins (64 and 128 nm, respectively) were applied onto 6% native polyacrylamide gel, and proteins were visualized by silver staining. The positions of molecular weight standards are indicated on the left of the gel. Swi1-Swi3 (lane 2), Swi1 (E662K)-Swi3 (lane 3), or Mrc1 (lane 4) protein and a combination of both proteins (lanes 5–8) were loaded without or with 40 nm Y-fork DNA substrate (lanes indicated with plus signs). Black and gray arrows indicate Mrc1 and Swi1-Swi3 proteins, respectively. The white arrows indicate the Mrc1-Swi complex, the amount of which increases in the presence of DNA (lane 6). The complex is partially disrupted in the Swi1 E662K mutant (lanes 7 and 8; the smeared band indicated by the bracket).
Mrc1 and Swi1 Mutants Cannot Recruit Mrc1 onto DNA Due to Loss of Protein Interaction
We next examined the Swi-Mrc1 interactions with these mutants. Neither the K235E,K236E nor the Δ221–228 mutant of Mrc1 was coimmunoprecipitated with Swi1-Swi3 (Fig. 6C, lanes 9 and 10), indicating that these mutations abolish the interaction with Swi1-Swi3 as well. This result strongly suggests that loss of the ternary complex formation with the mutant Mrc1 is due to its inability to interact with Swi1-Swi3.
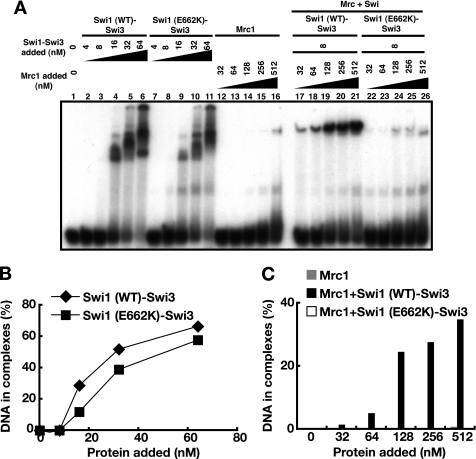
The E662K mutant of swi1 is deficient in polar fork arrest at a site called RTS (17). The Swi1 (E662K)-Swi3 complex was purified (supplemental Fig. S1A, lane 3), and its DNA binding activity on A-fork (3′) was examined (Fig. 8A). Swi1 (E662K)-Swi3 binds to DNA as efficiently as the wild type (Fig. 8, A (lanes 7–11) and B). However, it cannot enhance the binding of Mrc1 (Fig. 8, A (lanes 22–26) and C). This is due to the deficiency of the E662K complex to interact with Mrc1 because the amount of the slowly migrating band representing the Mrc1-Swi complex decreased in a native gel (Fig. 7, compare lanes 6 and 8). The smeared bands above the Mrc1 E662K band (bracket) may indicate that the complex may be unstable and dissociate during the run. These results indicate that E662K is at least partially defective in interaction with Mrc1, which leads to loss of the ability to facilitate binding of Mrc1 onto DNA.
FIGURE 8.
The Swi1-Swi3 complex containing a swi switching mutant, Swi1 E662K, binds to DNA but fails to facilitate DNA binding of Mrc1. Gel shift assays were conducted on A-fork (3′) with Mrc1 and wild-type or mutant Swi1-Swi3 complex. A, proteins were added at the concentrations indicated. Swi1 (wild type)-Swi3 (lanes 1–6), Swi1 (E662K)-Swi3 (lanes 7–11), Mrc1 (lanes 12–16), and combination of Mrc1 with Swi1 (wild type)-Swi3 (lanes 17–21) or with Swi1 (E662K)-Swi3 (lanes 22–26). B, quantification of DNA binding activity of wild-type or mutant Swi1-Swi3 on the basis of results in A. C, quantification of DNA binding activity of Mrc1 in the presence of wild-type or mutant Swi1-Swi3 on the basis of results in A.
Effect of Various swi1 or swi3 Mutations on DNA Replication Checkpoint Reactions
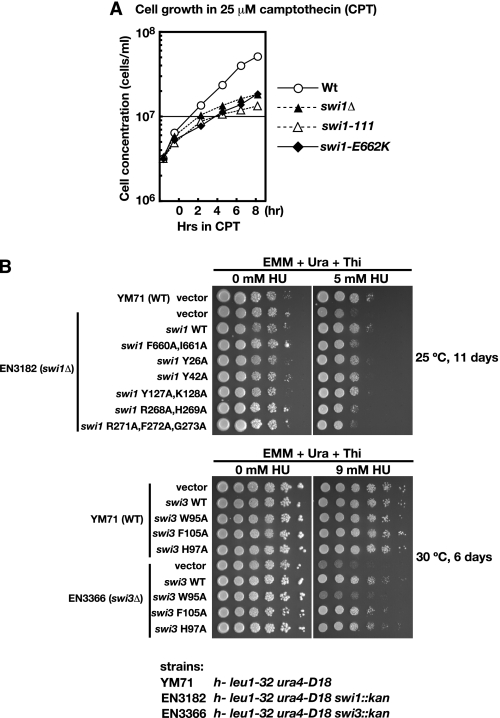
Sensitivity of the K235E,K236E mutant of Mrc1 to HU (15) suggests that the ability of the Swi complex to load Mrc1 onto DNA plays an important role in the DNA replication checkpoint. We examined checkpoint functions of other mutant proteins by expressing them in swi1Δ cells and examining the sensitivity to HU. We scored the growth on EMM plates with or without thiamine because the plasmid contains nmt1 promoter, which is suppressed in the presence of thiamine. The K662E mutant of Swi1 showed slow growth compared with the wild type in the presence of 25 μm camptothecin (Fig. 9A), supporting our conclusion regarding the functional importance of the Swi-Mrc1 interaction and facilitated binding of the ternary complex on DNA.
FIGURE 9.
Characterization of various Swi1 or Swi3 mutants. A, growth of swi1 E662K mutant in the presence of 25 μm camptothecin (CPT). WT (Y393), swi1Δ (Y400), swi1-111 (E111), and swi1-E662K (JZ277) cells exponentially growing in YES at 30 °C were challenged with 25 μm CPT at time 0 and incubated further at 30 °C. Cell concentrations were measured at the times indicated. B, wild-type and various mutant forms of Swi1 or Swi3 were expressed on pREP41-based vector in swi1Δ or swi3Δ cells, respectively. Cells were serially diluted and spotted on EMM + uracil + thiamine plates in the absence or presence of HU (5 mm for swi1 series and 9 mm for swi3 series). Plates were incubated at 25 °C for 11 days (for swi1) or at 30 °C for 6 days (for swi3), and photographs were taken. For swi1 series, the wild-type cells harboring the vector were also spotted. For swi3 series, the wild-type cells harboring all of the mutants constructed were also spotted.
We also generated other mutants of Swi1 and Swi3 by introducing amino acid substitutions at the evolutionarily conserved amino acids (supplemental Fig. S6). The results are summarized in Table 3. Among the Swi1 mutants examined, swi1 F660A,I661A, swi1 R268A,H269A, and swi1 R271A,F272A,G273A mutants exhibited significant sensitivity to HU on the plate containing thiamine. Among the swi3 mutants, W95A was most sensitive to HU under the same conditions (Fig. 9B). Interaction of swi3 W95A mutant with Mrc1 was examined by a native gel assay. The swi3 W95A mutant could not form a ternary complex with Mrc1 (supplemental Fig. S7, lane 5). These results further reinforce our conclusion that interaction of Swi1-Swi3 with Mrc1 is important for their functions in cellular responses to replication stress of fission yeast cells.
TABLE 3.
Summary of the characterization of Swi1 and Swi3 mutant proteins
| Name of clone | Complex formation between Swi1 and Swi3 | HU sensitivitya |
|---|---|---|
| Swi1 WT | Stable | R |
| Swi1 E662K | Stable | R |
| Swi1 F660A,I661A | Unstable | S |
| Swi1 Y26A | Unstable | s |
| Swi1 Y42A | Unstable | s |
| Swi1 Y127A,K128A | Unstable | s |
| Swi1 R268A,H269A | Unstable | S |
| Swi1 R271A,F272A,G273A | Stable | S |
| Swi3 WT | Stable | R |
| Swi3 W95A | Unstable | S |
| Swi3 F105A | Unstable | R |
| Swi3 H97A | Stable | R |
a R, resistant; S, sensitive; s, weakly sensitive.
DISCUSSION
In this paper, we have biochemically characterized the fission yeast Mrc1 and Swi1-Swi3 proteins, which play key roles in the maintenance of replication fork integrity. Our results show that the two proteins cooperate to facilitate their binding to DNA.
Swi1-Swi3 Complex Facilitates Binding of Mrc1 onto DNA and Generates a Ternary Complex
As previously reported for the N-terminal polypeptide of Mrc1 (15), the full-length fission yeast Mrc1 protein binds to arrested fork or D-loop-like structure more preferentially than other non-structured DNA (Fig. 1) (data not shown). The Swi1-Swi3 complex also binds to DNA but with much less specificity. It binds to linear duplex DNA as efficiently as arrested fork like structures but not to single-stranded DNA (Fig. 3). The affinity of Swi1-Swi3 to DNA is significantly higher than that of Mrc1. Furthermore, binding of Mrc1 to DNA was significantly stimulated by the presence of Swi1-Swi3 (Fig. 4). Mrc1 interacts with the Swi1-Swi3 complex, and this interaction is required for Swi-mediated facilitated binding of Mrc1 on DNA.
DNA Binding Mutants of Mrc1 Are Deficient in Formation of the Ternary Complex
Zhao and Russell (15) previously identified the DNA binding domain of Mrc1 as well as mutations that impair its DNA binding activity. These mutations cause mild sensitivity to HU. We purified a full-length Mrc1 protein containing these mutations (K235E,K236E and Δ221–283). The mutant proteins were defective in DNA binding in gel shift assays and could not generate the ternary complex on DNA with Swi1-Swi3 (Fig. 5). Unexpectedly, they failed to interact with Swi1-Swi3 in the coimmunoprecipitation assays (Fig. 6C). This strongly indicates that the failure of these mutants to generate the ternary complex may be due to the loss of interaction of Mrc1 with Swi1-Swi3. The results also suggest the possibility that the DNA binding domain and Swi1-Swi3 interacting domain may overlap on Mrc1 protein. However, it is also possible that the DNA binding activity of Mrc1 may be required for stable association of the ternary complex on DNA, which can be detected in gel shift assays. It should be noted that we cannot completely rule out the possibility that the mutations cause structural perturbation in the Mrc1 mutants that may be responsible for these defects, although the wild-type and mutant proteins behaved similarly during purification steps.
Interaction of Siw1-Swi3-Mrc1 with DNA
Because the interaction between Swi1-Swi3 and Mrc1 is independent of DNA (Fig. 6, A and B), it is likely that the Swi1-Swi3 complex recruits Mrc1 protein onto DNA through direct interaction to generate the ternary complex. Interaction of Mrc1 or Claspin with Swi1-Swi3/Tof1-Csm3 or Tim1-Tipin in yeast or Xenopus egg extracts, respectively, was previously reported (24, 29, 30, 32, 35, 41). High affinity of Swi1-Swi3 to DNA explains the high affinity binding of the Swi1-Swi3-Mrc1 (Swi-Mrc1) complex to DNA (Fig. 4). The failure to generate the ternary complex on single-stranded DNA is also consistent with the role of Swi1 protein as a loader of Mrc1 protein because Swi1-Swi3 does not bind to single-stranded DNA very efficiently (Fig. 3). This is also consistent with the in vivo observation that chromatin loading of Mrc1 or Claspin is facilitated by Swi1-Swi3/Tof1-Csm1 or Tim-Tipin complex, respectively (23, 29, 30, 35).
The absence of strong protection of Mrc1-, Swi1-Swi3-, or Swi-Mrc1-bound DNA from nuclease digestion was unexpected, considering the generation of a distinct complex with the template DNA in gel shift assays (Figs. 2 and 4 and supplemental Fig. S2). This suggests that the interaction with DNA may not be very tight or may not be strictly localized on the template. This may reflect the translocating nature of these proteins along with the replication fork or the intrinsic nature of its unique interaction. Indeed, challenge experiments with a cold substrate in gel shift assays indicate that the off-rate of the Mrc1-Swi complex on DNA is quite fast (supplemental Fig. S4). Although Mrc1 shows higher affinity to structured DNAs, such as branched DNA (Fig. 1) (data not shown), the Swi1-Swi3 protein does not show significant preference for structured DNAs (Fig. 3). The Swi-Mrc1 complex, on the other hand, exhibits some preference for branched molecules (Fig. 4). This may indicate that the Swi-Mrc1 complex may have slightly altered DNA binding specificity compared with the Swi1-Swi3 complex, presumably due to the interaction of Mrc1 with the template DNA.
Roles of Swi-Mrc1 Complex in Regulation of Replication Fork
The E662K mutant of Swi1, defective in polar fork arrest at a site called RTS1 (17), was shown to be proficient in binding to DNA but to be defective in formation of the ternary complex (Fig. 8). This is due to decreased interaction with Mrc1. A number of other mutants of Swi1 and Swi3 were generated and analyzed. Many of them (swi1 F660A,I661A, swi1 R268A,H269A, swi1 R271A,F272A,G273A, and swi3 W95A; see Fig. 9B and Table 3) were sensitive to HU. Swi3 W95A was found to be deficient in interaction with Mrc1 (supplemental Fig. S7). Thus, interaction between Swi1-Swi3 and Mrc1 and their ability to generate a ternary complex on DNA appear to be functionally important in cellular responses to replication stress.
Swi1-Swi3 as well as Mrc1 have been implicated in efficient fork progression as well as in stabilization of stalled replication forks as integral factors for an active replication fork. Fork progression rate is reduced in swi1Δ or mrc1Δ mutant of budding yeast, and in the same mutants, replication fork progression was uncoupled from DNA synthesis upon inhibition of replication fork progression, leading to aberrant fork unwinding (24). In the rDNA repeats, Swi1-Swi3 is required for replication fork arrest at replication fork barriers in the spacer regions of rDNA (19). This may reflect the roles of Swi and Mrc1 in stably arresting the replication fork when DNA synthesis is blocked at natural fork arrest sites. It would be interesting to examine the effect of Swi1-Swi3 or Swi-Mrc1 complex on the helicase activity of the MCM or CMG (Cdc45-MCM-GINS) complex in vitro. The ability of Swi1-Swi3 to facilitate the binding of Mrc1 to DNA and generation of a ternary complex on DNA is likely to be an important step for formation of a replication fork that is highly efficient and responsive to various fork stall signals.
Supplementary Material
Acknowledgments
We thank Dr. Paul Russell for generously providing the expression vectors for mutant Mrc1 proteins. We also thank Dr. Jacob Dalgaard for swi1 strains. We thank all of the members of our laboratory for helpful discussions.
This work was supported in part by a grant-in-aid for scientific research on priority area “Chromosome Cycle” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Takeda Science Foundation; and the Astellas Foundation for Research on Metabolic Disorders (to H. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- rDNA
- ribosomal DNA
- HU
- hydroxyurea
- EMM
- Edinburgh Minimal Medium.
REFERENCES
- 1.Greenfeder S. A., Newlon C. S. (1992) Mol. Cell. Biol. 12, 4056–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivessa A. S., Lenzmeier B. A., Bessler J. B., Goudsouzian L. K., Schnakenberg S. L., Zakian V. A. (2003) Mol. Cell. 12, 1525–1536 [DOI] [PubMed] [Google Scholar]
- 3.Andreassen P. R., D'Andrea A. D., Taniguchi T. (2004) Genes Dev. 18, 1958–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartek J., Lukas C., Lukas J. (2004) Nat. Rev. Mol. Cell. Biol. 5, 792–804 [DOI] [PubMed] [Google Scholar]
- 5.Cobb J. A., Shimada K., Gasser S. M. (2004) Curr. Opin. Genet. Dev. 14, 292–300 [DOI] [PubMed] [Google Scholar]
- 6.Boddy M. N., Russell P. (2001) Curr. Biol. 11, R953–956 [DOI] [PubMed] [Google Scholar]
- 7.Kumar S., Huberman J. A. (2009) Mol. Cell. Biol. 29, 602–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka K., Russell P. (2001) Nat. Cell. Biol. 3, 966–972 [DOI] [PubMed] [Google Scholar]
- 9.Kumagai A., Dunphy W. G. (2000) Mol. Cell. 6, 839–849 [DOI] [PubMed] [Google Scholar]
- 10.Alcasabas A. A., Osborn A. J., Bachant J., Hu F., Werler P. J., Bousset K., Furuya K., Diffley J. F., Carr A. M., Elledge S. J. (2001) Nat. Cell. Biol. 3, 958–965 [DOI] [PubMed] [Google Scholar]
- 11.Chini C. C., Chen J. (2003) J. Biol. Chem. 278, 30057–30062 [DOI] [PubMed] [Google Scholar]
- 12.Nitani N., Nakamura K., Nakagawa C., Masukata H., Nakagawa T. (2006) Genetics 174, 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin L., Locovei A. M., D'Urso G. (2008) Mol. Biol. Cell. 19, 4374–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sar F., Lindsey-Boltz L. A., Subramanian D., Croteau D. L., Hutsell S. Q., Griffith J. D., Sancar A. (2004) J. Biol. Chem. 279, 39289–39295 [DOI] [PubMed] [Google Scholar]
- 15.Zhao H., Russell P. (2004) J. Biol. Chem. 279, 53023–53027 [DOI] [PubMed] [Google Scholar]
- 16.Noguchi E., Noguchi C., McDonald W. H., Yates J. R., 3rd, Russell P. (2004) Mol. Cell. Biol. 24, 8342–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalgaard J. Z., Klar A. J. (2000) Cell 102, 745–751 [DOI] [PubMed] [Google Scholar]
- 18.Noguchi E., Noguchi C., Du L. L., Russell P. (2003) Mol. Cell. Biol. 23, 7861–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krings G., Bastia D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou D. M., Elledge S. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18143–18147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotter A. L., Suppa C., Emanuel B. S. (2007) J. Mol. Biol. 366, 36–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ünsal-Kaçmaz K., Chastain P. D., Qu P. P., Minoo P., Cordeiro-Stone M., Sancar A., Kaufmann W. K. (2007) Mol. Cell. Biol. 27, 3131–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshizawa-Sugata N., Masai H. (2007) J. Biol. Chem. 282, 2729–2740 [DOI] [PubMed] [Google Scholar]
- 24.Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K. (2003) Nature 424, 1078–1083 [DOI] [PubMed] [Google Scholar]
- 25.Hodgson B., Calzada A., Labib K. (2007) Mol. Biol. Cell 18, 3894–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foss E. J. (2001) Genetics 157, 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tourrière H., Versini G., Cordón-Preciado V., Alabert C., Pasero P. (2005) Mol. Cell. 19, 699–706 [DOI] [PubMed] [Google Scholar]
- 28.Voineagu I., Surka C. F., Shishkin A. A., Krasilnikova M. M., Mirkin S. M. (2009) Nat. Struct. Mol. Biol. 16, 226–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Errico A., Costanzo V., Hunt T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14929–14934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimmoto M., Matsumoto S., Odagiri Y., Noguchi E., Russell P., Masai H. (2009) Genes Cells 14, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J., Gold D. A., Shevchenko A., Shevchenko A., Dunphy W. G. (2005) Mol. Biol. Cell. 16, 5269–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) Nat. Cell. Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 33.Lou H., Komata M., Katou Y., Guan Z., Reis C. C., Budd M., Shirahige K., Cambell J. L. (2008) Mol. Cell. 106, 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komata M., Bando M., Araki H., Shirahige K. (2009) Mol. Cell. Biol. 29, 5008–5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bando M., Katou Y., Komata M., Tanaka H., Itoh T., Sutani T., Shirahige K. (2009) J. Biol. Chem. 284, 34355–34365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGlynn P., Al-Deib A. A., Liu J., Marians K. J., Lloyd R. G. (1997) J. Mol. Biol. 270, 212–221 [DOI] [PubMed] [Google Scholar]
- 37.Tanaka T., Mizukoshi T., Taniyama C., Kohda D., Arai K., Masai H. (2002) J. Biol. Chem. 277, 38062–38071 [DOI] [PubMed] [Google Scholar]
- 38.Tanaka T., Mizukoshi T., Sasaki K., Kohda D., Masai H. (2007) J. Biol. Chem. 282, 19917–19927 [DOI] [PubMed] [Google Scholar]
- 39.Mizukoshi T., Tanaka T., Arai K., Kohda D., Masai H. (2003) J. Biol. Chem. 278, 42234–42239 [DOI] [PubMed] [Google Scholar]
- 40.Masai H., Tanaka T., Kohda D. (2010) BioEssays 32, 687–697 [DOI] [PubMed] [Google Scholar]
- 41.Tanaka H., Kubota Y., Tsujimura T., Kumano M., Masai H., Takisawa H. (2009) Genes Cells 14, 949–963 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.