Abstract
The Myc proto-oncoproteins are transcription factors that recognize numerous target genes through hexameric DNA sequences called E-boxes. The mechanism by which they then activate the expression of these targets is still under debate. Here, we use an RNAi screen in Drosophila S2 cells to identify Drosophila host cell factor (dHCF) as a novel co-factor for Myc that is functionally required for the activation of a Myc-dependent reporter construct. dHCF is also essential for the full activation of endogenous Myc target genes in S2 cells, and for the ability of Myc to promote growth in vivo. Myc and dHCF physically interact, and they colocalize on common target genes. Furthermore, down-regulation of dHCF-associated histone acetyltransferase and histone methyltransferase complexes in vivo interferes with the Myc biological activities. We therefore propose that dHCF recruits such chromatin-modifying complexes and thereby contributes to the expression of Myc targets and hence to the execution of Myc biological activities.
Keywords: Development, Drosophila, Drosophila Genetics, Myc, Oncogene, Transcription, Transcription coactivators, Transcription Factors, Growth
Introduction
Myc genes were identified for their powerful transforming capabilities in vertebrate systems and later shown to be essential for normal development (reviewed in Refs. 1, 2). The molecular functions of Myc are evolutionarily conserved, and the single Myc homolog in Drosophila melanogaster (called Myc when referring to the protein, and diminutive or dm when referring to alleles) can substitute for vertebrate Myc (3) and vice versa (Ref. 4, for a recent review see Ref. 5). Drosophila Myc prominently controls cellular growth; null mutations prevent organismal growth and lead to death during early larval stages (6), whereas hypomorphic dm mutations prolong development and result in small adult flies, made up of smaller than wild-type cells (7). Conversely, overexpression of Myc results in bigger cells, but also stimulates apoptosis (7, 8). These effects of Myc are mediated by the transcriptional regulation of a large number of target genes (9, 10), including genes transcribed by RNA polymerases I (11) and III (12).
Activation of target genes is brought about by a complex of Myc with its partner protein Max, which recognizes hexameric DNA motifs called E-boxes and recruits a variety of transcriptional co-activators, including different histone acetyltransferases and chromatin-remodeling complexes, which are then thought to stimulate the expression of the affected genes (reviewed in Ref. 13). Only a subset of all E-boxes in the genome are bound by Myc, though. In Drosophila, Myc-dependent E-boxes are frequently located immediately downstream of transcription start sites (10, 14), and in vertebrates, Myc has been shown to only bind to euchromatic islands that are premethylated on lysines 4 and 79 of histone H3, as well as acetylated on histone H3 (15).
Interestingly, Drosophila Myc has recently been shown to interact physically and genetically with two trithorax group proteins that affect the methylation status of histones, the demethylase Lid and the Set1 histone methyltransferase component Ash2 (16), and these physical interactions are conserved in vertebrates (16, 17). However, the mechanistic consequences of the Myc:Lid and Myc:Ash2 interaction are currently unclear, and it is not known whether there is any connection to the observed association of vertebrate Myc with euchromatin islands, or whether such an association is even conserved and functionally relevant in Drosophila. Other aspects of transactivation by Myc also remain enigmatic, in particular how Myc preferentially recognizes pre-methylated histones, and why Myc-dependent E-boxes show such a positional bias, whether this bias reflects a particular mechanism of transactivation by Myc (e.g. a role in transcriptional elongation rather than initiation), and if so, which of the transcriptional co-activators that have been shown to physically interact with Myc are required for the activation of these target genes in vivo.
HCF4-1 (an acronym for “host cell factor”) was originally identified as a large cellular protein that is required for the transcription of viral genes in herpes simplex virus-infected cells, and it was later shown to be essential for cell cycle progression of normal, uninfected vertebrate cells (reviewed in 18). A single HCF homolog is also present in Drosophila (called dHCF), but no mutant phenotypes have been described so far (19, 20). In mammalian cells, HCF-1 is cleaved after synthesis into two parts that remain physically associated; the C terminus is required for passage through mitosis, whereas the N terminus is important for entry into S-phase (21). This N terminus bridges transcription factors, such as E2F family members that recognize HCF via the tetrapeptide (D/E)HXY (called “HCF1-binding motif” or HBM) (22), and transcriptional co-activators (a GCN5-containing histone acetyltransferase called ATAC, the Ash2 containing trithorax family of H3K4 methyltransferases (HMT)) (Refs. 23–26) or co-repressors (a Sin3 containing histone deacetylase (HDAC); Ref. 23). In the case of the E2F family, HCF-1 has been shown to recruit HDACs to the repressive E2F4, and H3K4 methyltransferases to the activating E2F1 (22), but it is currently unclear how these HCF complexes are differentially targeted to these transcription factors.
Here, we use a cell-based RNAi screen to identify dHCF as a novel co-factor for Myc. dHCF physically binds to Myc in vitro and in cultured cells, and the two proteins co-localize on shared target genes. Down-regulation of dHCF reduces the expression of Myc target genes in cultured cells, and impairs Myc ability to promote growth in vivo, thus confirming the importance of dHCF as a novel interaction partner for Myc.
EXPERIMENTAL PROCEDURES
Molecular Biology
Luciferase Reporters
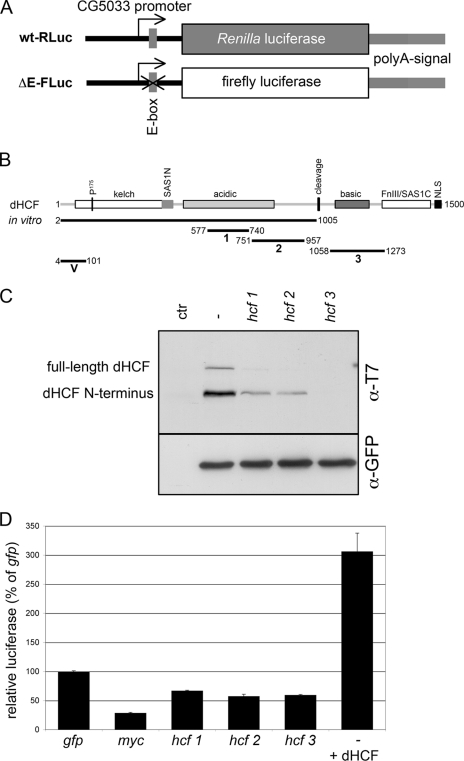
The wt-RLuc reporter contains sequences from −322 to +61 relative to the transcription start site of the Myc target CG5033 (E-box at +21, ATG at +56; 10), followed by the Renilla luciferase open-reading frame. In the ΔE-FLuc reporter the E-box is replaced by the sequence “GAATTC” and the the Renilla luciferase ORF by the firefly luciferase ORF.
Expression Plasmids
UAS-HA-Myc (27), pBSattB-UAS-HA-Myc (Myc overexpression in S2 cells; 28), pACXT-T7-dHCF (dHCF overexpression for Fig. 4C; 20), and pUAST-T7-dHCF-FLAG (dHCF overexpression for supplemental Fig. 4C; Ref. 26) have been described; pTub-GAL4 was used to drive expression of GAL4 in transiently transfected S2 cells (gift from K. Basler). pBSattB-UAS-HA-MycΔHBM is derived from pBSattB-UAS-HA-Myc, and directs the synthesis of a mutant Myc where the potential HCF-binding motif 387DHSY390 is replaced by the sequence AASA. For in vitro transcription/translation, coding sequences for HA-Myc or HA-MycΔHBM were inserted into the vector pRSetB. Sequences are available upon request.
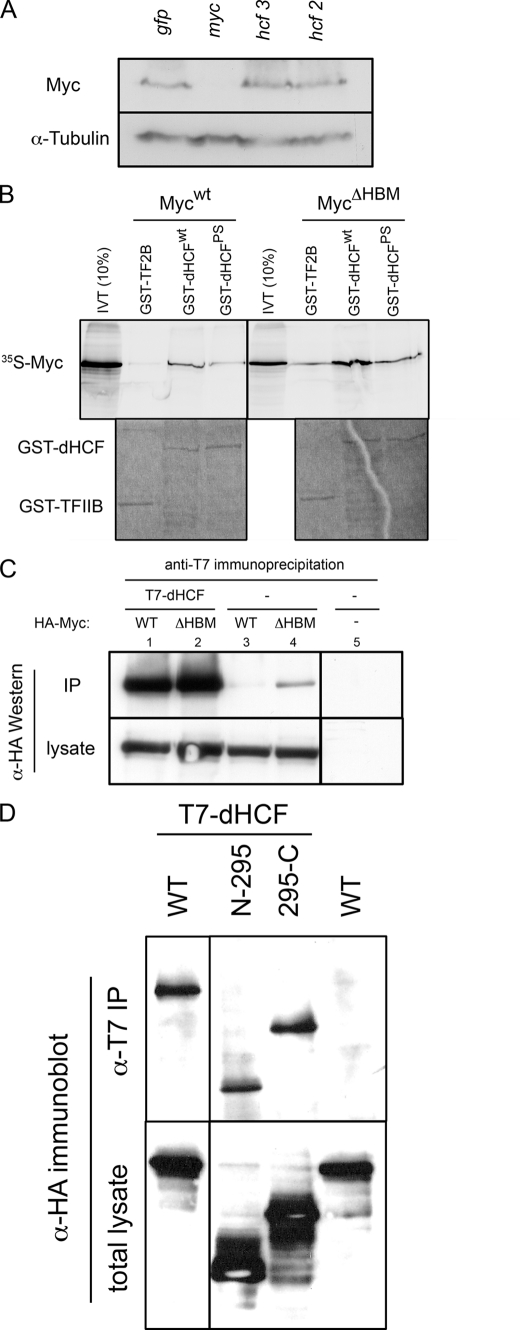
FIGURE 4.
dHCF physically interacts with Myc. A, dHCF knockdown does not affect Myc levels. S2 cells were transfected with the indicated dsRNAs and 48 h later processed for Western blotting against endogenous Myc and α-tubulin as a loading control. B, in vitro translated Mycwt binds to GST-dHCFwt and GST-dHCFPS, but not to the negative control protein GST-TFIIB; MycΔHBM binds preferentially GST-dHCF, but shows significant background binding to GST-TFIIB. The upper panel is a phosphorimager picture of input and bound 35S-labeled Myc proteins; each binding reaction contained 20% of in vitro synthesized Myc. The lower panel shows the same gel after Coomassie Blue staining to reveal the relative amounts of GST-fusion proteins. C, anti-T7-epitope antibodies precipitate HA-MycWT and HA-MycΔBHM when they are co-expressed with T7-dHCF, but not when they are expressed alone. Labels above the lanes indicate the transfected proteins (T7-dHCF and/or HA-MycWT/ΔHBM). As in B, HA-MycΔHBM shows some non-HBM-specific binding. Immunoprecipitates correspond to 50% of a 3-cm well, lysates to 20%. D, two non-overlapping fragments of Myc associate with dHCF. HA-tagged versions of Myc containing amino acids 1–295 or 295–717 (as indicated) both were co-precipitated with T7-dHCF, indicating that at least two regions of Myc mediate the interaction with dHCF.
dsRNA
Target sequences were subjected to BLAST analysis to ensure minimal homology with unrelated transcripts. Fragments of typically 600 bp in length were then amplified by PCR, using primers containing a 5′ T7 RNA polymerase binding site, followed by in vitro transcription. Primer sequences are listed in the supplemental materials.
Transgenic Flies
PCR fragments corresponding to the dsRNAs hcf2 and hcf3 (used for RNAi in S2 cells) were inserted as inverted repeats in the vector pWIZ, which contains GAL4-responsive UAS-sites (29). Using P-element mediated transgenesis, 5 independent lines, and one line were obtained for hcf2 and hcf3, respectively. Unless otherwise indicated, lines UAS-HCF-IR-22.9 (hcf2) and UAS-HCF-IR-33.1 (hcf3) were used for our experiments.
Tissue Culture
Drosophila S2 cells were cultured as described (Refs. 10, 12 and see supplemental materials).
RNAi Screen for Myc Cofactors
A total of 752 potentially transcription-associated proteins were selected in silico (supplemental Table S2), based on their Gene Ontology annotation. Gene-specific primer pairs were purchased from Eurogentec that allow synthesis of amplicons of typically 400 bp length (using Drosophila genomic DNA as template). Primary PCR-products were re-amplified with “universal tag” primers containing T7 polymerase promoter sequences. dsRNA was synthesized in vitro using the T7 Megascript kit (Ambion). For the screen, 50 ng of each luciferase reporter was mixed with 0.5 μl of Cellfectin in a total volume of 4.5 μl, incubated for 45 min at room temperature, mixed with 105 S2 cells in 45.5 μl of serum-free medium, and plated into single wells of 96-well plates. After 12 h, 50 μl of complete medium was added, the incubation continued for 48 h, and the cells processed for luciferase assays. The entire collection was screened at least twice. A total of 80 candidates with z-scores ≥ 1.5 (calculated from the difference between the ratio of each experimental well and the average ratio of all the samples within the same sample plate) were tested in another 96-well plate that also contained 8 wells with gfp-dsRNA (as a negative control) and 8 wells with myc-dsRNA (as a positive control). Among these candidates, 33 dsRNAs increased or decreased luciferase ratios by more than 1.5-fold as compared with gfp-dsRNA.
mRNA Expression Profiling
For dHCF mRNA expression profiling, total RNA from SL2 cells was extracted after 3 days of dHCF RNAi treatment using a combined TRIzol/RNeasy (Qiagen) protocol to be described in detail elsewhere.5 The labeling of mRNA and hybridization to the Affymetrix Drosophila Genome 2.0 array was also performed at the DNA Array Facility at the University of Lausanne. Expression values were measured using the RMA algorithm from the BioConductor Affy package.
Chromatin Immunoprecipitation
Ultra High Throughput DNA Sequencing (ChIP-seq)
SL2 cells were grown in suspension to a concentration of 2.5 × 106 cells/ml. After cross-linking, immunoprecipitation with anti-dHCF antibodies (25) and sonication, the size of fragments for ChIP-seq was 300–400 bp. To obtain 30 ng of DNA to create the DNA library for Ultra High Throughput (UHT) DNA sequencing, 12 ChIP samples were pooled into one sample. DNA was sequenced by Illumina, Inc. using an Ilumina Genome Analyzer, generating 33 bp reads for more than 8 millions fragments (∼250 Mb). A complete description of the results will be described elsewhere.5
Real-time PCR Quantification (q-PCR)
ChIP was performed after 4 days of RNAi against Myc or dHCF, using cells grown in 6-cm plates. 2.5–3 × 107 cells were harvested and formaldehyde cross-linked, DNA isolated and sonicated, samples immunoprecipitated, washed, and reverse cross-linked as described (30), except that, instead of RSB buffer, the cells were lysed in 5 mm PIPES (pH 8.0), 85 mm KCl, 0.5% Nonidet P-40, and the chromatin was sonicated for 12 cycles of 30 s pulses at maximum power using a Bioruptor (Diagenode) to obtain fragments of ∼700 bp. Rabbit anti-dHCFC antibody (25) was used to immunoprecipitate the chromatin. For comparison we used DNA (Input) from a chromatin preparation for which no immunoprecipitation was done. After DNA immunoprecipitation and reverse cross-linking, DNA was purified using a Qiagen PCR purification kit. This experiment was done in triplicate.
Real-time PCR of ChIP DNAs was performed in duplicate using a SYBR Green PCR Master Mix (Applied Biosystems) and Rotor-gene RG300A sequence detector (Corbet Research) under predetermined conditions for all primer sets. PCR quantification was done with delta relative CT quantification, in which the values are calculated relative to input as follows: ΔCT = CT (Input) − CT (sample); CT is the number of cycles when the level of fluorescence gives a signal over the background and in the linear portion of the amplification curve. The sample Input corresponded to 5% of total ChIP input DNA sample, %TI = (2ΔCT) × 20.
Analysis of Target Gene Expression
Luciferase assays and qRT-PCR experiments were carried out as described (Refs. 10, 12 and see supplemental materials).
Protein Analysis
Western blotting, in vitro interactions, and co-immunoprecipitations were carried out as described (Refs. 10, 12 and see supplemental materials).
Antibodies for Western Blotting
Primary antibodies were mouse anti-Myc hybridoma supernatant (diluted 1:5, Ref. 31), mouse anti-α-tubulin (1:25,000, Sigma), rabbit anti-HA-epitope (1:10,000, HA11, ICL), mouse anti-T7-epitope (1:10,000, Novagen), rat anti-dHCF-C terminus (1:3000, Ref. 25). Secondary antibodies were HRP-coupled goat anti-mouse (1:10,000, JacksonImmunoResearch), HRP-coupled goat anti-rat (1:3000, Amersham Biosciences), HRP-coupled donkey anti-rabbit (1:3000, Amersham Biosciences).
Drosophila Work
Flies were kept on standard Drosophila medium. Test crosses were performed in climate-controlled chambers (25 °C on a 12 h light/12 h dark cycle). Fly lines were from the Bloomington stock center, unless otherwise indicated: UAS-T7-dHCF-FLAG (Fig. 2A; Ref. 26), UAS-HCF-IR4353 [3rd] (Fig. 3D; Vienna Drosophila Resource Center [VDRC]), UAS-LacZ-IRM3–1 (12),6 UAS-Ash2-IR1374 [2nd] (Fig. 6B; VDRC), UAS-GCN5-IR11218-T2 [3rd] (Fig. 6B, VDRC). Detailed genotypes for each figure are listed in the supplemental materials.
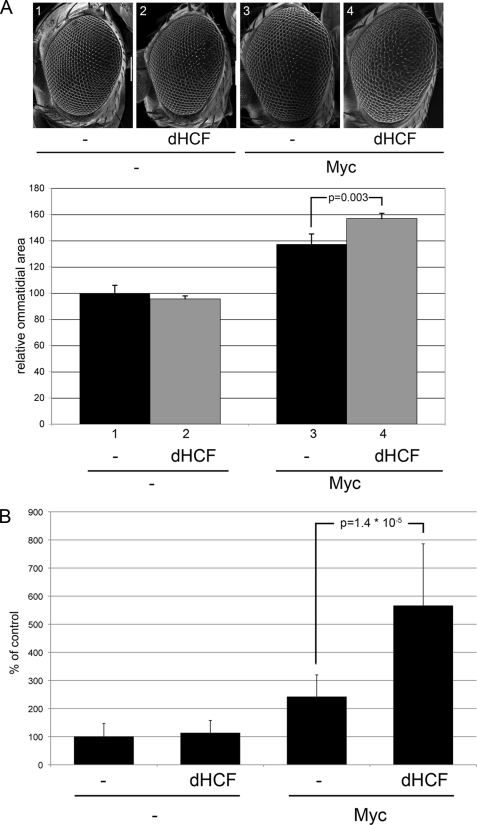
FIGURE 2.
dHCF synergizes with Myc in the induction of growth. A, top panel, scanning electron micrographs (S.E.) of representative eyes overexpressing Myc and/or dHCF under the control of GMR-GAL4; all eyes are at the same magnification. Bottom panel, ommatidial areas of eyes corresponding to the genotypes shown above. Shown are averages and standard deviations from 4 to 6 independent eyes per genotype. B, average areas of 45-h-old wing imaginal disc clones overexpressing Myc and/or dHCF (n = 25, except for the co-expression of Myc and dHCF: n = 17).
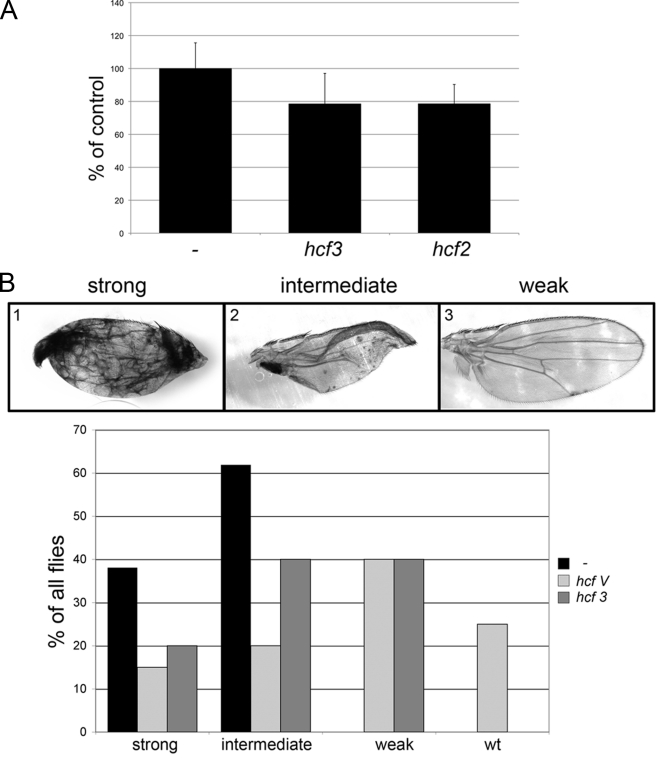
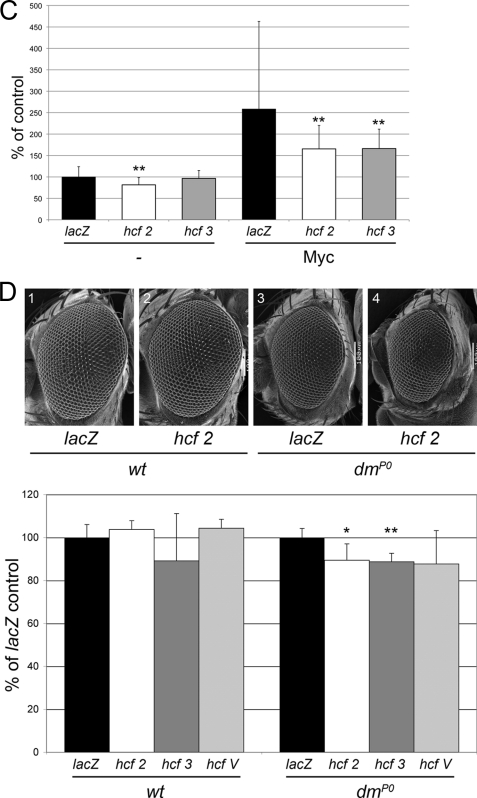
FIGURE 3.
Knockdown of dHCF in vivo impairs (Myc-dependent) growth. A, expression of dHCF-dsRNA in bristle precursor cells with sca-GAL4 reduces the average area of posterior scutellar bristles (n = 6, 7, 4, for control, hcf2, hcf3, respectively); the size reduction is borderline significant (p = 0.06 for both hcf-samples as compared with control). B, knockdown of dHCF alleviates wing defects caused by Myc overexpression under the control of ap-GAL4. Adult Myc-overexpressing wings were assigned to the phenotypic classes shown in the top panel, according to their appearance under a dissecting microscope (n = 21 for control and 25 for each of the dHCF samples). C, average area of salivary gland nuclei that have been overexpressing Myc and/or dHCF-dsRNA for 45 h (n = 34–48). D, expression of dHCF-dsRNA during eye development significantly reduces the area of dmP0 (hypomorphic Myc-mutant) ommatidia, but not of wild-type ommatidia (n = 4–7). * and ** indicate that comparisons with the corresponding control genotype (without dHCF-RNAi) are significant with p < 0.05 and p < 0.01, respectively.
FIGURE 6.
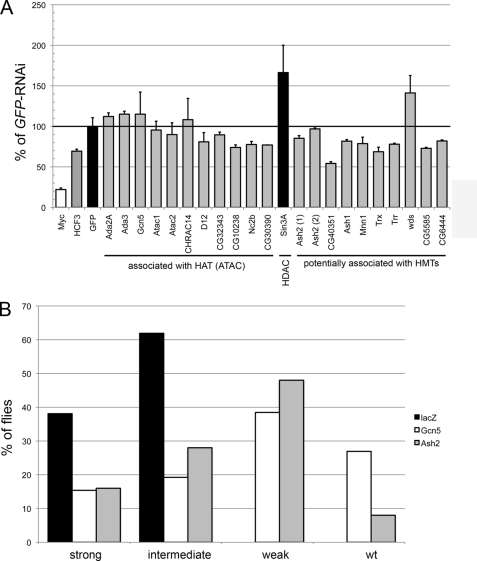
Histone methyltransferases and acetyltransferases affect Myc-dependent processes. A, down-regulation of potentially dHCF-associated HATs (ATAC complex) and HMTs moderately reduces the activity of the Myc-dependent luciferase reporter. Luciferase assays were carried out in duplicate 48 h after co-transfection of the indicated dsRNAs. B, suppression of Myc-induced wing defects by co-expression of ash2- or GCN5-dsRNAs. Neither of these RNAi-transgenes had a strong effect on wing morphology when expressed on their own (although a second independent UAS-GCN5-IR insertion, expressing the same dsRNA, produced strongly deformed wings on its own upon overexpression by ap-GAL4, and it also did not rescue the Myc overexpression defect, presumably because it down-regulated GCN5 too strongly). The experiment was carried out and evaluated as indicated in Fig. 3B (n = 21, 26, 31 for lacZ, GCN5, ash2, respectively). Note that the full composition of the Drosophila dHCF-HMT complex has not been published, and some of the listed proteins were included because of their similarity to vertebrate or yeast HMT-components (Ash2/Hs Ash2L; CG40531/Sc Set1; Ash1, Trx, Trr/SET-domain containing HMTs; Mnn1/Hs Men1; wds/Hs WDR5; CG5585/Hs RBBP5; CG6444/Sc Sdc1); the ATAC components are taken from Refs. 25, 45.
Assays for Myc Activity
Ommatidial areas were determined from scanning electron micrographs of adult eyes as described (27). To determine bristle area, adult scutella were removed, cleaned from attaching muscle tissue, and mounted in 10% glycerol on microscope slides. Pictures were recorded with a Zeiss Axiophot at a 5× magnification, and relative bristle areas were determined using Adobe Photoshop.
Larval imaginal disc and salivary gland clones were generated by heat-shocking 72 h-old larvae carrying “hs-FLP actin-FRT-CD2-FRT-GAL4 UAS-GFP”, as well as additional UAS transgenes, for 8 min at 37 °C. The tissues containing these clones were dissected, fixed, and photographed 45 h later, and the clones analyzed as described (12).
Statistics
All error bars correspond to standard deviations, and p values were calculated using the Student's t test.
RESULTS
dHCF Is Important for the Expression of Myc Targets in Cultured Cells
To identify Myc transcriptional co-factors based on their functional requirement for Myc-dependent gene activation, we developed a Myc-dependent luciferase reporter system. The regulatory sequences of the Myc target gene CG5033 (10), extending from the transcription start site of the upstream neighboring gene to the translation initiation codon, were cloned in front of the Renilla luciferase open reading frame (Fig. 1A). In a second reporter, a mutant derivative of this regulatory region (lacking the only Myc:Max binding E-box) drives the expression of firefly luciferase. We have previously shown that the former construct accurately reflects changes in Myc activity, whereas the latter is unresponsive to Myc (10). Therefore, the ratio of Renilla to firefly luciferase serves as a faithful reporter for Myc activity in cells that have been transfected with both constructs, but it should be insensitive to changes in other pathways that affect CG5033 expression through sequences outside of the E-box, such as alterations in the basal transcription machinery. To find potential co-activators of Myc, we established a list of 752 transcription-associated genes (see “Experimental Procedures” and supplemental Table S2) and targeted them individually with dsRNA. Using this approach, we found that dsRNA directed against dHCF significantly reduces Myc-dependent transcriptional activation.
FIGURE 1.
dHCF is required for the full expression of a Myc reporter. A, schematic of the Myc-responsive luciferase system. The dark line represents genomic sequences from −322 to +61 (start of the open-reading frame) of the Myc target CG5033, and the small gray rectangle shows the position of the single Myc binding site (E-box) in this region. B, schematic of dHCF and the constructs used in this work. The different domains of dHCF have been described before (P175: proline mutated in a temperature-sensitive mutant of vertebrate HCF-1; SAS1N: self-association domain 1N; FnIII/SAS1C: fibronectin-type III domain/self-association domain 1C; NLS: bipartite nuclear localization signal; Refs. 19, 20, 44). Numbers correspond to amino acid positions. In vitro shows the part of dHCF that was used for in vitro translation and in a fusion with GST. The short black lines indicate the extents of dsRNAs used for RNAi (see “Experimental Methods”): 1, from the original screen; 2 & 3, used for tissue culture experiments and in transgenic flies; V, for transgenic experiments. C, efficiency of dHCF knockdown. S2 cells were transfected with pTub-GAL4, UAST-T7-dHCF-FLAG, UAS-GFP either alone (lane −) or together with the indicated hcf dsRNAs; ctr lane lacks ectopically expressed proteins. 48 h later, cell lysates were analyzed by immunoblotting with anti-T7-epitope and anti-GFP antiserum. D, changes of dHCF levels affect the Myc-dependent reporter. S2 were co-transfected with the Myc-dependent luciferase reporters shown in A, together with the indicated dsRNAs or with pACXT-T7-dHCF, and assayed 48 h later for relative luciferase activity (wt-RLuc/ΔE-FLuc, with the gfp-dsRNA sample being set to 100%). Error bars show standard deviations from duplicate transfections.
To demonstrate the specificity of this effect and rule out RNAi off-target effects, we synthesized two additional dsRNA moieties that target non-overlaping sequences in dHCF (Fig. 1B). All three molecules strongly reduce the levels of co-transfected epitope tagged dHCF, and in the case of hcf3 no more protein could be detected (Fig. 1C). All three dsRNAs also significantly reduced the expression of the Myc reporter, albeit not to the same extent as down-regulation of Myc. Conversely, overexpression of dHCF increased the expression of this reporter (Fig. 1D). This demonstrates that dHCF is required for the full expression of the Myc reporter construct and is limiting in normal S2 cells, but it also suggests that Myc retains some activating potential in the absence of dHCF.
To extend these observations to endogenous Myc-regulated promoters, we targeted dHCF by RNAi and then used microarrays to examine the mRNA levels of 30 Myc targets (listed by Ref. 10 in supplemental Table S2, these genes are only moderately affected by Myc knockdown, as is typical for Myc targets, see e.g. 32); these genes include CG5033, from which the luciferase reporter system was derived. As shown in Table 1, 13 of the genes that require Myc for their full expression are also significantly decreased upon depletion of dHCF (43%); all of these genes (including the CG5033 luciferase reporter) contain consensus E-boxes, and therefore most likely correspond to “traditional” Myc targets that rely on Myc:Max heterodimers for their transactivation (12). This effect of dHCF was further confirmed by quantitative real-time PCR on a subset of targets, and it was also shown for a second non-overlapping dHCF-dsRNA fragment (supplemental Fig. S1). In general, knockdown of dHCF affects the Myc targets to a lesser extent than Myc depletion, consistent with our observations with the luciferase reporter system, and Myc targets transcribed by RNA polymerase III (12) are not significantly affected by knockdown of dHCF (supplemental Fig. S1). These results suggest that dHCF is involved in the transcriptional regulation of a subset of native Myc-regulated genes.
TABLE 1.
Many Myc-activated genes are also activated by dHCF in S2 cells
Relative expression levels of previously identified Myc target genes (Ref. 10, supplemental Table S2) at the indicated times after addition of Myc- or dHCF-dsRNA were determined with quantitative real-time PCR (column labeled “PCR”) or with microarrays (all other columns); the data for Myc-RNAi are taken from (Ref. 10, supplemental Table S2). As control, cells were treated with Luciferase-dsRNA (control for column “dHCF-RNAi 3 d”) or GFP-dsRNA (all other columns); the indicated values represent relative reductions of expression in response to dHCF- or Myc-RNAi. Genes printed in italics and boldface are significantly affected by dHCF-RNAi (adjusted p value <0.05). The column labeled “dHCF ChIP” shows the position of the closest dHCF binding site and the column “Myc E-box” indicates the position of consensus Myc binding sites, in nucleotides relative to the transcription start site. The dHCF-RNAi and -ChIP results are from whole genome analyses to be described elsewhere (Footnote 5).
| Gene | dHCF RNAi |
Myc RNAi |
dHCF ChIP | Myc E-box | |||
|---|---|---|---|---|---|---|---|
| 3 d | 6 h | 12 h | 2 d | 2 d PCR | |||
| Ribosome biogenesis/rRNA processing | |||||||
| Fib | 1.4 | 1.8 | 3.5 | 2.8 | 2.4 ± 0.1 | −93 | 26, 834 |
| hoip | 1.3 | 1.6 | 3.4 | 2.0 | 1.6 ± 0.4 | −23 | −15 |
| NHP2 | 1.3 | 2.1 | 2.5 | 2.3 | −1 | 41 | |
| nnp-1 | 1.7 | 2.4 | 2.5 | 1.9 ± 0.2 | 35 | 29 | |
| nop5 | 1.5 | 1.7 | 2.7 | 2.6 | 2.5 ± 0.6 | −23 | −509, −9 |
| nop56 | 1.4 | 1.6 | 2.5 | 2.5 | 2.1 ± 0.2 | −56 | 29 |
| nopp140 | 2.3 | 2.5 | 1.9 | 1 | 48, 385 | ||
| pit | 1.7 | 2.4 | 2.2 | −80 | |||
| CG1381 | 1.2 | 1.6 | 2.3 | 2.1 | −41 | 29 | |
| CG1542 | 2.1 | 2.3 | 2.0 | 11 | 78 | ||
| CG4038 | 1.8 | 2.6 | 2.3 | −14 | 36 | ||
| CG4364 | 1.6 | 1.8 | 2.3 | 2.2 | 4 | −421 | |
| CG5033 | 1.2 | 1.6 | 2.2 | 1.8 | −19 | 21 | |
| CG5728 | 1.9 | 2.4 | 1.7 | −51 | 31, 789 | ||
| CG6712 | 1.3 | 2.6 | 3.7 | 2.8 | 1.6 ± 0.4 | 7 | 46 |
| CG8939 | 1.2 | 1.8 | 2.7 | 2.0 | −34 | 15 | |
| CG9799 | 1.8 | 2.1 | 1.8 | no peak | 6, 44 | ||
| Other/unknown | |||||||
| FK506-bp1 | 1.7 | 1.9 | 1.7 | −43 | −114, 216 | ||
| I(2)09851 | 1.7 | 2.3 | 1.7 | −7 | 35, 830 | ||
| mbm | 2.4 | 3.7 | 2.7 | 423 | 416 | ||
| rrp46 | 2.2 | 2.1 | 2.2 | −18 | |||
| CG1234 | 2.0 | 3.0 | 2.2 | −9 | |||
| CG1785 | 1.9 | 2.4 | 2.1 | −16 | −454, 19 | ||
| CG6751 | 1.7 | 2.2 | 2.0 | 57 | −641, 29 | ||
| CG7845 | 1.3 | 2.9 | 3.5 | 2.0 | 58 | −697, 24 | |
| CG10341 | 1.3 | 1.5 | 2.3 | 1.7 | 1 | 17 | |
| CG10805 | 1.3 | 2.1 | 2.7 | 2.0 | −61 | −13 | |
| CG11660 | 2.3 | 2.1 | 2.0 | 21 | 224 | ||
| CG15019 | 2.0 | 2.0 | 1.7 | −31 | −85, 936 | ||
| CG30349 | 1.8 | 2.7 | 2.1 | 19 | 90, 236 | ||
Overexpressed dHCF Synergizes with Myc in the Control of Growth in Vivo
To determine to which extent the contribution of dHCF is physiologically relevant for the biological activities controlled by Myc, we turned to transgenic flies. It has previously been shown that overexpression of Myc in the developing eye (under the control of GMR-GAL4) results in larger adult ommatidia, reflecting the growth-promoting ability of Myc (8, 12, 28). Overexpression of dHCF under the same regime has no effect on ommatidial size. However, dHCF significantly enhances the growth-promoting activity of co-expressed Myc (Fig. 2A), indicating that dHCF levels indeed affect the rates of growth and that this effect is specifically mediated by Myc. This synergy between Myc and dHCF is reflected at the cellular level. Overexpression of Myc in wing imaginal disc clones is known to strongly stimulate clonal growth (7). Again, expression of dHCF alone does not alter the area of such clones, but it strongly potentiates the ability of Myc to enhance cellular and clonal growth (Fig. 2B).
dHCF Is Required for Myc-dependent Processes in Vivo
To complement this observation, we sought to reduce dHCF levels. Because no dHCF mutants have been published, we resorted to RNA interference to down-regulate dHCF; as in the tissue culture experiments, we expressed different dsRNAs targeting non-overlapping sequences in the dHCF mRNA to rule out RNAi off-target effects (Fig. 1B). When expressed in bristle precursor cells (with sca-GAL4), dsRNA against dHCF leads to a slight reduction in bristle size in adult females (Fig. 3A), although no such effect is seen in males (not shown). This is reminiscent of the prominent effect of Myc on adult bristle size (7, 33), consistent with the notion that dHCF is required for Myc activity in vivo as well. To show this more directly, we monitored the consequences of dHCF down-regulation in the presence of increased Myc levels. Myc was strongly overexpressed under the control of ap-GAL4 in the dorsal compartment of larval wing imaginal discs. As we have described earlier, this treatment promotes an overgrowth of the dorsal wing blade (as compared with the ventral one), resulting in adult flies with bent down wings. In addition, this regime also produces necrotic patches and dissociation of the dorsal and ventral wing surfaces, presumably as a consequence of Myc-driven apoptosis and effects on cell-cell adhesion (Ref. 28 and Fig. 3B). Expression of dHCF-dsRNAs with ap-GAL4 produces adult wings with milder defects: many of them are slightly bent up, as a result of reduced growth in the dorsal half, but only few show necrotic patches and dissociation of ventral and dorsal wing tissue. However, this down-regulation of dHCF significantly suppresses the defects associated with Myc overexpression (Fig. 3B). Importantly, dHCF knockdown does not generally reduce the activity of the GAL4 transcription factor in transgenic flies, as a GAL4-driven LacZ transgene is expressed at similar levels in control larvae and upon co-expression of dHCF-dsRNA (supplemental Fig. S2). Hence, the observed ability of dHCF-RNAi to suppress the Myc-induced wing malformations indicates an effect of dHCF on Myc activity; it suggests that overexpressed Myc requires sufficient quantitites of dHCF to trigger its biological effects.
This requirement can also be seen in the polyploid larval salivary glands. The ploidy of such cells is strongly influenced by Myc: Myc mutant salivary gland cells suffer from a dramatic deficit in endoreplication (6, 34), whereas Myc overexpression results in excessive DNA contents (6, 35). Whereas knockdown of dHCF has a mild effect on the nuclear size of these cells in a control situation (possibly mediated by Myc), depletion of dHCF strongly cripples Myc ability to stimulate endoreplication (Fig. 3C), again indicating that dHCF becomes limiting for Myc activity when Myc is overexpressed. An analogous experiment in clones of wing imaginal disc cells shows that dHCF knockdown also restricts Myc effect on clonal size in diploid tissues. However, depletion of dHCF restricts growth in this tissue even in a control situation, when Myc is not overexpressed, possibly because dHCF is already limiting for the activity of endogenous Myc, or because some of the dHCF effect is mediated by other partners (supplemental Fig. S3).
Finally, a genetic interaction between Myc and dHCF can also be documented that does not rely on overexpression of either of the two partners. To show this interaction, we again used ommatidial size as an indicator of growth. A hypomorphic Myc mutation allows the development of grossly normal eyes, which however are composed of 10% smaller ommatidia (27). Knockdown of dHCF does not significantly affect ommatidial area in a wild-type background, but clearly reduces the size of Myc mutant ommatidia (Fig. 3D). This suggests that dHCF activity becomes limiting not only under conditions of elevated, but also of reduced Myc levels.
dHCF Interacts with Myc to Activate the Expression of Myc Target Genes
These experiments show that dHCF and Myc synergize in different tissues in the induction of growth, that dHCF can become limiting upon overexpression of Myc, and that simultaneous reduction of both dHCF and Myc levels disproportionally affects growth. Together, they indicate that dHCF is physiologically relevant for Myc ability to transactivate its target genes and promote growth, and they raise the question as to the molecular basis of this effect.
Knockdown of dHCF in S2 cells does not affect Myc protein levels (Fig. 4A), suggesting that dHCF might influence the activity of Myc, possibly via a direct interaction. Indeed, Myc contains a sequence DHSY (amino acids 387–390), which conforms to the consensus sequence (D/E) HXY of the HCF-binding motif (HBM). To test for a physical interaction between Myc and dHCF, we in vitro translated Myc and incubated it with a bacterially produced fusion protein between GST and the N-terminal part of dHCF. As shown in Fig. 4B, IVT-Myc binds to GST-dHCFN; conversely, IVT-dHCF interacts with GST-Myc (supplemental Fig. 4B). Under the assay conditions, we detect only background binding of the IVT proteins to GST alone (supplemental Fig. 4, A and B), and a weak interaction with the negative control GST-TF2B (Fig. 4B, supplemental Fig. 4, A and B); this latter interaction may reflect nonspecific binding of the IVT-proteins, and in repeated experiments it is always considerably weaker than the specific interactions described above. A strong interaction is also observed when HA-epitope-tagged Myc and full-length T7-epitope-tagged dHCF are co-expressed in S2 cells: HA-Myc is efficiently co-immunoprecipitated with anti-T7 antibodies from extracts containing T7-dHCF, but not from control extracts, attesting further to the specificity of the interaction (Fig. 4C). In the converse experiment, T7-dHCF is efficiently retrieved in anti-HA immunoprecipitates from extracts containing HA-Myc, but not from control extracts (supplemental Fig. S4C). Despite several attempts, we have not been able to convincingly document the interaction between endogenous Myc and dHCF proteins; this presumably reflects the limitations of the available antibodies and the low expression levels (and possibly solubility) of the proteins.
Unexpectedly, the putative HBM in Myc is not required for its association with dHCF. The Myc derivative MycΔHBM, in which the the potential HBM has been mutated to the sequence “AASA”, efficiently interacts with dHCF, both in vitro (Fig. 4B and supplemental Fig. S4B) and in S2 cells (Fig. 4C). Consistent with this observation, a mutation of the dHCF sequence 175PCPRLG to 175SCPRLG does not abolish binding to either MycWT or MycΔHBM in vitro (Fig. 4B and supplemental Fig. 4, A and B), although mutation of the analogous P134 in vertebrate HCF is known to interfere with binding to HBM-containing vertebrate proteins (18, 36).
These data demonstrate that dHCF directly interacts with Myc, and that this interaction does not require the consensus HBM motif in Myc. To our knowledge, this is the first report of a transcription factor that does not rely on an HBM for its interaction with HCF (see “Discussion”). Additional experiments reveal that none of the previously characterized motifs in Myc (MB1/N terminus, MB2, MB3, BHLHZ) are individually required for binding to dHCF, as all Myc derivatives lacking single motifs still interact with dHCF in transiently transfected S2 cells (supplemental Fig. S4D). However, the experiment shown in Fig. 4D demonstrates that Myc contains at least two independent contact surfaces for dHCF, since two non-overlapping Myc fragments (comprising either the first 295 amino acids, containing the N terminus and MB2, or the amino acids 295–719, containing MB3 and the BHLHZ domain) both interact with dHCF in transiently transfected S2 cells. Hence it remains possible that the protein motifs listed above (including the HBM) contribute to the Myc:dHCF interaction. Interestingly, vertebrate c-Myc has previously been shown to interact with several proteins through both its N terminus and its C terminus (e.g. Skp2, CBP/p300, PARP-10, Refs. 37, 38–41), suggesting that such a bipartite interaction domains may constitute a general feature of Myc proteins.
Interaction of dHCF with Myc on Their Target Genes
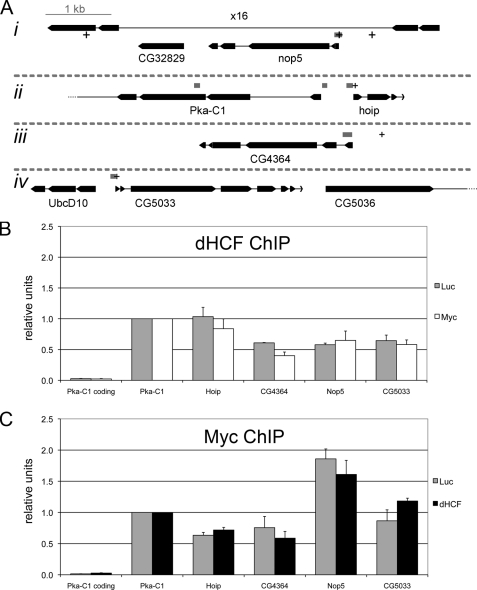
We next looked at the dHCF:Myc interaction in the context of their common target genes. Orian et al. (9) had identified 296 genes that can be bound by Myc in Drosophila Kc cells, using DamID in combination with Drosophila cDNA arrays. Among these genes, we focused on the genes with associated CG identifiers; 159 of these 228 genes (75%) were found also to be bound by dHCF at their promoters in S2 cells by chromatin-immunoprecipitation followed by deep sequencing (ChIP-seq).7 Indeed, all Myc-regulated genes shown in Table 1 (except CG9799) are bound by dHCF, most often close to the transcription start site (Table 1). Four of these Myc/dHCF-regulated genes were selected for further analysis: hoip, nop5, CG4364, and CG5033; a fifth gene, Pka-C1, was chosen for comparison, as it shows associated Myc-binding as well as dHCF at its promoter, but does not change in expression upon depletion of either Myc or dHCF (data not shown). Myc and dHCF promoter occupancy was measured by anti-Myc and anti-dHCF ChIP followed by quantitative PCR with primer pairs directed toward each specific dHCF binding site at the promoter region (see “Experimental Procedures” and Fig. 5A). As expected, all five promoter regions were positive for dHCF binding in this assay (Fig. 5B). Consistent with a close association of dHCF with Myc on these promoters, all five were also positive for Myc binding (Fig. 5C); as a negative control, a PCR fragment located in the Pka-C1 coding region did not interact with either dHCF or Myc (Fig. 5, B and C).
FIGURE 5.
dHCF and Myc co-localize at the promoters of Myc-regulated genes. A, schematic representation of the fragments probed in chromatin immunoprecipitations (ChIP assays). Exons are depicted as boxes and introns as thin lines. Gray boxes show positions of the PCR fragments and + signs consensus E-boxes. The fragment located in the coding region of Pka-C1 (3rd exon) serves as a negative control. All 4 chromosomal regions (i-iv) are depicted at the same scale. B and C, relative binding of dHCF (B) and Myc (C) to the indicated regions. SL2 cells were incubated with the indicated dsRNAs (Myc, dHCF, or Luciferase, which served as a negative control), and chromatin extracts prepared 3 days later for ChIP with polyclonal antibodies against dHCF or Myc. The diagrams represent the average of three biological replicates, and standard errors are shown as error bars.
To determine whether Myc is involved in the recruitment of dHCF to Myc target promoters (or vice versa), we repeated the ChIP analysis after knockdown of either Myc or dHCF. Surprisingly, although Myc was efficiently depleted, dHCF interaction with promoters was little affected, and vice versa (Fig. 5, B and C; supplemental Fig. S5), suggesting that Myc and dHCF bind to their common target genes independently. For Myc, this binding is presumably mediated by direct interaction with specific DNA sequences, in particular consensus E-boxes (Fig. 5A) or variants thereof (not shown). In contrast, HCF proteins are not known to bind DNA directly (18), suggesting that dHCF is recruited by another mechanism, possibly by interaction with other sequence-specific transcription factors such as the E2F proteins (22).
Involvement of dHCF-associated Chromatin-modifying Complexes
The recruitment of dHCF can in turn bring different transcriptional co-factor complexes to these target genes, notably the GCN5 containing ATAC histone acetyltransferase and an Ash2 containing histone methyltransferase complex (see Introduction). We therefore tested whether these complexes are also involved in Myc-dependent processes. Knockdown of several components of the ATAC complex, as well as of several potentially HMT-associated proteins mildly reduce the activity of the Myc-dependent luciferase reporter, consistent with an involvement of such complexes in dHCF-dependent activation of Myc targets (Fig. 6A). Furthermore, knockdown of either Ash2 or GCN5 strongly ameliorated the wing defects caused by Myc overexpression (Fig. 6B), which is consistent with a role for dHCF-HMT and dHCF-ATAC complexes in contributing to the activation of growth-relevant targets of Myc.
DISCUSSION
Here, we identify dHCF as a novel important co-factor for Drosophila Myc. dHCF physically interacts with Myc, and it is required for the full expression of Myc target genes in tissue culture cells, as well as for Myc biological activities in vivo. dHCF acts as a co-activator for Myc-dependent transcription, but clearly it is not the only factor that contributes to Myc-dependent transcription. Several other such co-activators have previously been identified in vertebrate systems, and even near-complete elimination of dHCF from S2 cells only reduces, but does not eliminate, Myc ability to transactivate. Furthermore, dHCF only contributes to the expression of RNA polymerase II-transcribed Myc targets, but does not seem to play an important role for RNA polymerase III-transcribed Myc targets. It is also likely that the extent of dHCF contribution differs for different cell types, although we find dHCF to be important in all the tissues we have investigated so far (eye and wing imaginal discs, salivary glands, adult bristles, S2 tissue culture cells).
How then does dHCF influence the expression of Myc targets? Our data suggest that dHCF reaches these genes independently of Myc. This could occur through one of the other sequence-specific transcription factors that HCF has been shown to bind to. The combined action of these HCF-associated proteins is expected to target HCF to a large fraction of the genome, including many Myc-regulated genes; for example, the dHCF-interacting transcriptonal repressor dE2F2 (22) alone binds to more than 4000 gene promoters (42). Such transcription factors use their HBM to contact a region around proline[134] in human HCF-1; in contrast, the Myc:dHCF interaction requires neither Myc HBM nor the analogous proline[175] in dHCF. Thus, dHCF could potentially bind both Myc and another transcription factor at the same time. Such a trimeric interaction might stabilize the association of either of the three proteins with their common target genes. Alternatively, such an interaction might occur only transiently, to allow the handing over of dHCF from one transcription factor to another one. In this scenario, the Myc putative HBM might directly compete with the HBM of another transcription factor, and the presence of two dHCF-interaction surfaces would favor Myc in this tug-of-war for dHCF. Finally, the HBM could not be involved in the Myc:dHCF interaction at all, a particularly intriguing possibility as vertebrate Myc proteins do not contain a detectable HBM. To address these possibilities, it will be necessary to identify the mechanism by which dHCF binds to target genes in the absence of Myc, and to map the Myc-dHCF interaction surfaces on both proteins.
Irrespective of how dHCF finds these target genes, it is likely that it is not the presence of dHCF per se, but one of its associated protein complexes that enhances the expression of Myc target genes. Three such complexes have been described (see Introduction). The HDAC complex is generally associated with repression and is known to interact with the Myc antagonists of the Mxi/Mnt family; consistent with this, knockdown of the HDAC-component Sin3 increases the expression of the Myc-dependent luciferase reporter. In contrast, knockdown of several putative HMT or ATAC components reduces Myc-dependent transcription and/or suppresses Myc overexpression phenotypes in vivo. The effects on Myc reporter expression are mild, possibly because of inefficient knockdowns. Nevertheless, such a moderate effect of these complexes is consistent with the moderate influence of dHCF on Myc target gene expression. Interestingly, the ATAC component GCN5 (43) and the HMT member Ash2 have previously been shown to physically associate with Myc (Ref. 16 for the Drosophila proteins, Ref. 17 for the vertebrate proteins). It is conceivable that these documented interactions are in part mediated by HCF, and that the interaction between Myc and HCF is conserved in vertebrates. An HBM is not found in vertebrate Myc proteins, but it will be interesting to determine whether the other HCF binding surface(s) on Myc are evolutionarily conserved, and to which extent HCF contributes to the multiple activities of vertebrate Myc proteins in normal situations and in cancer.
Supplementary Material
Acknowledgments
We thank many colleagues for the gifts of reagents (A. Orian, P. Bellosta, D. Stein & J. Workman for antibodies; F. Serras for plasmids and flies; P. Zipperlen, R. Städeli & K. Basler for RNAi reagents) and for advice (L. Atanasyan, P. Zipperlen, G. Hausmann, R. Städeli, C. Mosimann & K. Basler). We also thank M. Moser, R. Grunder, and R. Perez for experimental support.
These studies were supported by grants from the Swiss National Science Foundation (to P. G. and W. H.), from the DFG (to P. G.), and by the University of Lausanne (W. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables S1 and S2.
M. Albarca-Aguilera and W. Herr, unpublished results.
D. Steiger and P. Gallant, unpublished results.
M. Albarca-Aguilera, F. Schütz, V. Praz, M. Delorenzi, and W. Heu, manuscript in preparation.
- HCF
- host cell factor
- dHCF
- Drosophila HCF
- HMT
- histone methyltransferase
- HDAC
- histone deacetylase.
REFERENCES
- 1.Oster S. K., Ho C. S., Soucie E. L., Penn L. Z. (2002) Adv. Cancer Res. 84, 81–154 [DOI] [PubMed] [Google Scholar]
- 2.Pirity M., Blanck J. K., Schreiber-Agus N. (2006) Curr. Top Microbiol. Immunol. 302, 205–234 [DOI] [PubMed] [Google Scholar]
- 3.Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G. R., Bishop J. M. (2001) Nature 414, 768–773 [DOI] [PubMed] [Google Scholar]
- 4.Benassayag C., Montero L., Colombié N., Gallant P., Cribbs D., Morello D. (2005) Mol. Cell Biol. 25, 9897–9909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallant P. (2009) Adv. Cancer Res. 103, 111–144 [DOI] [PubMed] [Google Scholar]
- 6.Pierce S. B., Yost C., Britton J. S., Loo L. W., Flynn E. M., Edgar B. A., Eisenman R. N. (2004) Development 131, 2317–2327 [DOI] [PubMed] [Google Scholar]
- 7.Johnston L. A., Prober D. A., Edgar B. A., Eisenman R. N., Gallant P. (1999) Cell 98, 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montero L., Müller N., Gallant P. (2008) Genesis 46, 104–111 [DOI] [PubMed] [Google Scholar]
- 9.Orian A., Van Steensel B., Delrow J., Bussemaker H. J., Li L., Sawado T., Williams E., Loo L. W., Cowley S. M., Yost C., Pierce S., Edgar B. A., Parkhurst S. M., Eisenman R. N. (2003) Genes Dev. 17, 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulf T., Bellosta P., Furrer M., Steiger D., Svensson D., Barbour A., Gallant P. (2005) Mol. Cell Biol. 25, 3401–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grewal S. S., Li L., Orian A., Eisenman R. N., Edgar B. A. (2005) Nat. Cell Biol. 7, 295–302 [DOI] [PubMed] [Google Scholar]
- 12.Steiger D., Furrer M., Schwinkendorf D., Gallant P. (2008) Nat. Genet. 40, 1084–1091 [DOI] [PubMed] [Google Scholar]
- 13.Cowling V. H., Cole M. D. (2006) Sem. Cancer Biol. 16, 242–252 [DOI] [PubMed] [Google Scholar]
- 14.Teleman A. A., Hietakangas V., Sayadian A. C., Cohen S. M. (2008) Cell Metabol. 7, 21–32 [DOI] [PubMed] [Google Scholar]
- 15.Guccione E., Martinato F., Finocchiaro G., Luzi L., Tizzoni L., Dall' Olio V., Zardo G., Nervi C., Bernard L., Amati B. (2006) Nat. Cell Biol. 8, 764–770 [DOI] [PubMed] [Google Scholar]
- 16.Secombe J., Li L., Carlos L., Eisenman R. N. (2007) Genes Dev. 21, 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lüscher-Firzlaff J., Gawlista I., Vervoorts J., Kapelle K., Braunschweig T., Walsemann G., Rodgarkia-Schamberger C., Schuchlautz H., Dreschers S., Kremmer E., Lilischkis R., Cerni C., Wellmann A., Lüscher B. (2008) Cancer Res. 68, 749–758 [DOI] [PubMed] [Google Scholar]
- 18.Wysocka J., Herr W. (2003) Trends Biochem. Sci. 28, 294–304 [DOI] [PubMed] [Google Scholar]
- 19.Izeta A., Malcomber S., O'Hare P. (2003) Gene 305, 175–183 [DOI] [PubMed] [Google Scholar]
- 20.Mahajan S. S., Johnson K. M., Wilson A. C. (2003) J. Cell Physiol. 194, 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Julien E., Herr W. (2003) EMBO J. 22, 2360–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyagi S., Chabes A. L., Wysocka J., Herr W. (2007) Mol. Cell 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 23.Wysocka J., Myers M. P., Laherty C. D., Eisenman R. N., Herr W. (2003) Genes Dev. 17, 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokoyama A., Wang Z., Wysocka J., Sanyal M., Aufiero D. J., Kitabayashi I., Herr W., Cleary M. L. (2004) Mol. Cell Biol. 24, 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guelman S., Suganuma T., Florens L., Swanson S. K., Kiesecker C. L., Kusch T., Anderson S., Yates J. R., 3rd, Washburn M. P., Abmayr S. M., Workman J. L. (2006) Mol. Cell Biol. 26, 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beltran S., Angulo M., Pignatelli M., Serras F., Corominas M. (2007) Genome Biol. 8, R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellosta P., Hulf T., Balla Diop S., Usseglio F., Pradel J., Aragnol D., Gallant P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11799–11804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwinkendorf D., Gallant P. (2009) Gene 436, 90–100 [DOI] [PubMed] [Google Scholar]
- 29.Lee Y. S., Carthew R. W. (2003) Methods 30, 322–329 [DOI] [PubMed] [Google Scholar]
- 30.Wells J., Boyd K. E., Fry C. J., Bartley S. M., Farnham P. J. (2000) Mol. Cell Biol. 20, 5797–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prober D. A., Edgar B. A. (2000) Cell 100, 435–446 [DOI] [PubMed] [Google Scholar]
- 32.Cole M. D., Cowling V. H. (2008) Nat. Rev. Mol. Cell Biol. 9, 810–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallant P., Shiio Y., Cheng P. F., Parkhurst S. M., Eisenman R. N. (1996) Science 274, 1523–1527 [DOI] [PubMed] [Google Scholar]
- 34.Maines J. Z., Stevens L. M., Tong X., Stein D. (2004) Development 131, 775–786 [DOI] [PubMed] [Google Scholar]
- 35.Britton J. S., Lockwood W. K., Li L., Cohen S. M., Edgar B. A. (2002) Dev. Cell 2, 239–249 [DOI] [PubMed] [Google Scholar]
- 36.Goto H., Motomura S., Wilson A. C., Freiman R. N., Nakabeppu Y., Fukushima K., Fujishima M., Herr W., Nishimoto T. (1997) Genes Dev. 11, 726–737 [DOI] [PubMed] [Google Scholar]
- 37.Kim S. Y., Herbst A., Tworkowski K. A., Salghetti S. E., Tansey W. P. (2003) Mol. Cell 11, 1177–1188 [DOI] [PubMed] [Google Scholar]
- 38.Yu M., Schreek S., Cerni C., Schamberger C., Lesniewicz K., Poreba E., Vervoorts J., Walsemann G., Grötzinger J., Kremmer E., Mehraein Y., Mertsching J., Kraft R., Austen M., Lüscher-Firzlaff J., Lüscher B. (2005) Oncogene 24, 1982–1993 [DOI] [PubMed] [Google Scholar]
- 39.Faiola F., Liu X., Lo S., Pan S., Zhang K., Lymar E., Farina A., Martinez E. (2005) Mol. Cell Biol. 25, 10220–10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von der Lehr N., Johansson S., Wu S., Bahram F., Castell A., Cetinkaya C., Hydbring P., Weidung I., Nakayama K., Nakayama K. I., Söderberg O., Kerppola T. K., Larsson L. G. (2003) Mol. Cell 11, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 41.Vervoorts J., Luscher-Firzlaff J. M., Rottmann S., Lilischkis R., Walsemann G., Dohmann K., Austen M., Luscher B. (2003) EMBO Rep. 4, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Georlette D., Ahn S., MacAlpine D. M., Cheung E., Lewis P. W., Beall E. L., Bell S. P., Speed T., Manak J. R., Botchan M. R. (2007) Genes Dev. 21, 2880–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMahon S. B., Wood M. A., Cole M. D. (2000) Mol. Cell Biol. 20, 556–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capotosti F., Hsieh J. J. D., Herr W. (2007) Mol. Cell Biol. 27, 7063–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suganuma T., Gutiérrez J. L., Li B., Florens L., Swanson S. K., Washburn M. P., Abmayr S. M., Workman J. L. (2008) Nat. Struct. Mol. Biol. 15, 364–372 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.