Abstract
Peptidylarginine deiminases (PADs) are enzymes that convert arginine to citrulline in proteins. In this study, we examined PAD-mediated citrullination and its effect on pro-inflammatory activity in the macrophage cell line RAW 264.7. Citrullination of 45–65-kDa proteins was induced when cells were treated with lipopolysaccharide (LPS; 1 μg/ml). Protein citrullination was suppressed by the intracellular calcium chelator BAPTA/AM (30 μm). LPS treatment up-regulated COX-2 levels in cells. Interestingly, overexpressing PAD2 reduced LPS-mediated COX-2 up-regulation by 50%. PAD2 overexpression also reduced NF-κB activity, determined by NF-κB-driven luciferase activity. The effect of PAD2 on NF-κB activity was further examined by using HEK 293 cells transfected with NF-κB luciferase, IκB β/γ kinase (IKKβ/γ) subunits, and PAD2. IKKβ increased NF-κB activity, but this increase was markedly suppressed when PAD2 was present in cells. IKKβ-mediated NF-κB activation was further enhanced by IKKγ in the presence of calcium ionophore A23187. However, this stimulatory effect of IKKβ/γ was abolished by PAD2. Coimmunoprecipitation of cell lysates showed that IKKγ and PAD2 can coimmunoprecipitate in the presence of the Ca2+ ionophore. IKKγ coimmunoprecipitated truncation mutants, PAD2(1–385) and PAD2(355–672). The substitution of Gln-358 (a putative ligand for Ca2+ binding) with an Ala abolished coimmunoprecipitation. Conversely, PAD2 coimmunoprecipitated truncation mutants IKKγ(1–196) and IKKγ(197–419). In other experiments, treating RAW 264.7 cells with LPS induced citrullination in the immunoprecipitates of IKKγ. In vitro citrullination assay showed that incubation of purified PAD2 and IKKγ proteins in the presence of Ca2+ citrullinated IKKγ. These results demonstrate that PAD2 interacts with IKKγ and suppresses NF-κB activity.
Keywords: Endotoxin, Inflammation, Lipopolysaccharide (LPS), Macrophage, NF-Kappa B, Citrullination, Peptidylarginine Deiminase (PAD)
Introduction
Peptidylarginine deiminases (PADs)2 (E.C.3.5.3.15) are Ca2+-dependent enzymes that catalyze the conversion of arginine in proteins to citrulline with concomitant production of ammonia (1, 2). Because the conversion results in a change in charge (positive to neutral), protein citrullination or deimination can affect both intramolecular and intermolecular interactions, potentially altering protein structure and function (3–5). Citrullination can engender autoimmune responses such as those seen in rheumatoid arthritis (6, 7), multiple sclerosis (8), and psoriasis (9).
Protein citrullination can be induced by inflammatory stimuli such as LPS and TNF (10). Citrullination of proteins such as histones regulates gene transcription (11, 12). Smoking increases PAD expression and citrullination in human lung (13). Among five different PAD isotypes identified in mammals (2), PAD2 and PAD4 are responsible for citrullination in macrophages (14, 15). PAD2 protein is observed in macrophages, although the mRNA is expressed in monocytes and monocytes-derived macrophages (14). PAD4 is expressed mainly in the granulocytes, lymphocytes, endothelial cells, monocytes, and macrophages (16–18). PAD4 is predominantly found in nuclei in these cells. However, the precise role of the citrullination in diseases and inflammation remains unclear. Moreover, the physiological role of PADs in macrophages has yet to be fully determined.
Macrophages are effector cells that regulate inflammatory responses in a variety of tissues including lung. The lung harbors abundant alveolar macrophages and is constantly challenged by inflammatory stimuli. Macrophages are the major responders to LPS (endotoxin) (19, 20), and LPS-activated macrophages produce pro-inflammatory proteins including cyclooxygenase-2 (COX-2) (21). COX-2 is responsible for formation of prostaglandins (21–23), which promote inflammation through a variety of mechanisms. The synthesis of COX-2 is mediated by nuclear translocation and activation of NF-κB that normally resides in the cytoplasm and forms a protein complex with inhibitory κB (IκB). The dissociation of NF-κB from IκB is mediated by IκB kinase (IKK) signalosome, which consists of two catalytic subunits, IKKα and IKKβ, and a regulator, IKKγ. IKKγ (also known as NF-κB essential modulator; NEMO) is an important regulatory component of the IKK complex. When cells are stimulated by LPS, IKK signalosome is activated to cause NF-κB dissociation from IκBα, and NF-κB is translocated to nucleus, where it activates target genes (24, 25).
In this study, we tested whether PAD2-mediated citrullination affects NF-κB activity in the mouse leukemic monocyte macrophage cell line RAW 264.7. We examined LPS-induced citrullination, NF-κB luciferase activity, and in vitro interaction between PAD2 and IKKγ. Our data demonstrate that PAD2 suppresses NF-κB activity probably by interacting with IKKγ.
EXPERIMENTAL PROCEDURES
Reagents
Ultrapure TLR4-specific LPS from Escherichia coli (Alexis Biochemicals, San Diego, CA) was directly added to culture medium. The intracellular calcium chelator BAPTA/AM (Calbiochem) was dissolved in dimethyl sulfoxide (DMSO), and the calcium ionophore A23187 (Sigma-Aldrich) was dissolved in ethanol.
RT-PCR and Full-length cDNA for PAD2
Total RNA was extracted from untreated RAW 264.7 cells using the RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA was synthesized from total RNA by using the SuperScriptTM first-strand synthesis system for RT-PCR (Invitrogen). To confirm the presence of PAD2 and PAD4 in RAW 264.7 cells, we performed PCR. For PCR, two sets of primers were designed based on the published sequence of mouse PAD2 (GenBankTM accession number: NM_008812.1). The forward primer was 5′-CTTCAAGATGGATGAAAATCACCAGG-3′, and the reverse primer was 5′-CACCATGTGCCACCACTTGAAGGC-3′, which amplify a 277-bp PAD2 product. The forward primer of the other sets was 5′-GTTATGTTCAAGGGCCTGGGAGGCATG-3′, and the reverse primer was 5′-TAGCACGATCATGTTCACCATGTTAGG-3′, which amplify a 246-bp PAD2 product. PCR was performed for 30 cycles with Pfu DNA polymerase (Promega, Madison, WI) at 94 °C for 40 s, 60 °C for 30 s, and 72 °C for 40 s. The final extension time at 72 °C was 7 min. The products were sequenced in the DNA Sequencing Facility at Vanderbilt University.
The full-length PAD2 cDNA was cloned by PCR with primers corresponding to the 5′and 3′ ends of mouse PAD2. The sequence of the full-length PAD cDNA was identical to the published PAD2 of mouse (the amino acid sequence of cloned PAD2 is available in the supplemental material).
Plasmids
FLAG-tagged PAD2 was constructed by excising out full-length PAD2 from the plasmid pBlescript/PAD2 with EcoRI and SalI and subcloning into pCMV-Tag 2C Vector, FLAG-tagged truncation mutants PAD(1–385) and PAD(355–672) were made by deletion of corresponding nucleotides. The point mutant PAD(355–672)/Q358A was generated using a site-directed mutagenesis kit (Agilent Technologies). The N-terminal hemagglutinin (HA)-tagged IKKγ was made by ligating IKKγ into pCDNA 3.1(+) plasmid (Invitrogen). HA-tagged truncation mutants IKKγ(1–196) and IKKγ(197–419) were constructed by deletion of corresponding nucleotides. FLAG-tagged IKKβ plasmid was kindly provided by F. Mercurio (Celgene Corp., Summit, NJ), and T7-tagged NEMO/IKKγ plasmid was a gift from E. S. Alnemri (Thomas Jefferson University). The luciferase reporter plasmid NF-κB-Luc was purchased from Agilent Technologies.
Cell Transfection
The murine macrophage cell line RAW 264.7 and human embryonic kidney (HEK) 293 cells were purchased from the American Type Culture Collection (Manassas, VA). Cells were cultured in DMEM (Invitrogen) supplemented with 10% endotoxin-free, heat-inactivated fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml) in a 5% CO2 atmosphere at 37 °C in a humidified incubator. Transfection was achieved with GeneporterTM 2 transfection reagent (Gene Therapy Systems, San Diego, CA) for RAW 264.7 cells and Lipofectamine 2000 (Invitrogen) for HEK 293 cells according to the manufacturer's instructions. Cells at 24 h after transfection were used for experiments.
Detection of Citrullinated Proteins
Protein citrullination was analyzed by immunoblot with an antibody specific to modified peptidylcitrulline residues. Cells were lysed with radioimmunoprecipitation assay cell lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% sodium orthovanadate, 1% Triton X-100, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate) supplemented with EDTA-free protease inhibitor cocktails (Roche Diagnostics). Cell debris was removed by microcentrifugation for 10 min, and protein concentration was determined by the Bradford assay reagent (Bio-Rad). Equal amounts of proteins were separated by SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and then cross-linked by formaldehyde incubation to improve protein retention. Citrullinated proteins were detected using the anti-citrulline (modified) detection kit (Millipore, Billerica, MA) with modified anti-citrulline rabbit polyclonal antibody (1:1000) and a goat anti-Rabbit IgG antibody conjugated with horseradish peroxidase (GAR-HRP) (1:5000; Millipore). Citrullinated protein bands were detected using the enhanced chemiluminescence ECL Plus (GE Healthcare). As an internal control, the membrane was stripped and reprobed with β-actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunoblot for COX-2 in RAW 264.7 Cells
Equal amounts of proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. The membrane was preincubated in phosphate-buffered saline (PBS) containing 0.05% Tween 20 and 5% nonfat dry milk and then incubated with polyclonal anti-COX-2 (Cayman Chemical) at room temperature for 2 h. The blot was washed with PBS containing 0.05% Tween 20 for 20 min (4 × 5 min) and then incubated with GAR-HRP (1:5000) for 1 h. Tubulin (Santa Cruz Biotechnology) was used as an internal control.
Immunoprecipitation
HEK 293 cells were co-transfected with (i) HA-tagged IKKγ (WT) and FLAG-tagged PAD2 (WT) or truncation mutants or (ii) FLAG-tagged PAD2 (WT) and HA-tagged IKKγ (WT) or truncation mutants for determining domains responsible for interaction. At 24 h after transfection, cells were treated with either 1 μm A23187 or no A23187 in serum-free medium for 1 h. Cell lysates were prepared in immunoprecipitation buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1.5 mm EDTA, 3% glycerol, 0.5% Nonidet P-40, 1 mm NaF, and 1 mm Na3VO4) supplemented with 1 mm dithiothreitol and EDTA-free protease inhibitor cocktails (Roche Diagnostics). Lysates were incubated with the monoclonal anti-HA antibody (1–2 μg) (Cell Signaling, Danvers, MA) at 4 °C overnight and then with protein A-Sepharose beads (Zymed Laboratories Inc., San Francisco, CA) at 4 °C for 30 min. The beads were washed with immunoprecipitation buffer containing 250 mm NaCl more than five times, and the immunoprecipitates were dissociated from the beads by adding SDS-sample buffer and boiling. The immunoprecipitates were subjected to immunoblotting with monoclonal anti-FLAG M2 antibody (Sigma-Aldrich). Cell lysates for input were immunoblotted with monoclonal anti-FLAG M2 antibody or monoclonal anti-HA antibody and with a goat horseradish peroxidase-conjugated anti-mouse IgG antibody (1:5000; Millipore). For immunoprecipitation of endogenous IKKγ from RAW 264.7 cells, cells were treated with BAPTA/AM (30 μm) for 1 h and then with LPS (1 μg/ml) for 4 h. Cells were then lysed with immunoprecipitation buffer and subjected to immunoprecipitation with anti-IKK-γ antibody (BD Pharmingen). After washes, protein citrullination was determined. To separate immunoprecipitated IKKγ (48 kDa) from IgG heavy chains (50 kDa), the SDS sample buffer without β-mercaptoethanol was used for SDS-PAGE.
NF-κB Reporter Gene Assay
The NF-κB reporter assay was performed by the Dual-Luciferase assay kit (Promega) as described previously (22). Briefly, RAW 264.7 cells were transfected with NF-κB-Luc reporter plasmid, and 24 h after transfection, cells were treated with LPS (1 μg/ml) for 4 h. For HEK 293 cells, cells were transfected in combination with NF-κB-Luc, FLAG-tagged PAD2, FLAG-tagged-IKKβ, and HA-tagged IKKγ. Cells at 24 h after transfection were then treated with A23187 (1 μm) for 1 h to induce PAD2 activation. After washes with PBS, cell extracts were prepared with lysis buffer, and 20-μl aliquots of the supernatant were mixed with 100 μl of luciferase assay solution provided by the company. Luciferase activity was measured using the Synergy 2 SL luminescence microplate reader (BioTek, Winooski, VT).
In Vitro Citrullination of IKKγ
To purify IKKγ protein, HEK 293 cells were transfected with T7-IKKγ plasmid and lysed 24 h later. T7-IKKγ proteins were purified using the T7·Tag affinity purification kit (EMD Chemicals, Gibbstown, NJ). After washing away unbound proteins, T7-IKKγ was eluted from beads at pH 2.2. The protein fractions were analyzed by SDS-PAGE followed by silver staining. For in vitro citrullination assay, 0.1 mg/ml purified IKKγ was mixed with 1 unit of rabbit muscle PAD2 (Sigma-Aldrich) in reaction buffer (0.1 m Tris-HCl, pH 8.0, 10 mm CaCl2, 2 mm DTT) at 55 °C for 2 h. Reaction was stopped by adding 0.5 m EDTA. Citrullination was determined by the immunoblot described above. Silver staining of the proteins loaded on the gel was performed using the Silver Stain Plus kit (Bio-Rad).
Statistical Analysis
Data were reported as means ± S.E. Levels of significance were assessed using the unpaired, two-tailed Student's t test for comparison between two samples and one-way analysis of variance with Bonferroni post test for comparison among more than two samples. The p value of less than 0.05 was considered significant. Data were analyzed using Origin 8.1 software (OriginLab Corp., Northampton, MA).
RESULTS
PAD Activity Is Induced by LPS Treatment in RAW 264.7 Cells
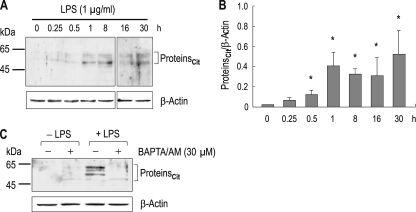
To examine PAD activity in RAW 264.7 cells, we treated cells with LPS (1 μg/ml in DMEM containing 2 mm Ca2+) for 0.25–30 h and determined protein citrullination by immunoblot with the antibody to modified citrulline. Fig. 1A shows the time course of citrullination after LPS treatment (n = 4). Proteins with the molecular mass of 45–65 kDa were prominently citrullinated, comparable with a previous report (26). Most of time, two or three citrullinated major bands appeared on the Western blot. The reason for this is unclear. Vertical stripes or dots at 0.25, 0.5, 16, and 30 h are immunoblot artifacts. Citrullination was gradually observed over time. When compared with control (0 h), substantial levels of citrullination were found at 1 h determined by densitometric measurements of citrullinated proteins normalized to β-actin (p < 0.05; one-way analysis of variance with Bonferroni post test) (Fig. 1B). Activation of PAD enzymes requires Ca2+ (1, 2, 4), and thus, to further characterize citrullination, we applied LPS to cells for 4 h after pretreating with the Ca2+ chelator BAPTA/AM (30 μm) for 1 h. Fig. 1C shows a representative immunoblot result. Control cells without LPS had negligible citrullination regardless of BAPTA/AM in the medium (Fig. 1C, lanes 1 and 2). A slight citrullination in the presence of BAPTA/AM is often observed, the reason for which is unclear. Cells with LPS had typical citrullination in the absence of BAPTA/AM (lane 3). However, the citrullination was suppressed by BAPTA/AM (lane 4). These results demonstrate that LPS induces protein citrullination in RAW 264.7 cells and that citrullination is calcium-dependent.
FIGURE 1.
Lipopolysaccharide-induced citrullination in RAW 264.7 cells. A, time course of citrullination after LPS treatment. Cells were treated with LPS (1 μg/ml) for 0.25–30 h and lysed for immunoblot analysis with the anti-citrulline antibody (n = 4). β-Actin served as an internal control. Vertical stripes or dots at 0.25 (lane 2), 0.5 (lane 3), 16 (lane 6) and 30 h (lane 7) are artifacts. Proteincit, citrullinated proteins. B, densitometric measurements of citrullinated proteins. The intensity of immunoreactive bands was normalized to the intensity of β-actin (n = 4). Artifacts were not included for quantitation. C, effect of the Ca2+ chelator BAPTA/AM on citrullination. Cells were incubated with 30 μm BAPTA/AM or none for 1 h before LPS treatment (1 μg/ml; 4 h). *, p < 0.05.
PAD2 Reduces COX-2 Expression in LPS-treated Cells
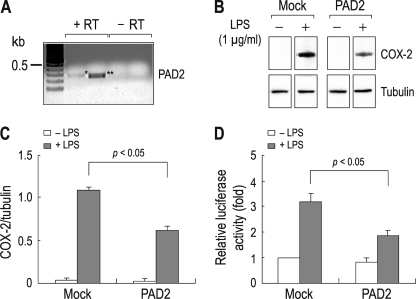
Among five PAD isotypes characterized in mammals (2), PAD2 and PAD4 are closely associated with rheumatoid arthritis (7). PAD2 is expressed in cytosol, whereas PAD4 is predominantly found in nucleus (27), and thus, we focused on PAD2 in this study to determine the effect of cytosolic citrullination on inflammatory gene expression in macrophage cells. RT-PCR with primers specific to mouse PAD2 detected PAD2 mRNA in RAW 264.7 cells (Fig. 2A). PCR without reverse transcriptase detected none. We also performed RT-PCR with primers for PAD4 but could not detect its mRNA (data not shown). We then cloned the full-length PAD2 cDNA by PCR with primers corresponding to mouse PAD2 (amino acid sequence comparison of the cloned mouse PAD2 with human PAD4 is provided in the supplemental material).
FIGURE 2.
Suppression of NF-κB activity by PAD2 overexpressed in RAW 264.7 cells. A, RT-PCR for PAD2. Expression of PAD2 mRNA was determined by PCR with two sets of primers amplifying 277 bp (*; lane 2) and 246 bp (**; lane 3) PAD2 cDNA fragments. Total RNA extracted from untreated cells was used. PCR without RT reaction (−RT) served as negative controls. PCR products were sequenced to confirm the specificity of the products. B, effect of PAD2 overexpression on COX-2 production. Cells transfected with the full-length mouse PAD2 were treated with LPS (1 μg/ml) for 4 h. COX-2 production was determined by immunoblot. Cells transfected with vector only (Mock) served as controls. Tubulin served as an internal control for immunoblot. C, comparison of COX-2 expression between control cells and PAD2-expressing cells. Densitometric measurements of immunoreactive bands were done (n = 3). The intensity of COX-2 was normalized to the intensity of tubulin. D, NF-κB activity in control cells and PAD2-expressing cells. Cells transfected with the NF-κB reporter plasmid and PAD2 or none were incubated for 24 h and then treated with LPS for 4 h. NF-κB activity was measured by luciferase assay (n = 4) as described previously (22). The activity was presented relative to the value in mock cells without PAD2 and LPS (leftmost bar).
RAW 264.7 cells were transfected with PAD2 and treated with LPS for 4 h, and an immunoblot was performed to determine COX-2 protein expression. Fig. 2B shows that COX-2 expression was induced by LPS, consistent with previous reports (21–23). The induction occurred in both groups of cells transfected with PAD2 or vector only (Mock in the figure), but the COX-2 expression level was substantially lower in cells transfected with PAD2. This decrease corresponded to 50% of the controls (p < 0.05; n = 3; Fig. 2C). The internal control tubulin had negligible change.
This finding suggests that PAD2 can suppress COX-2 expression induced by LPS. To further characterize this effect, we compared NF-κB activity between control cells and PAD2-overexpressing cells. Cells were transiently transfected with PAD2 and the NF-κB luciferase construct, and 24 h later, luciferase activity in cells was measured after 4 h of treatment with LPS (Fig. 2D). Controls were cells without PAD2 (Mock in the figure). The application of LPS increased luciferase activity by 3.2- ± 0.3-fold (n = 4) in control cells. However, in PAD2-expressing cells, LPS increased luciferase activity by 1.7- ± 0.2-fold (n = 4). The decrease corresponded to 47% of the control (p < 0.05).
PAD2 Inhibits IKK Complex Activity
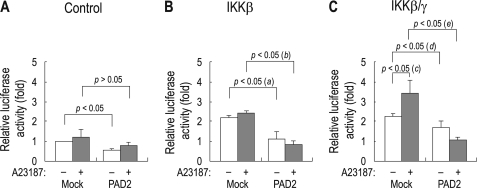
We tested whether PAD2 inhibits IKK complex from activating NF-κB. For this analysis, we used HEK 293 cells to ensure robust expression of wild type and truncated mutant DNA plasmids. HEK 293 cells were transiently transfected with IKKβ and IKKγ subunits, PAD2, and NF-κB-luciferase construct. PAD2 activation was achieved by inducing Ca2+ influx using 1 μm calcium ionophore A23187 (18). Fig. 3A shows NF-κB activity in cells transfected with or without PAD2 (no subunits of IKK complex). The values were presented relative to the value for cells without PAD2. Overall, NF-κB activities were low in these control cells regardless of PAD2 expression and A23187 treatment (n = 3 for each).
FIGURE 3.
Suppression of IKK activity by PAD2. A, NF-κB activity in HEK 293 cells transfected with the NF-κB reporter and either PAD2 or vector only (Mock). B and C, NF-κB activity in HEK 293 cells transfected with IKKβ (B) or IKKβ/γ (C) in addition to the plasmids described in A. The values were presented relative to the value in mock cells that have no PAD2 and the calcium ionophore A23187 (leftmost bar in panel A). Twenty-four hours after transfection, cells were treated with the calcium ionophore A23187 (1 μm) for 1 h to induce PAD2 activation. All data were obtained from three independent experiments. (a), p < 0.05; (b), p < 0.05; (c), p < 0.05; (d) and (e), p < 0.05.
Fig. 3B shows NF-κB activity in cells transfected with IKKβ with or without PAD2. IKKβ increased NF-κB activity by 2.1-fold when compared with control cells without IKKβ (p < 0.05). A23187 treatment had negligible effect. However, the increased NF-κB activity by IKKβ was significantly suppressed in cells expressing PAD2 (p < 0.05; (a) in the figure). The suppression also occurred in the presence of A23187 (p < 0.05; (b) in the figure), thus indicating that PAD2 reduces IKKβ activity regardless of Ca2+ in the bath. In the third set of cells transfected with IKKβ/γ with or without PAD2 (Fig. 3C), IKKβ/γ coexpression increased NF-κB activity by 2.4-fold (p < 0.05). The activity was further stimulated in the presence of A23187 (p < 0.05; (c) in the figure), consistent with the report (28) that nuclear transport of IKKγ requires Ca2+. However, this IKKβ/γ-mediated effect was suppressed by PAD2 (p < 0.05; (d) and (e) in the figure). The suppression was more substantial in the presence of A23187 as the NF-κB activity was substantially reduced to the basal level for control cells.
PAD2 Interacts with IKKγ
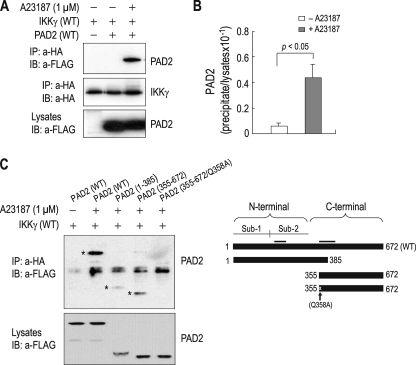
The above finding suggests that PAD2 suppresses NF-κB activity by affecting the IKK complex. Given the fact that IKKγ interacts with many different proteins (29, 30), we then hypothesized that PAD2 might also interact with IKKγ to achieve suppression. To test this hypothesis, we performed coimmunoprecipitation of HEK 293 cells expressing PAD2 and IKKγ. Cells were transfected with FLAG-tagged PAD2 and HA-tagged IKKγ, and lysates were immunoprecipitated with the anti-HA antibody and immunoblotted with anti-FLAG antibody. Fig. 4A shows that PAD2 was detected in the immunoprecipitates from cells. The detection required Ca2+ as it was absent under A23187-free conditions. This also indicates that the detection is not due to incomplete washing of protein A-Sepharose beads. PAD2 levels are similar in the two groups of cells treated with and without the ionophore (bottom panel; one-tenth of the lysates used for coimmunoprecipitation was loaded on the gel). Densitometric measurements of PAD2 signal intensity in the immunoprecipitates versus lysates (one-tenth) were estimated to be 4.3% (n = 4) of PAD2 interacting with IKKγ (Fig. 4B).
FIGURE 4.
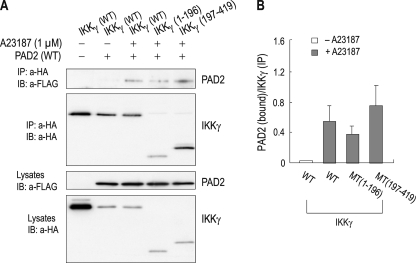
Interaction of PAD2 and IKKγ. A, coimmunoprecipitation of PAD2 and IKKγ. HEK 293 cells were transfected with the plasmid containing HA-tagged IKKγ and either FLAG-tagged PAD2 or vector only. Twenty-four hours later, cells were treated with 1 μm A23187 for 1 h, lysed, and immunoprecipitated with the anti-HA antibody (a-HA). The immunoprecipitates (IP) were then immunoblotted (IB) with the anti-FLAG antibody (a-FLAG). One-tenth of lysates was loaded as input controls (bottom panel). B, densitometric measurements of PAD2 bound to IKKγ. The intensity of PAD2 in the immunoprecipitates was normalized to the corresponding intensity in the lysates (n = 4). C, coimmunoprecipitation of truncation mutants of PAD2 with IKKγ. PAD2(1–385) contains the N-terminal 385 amino acids, whereas PAD2(355–672) contains the C-terminal 323 amino acids. PAD2(355–672)/Q358A has an Ala instead of Gln-358 (a putative ligand for Ca2+ binding). The molecular masses of PAD2(1–385) and PAD2(355–672) are 42 and 35 kDa, respectively. The 50-kDa bands in all lanes are IgG heavy chains. *, PAD2 coimmunoprecipitated with IKKγ. The schematic diagram for truncation mutants is shown in the right panel. Putative ligand sites for Ca2+ binding are clustered in two areas (bars). Sub-1, Subdomain-1; Sub-2, Subdomain-2.
To further characterize PAD2/IKKγ interaction, we made truncation mutants: PAD2(1–385), which has the N-terminal 385 amino acids of the protein, and PAD2(355–672), which has the C-terminal 318 amino acids. In addition, a point mutation replacing Gln-358 (a putative ligand for Ca2+ binding; see supplemental material) with an Ala was introduced on PAD2(355–672) to evaluate the effect of Ca2+ on PAD2/IKKγ interaction. Fig. 4C shows both PAD2(1–385) and PAD2(355–672) being coimmunoprecipitated with IKKγ (the 50-kDa band in all lanes is IgG heavy chain). The signal intensity of PAD2(1–385) was relatively weak when compared with those for wild type PAD2 and PAD2(355–672), but PAD2(1–385) protein expression in lysates also appeared to be low. Thus, the weak PAD2(1–385) signal in the immunoprecipitates does not represent weak interaction between this mutant and IKKγ. In contrast, the point mutant PAD2(355–672)/Q358A was not detected. There was no immunoreactive band corresponding to the truncation protein that serves as the template protein. Thus, the mutation at Gln-358 completely abolished PAD2/IKKγ interaction despite the presence of A23187 in the medium.
In other experiments, we also constructed truncation mutants of IKKγ: IKKγ(1–196), which contains the N-terminal 196 amino acids of the protein, and IKKγ(197–419), which contains the remaining amino acids. Coimmunoprecipitation was performed with cells transfected with PAD2 and these mutants as well as wild type IKKγ. PAD2 was coimmunoprecipitated with these truncation mutants (Fig. 5A). The amount of PAD2 being coimmunoprecipitated with IKKγ(1–196) appeared to be lower, but the amount of IKKγ(1–196) bound to protein A-Sepharose beads was also low. Thus, densitometric measurements of PAD2 relative to IKKγ (Fig. 5B) exhibited similar values among different immunoprecipitates (p > 0.05; n = 4). These results indicate that the interaction of PAD2 and IKKγ involves multiple bindings, at least one in the region of 1–196 residues of IKKγ and another in the region of 197–419 residues.
FIGURE 5.
Binding assay of PAD2 and truncation mutants of IKKγ. A, coimmunoprecipitation of PAD2 and truncation mutants of IKKγ. IKKγ(1–196) contains the N-terminal 196 amino acids of the protein, whereas IKKγ(197–419) contains the remaining 223 amino acids. The molecular masses of IKKγ(1–196) and IKKγ(197–419) are 22 and 25 kDa, respectively. Coimmunoprecipitation was performed with the protocol described in the legend for Fig. 4. For control cells (lane 1), a 5-fold higher concentration of IKKγ was used for transfection to ensure that the lack of PAD2 in the immunoprecipitates (IP) is not due to insufficient amounts of IKKγ. IB, immunoblot; a-HA, anti-HA antibody; a-FLAG, anti-FLAG antibody. B, relative ratio of PAD2 to IKKγ in the immunoprecipitates. The pixel intensity of PAD2 was normalized to the intensity of IKKγ in the same immunoprecipitates (n = 4). MT, truncation mutant of IKKγ.
PAD2 Can Citrullinate IKKγ
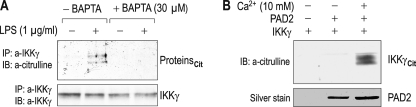
The finding of the ability of PAD2 to suppress IKKβ/γ activity and interact with IKKγ led us to postulate that PAD2 might citrullinate IKKγ and/or its associated proteins. To test this hypothesis, we performed two sets of experiments. First, RAW 264.7 cells were treated with LPS after pretreatment with BAPTA/AM (30 μm), and immunoprecipitation was performed with an anti-IKKγ antibody. The immunoprecipitates were then immunoblotted with anti-citrulline antibody. Fig. 6A shows representative results. LPS-induced citrullination was observed in IKKγ immunoprecipitates (lane 2) but abolished by BAPTA/AM (lane 4), consistent with the Ca2+-dependent PAD activity. The detection of citrullinated proteins indicates that either IKKγ or its binding partners, or both, is citrullinated. The anti-citrulline antibody recognized double bands, whereas the anti-IKKγ antibody recognized a single IKKγ band in a sister immunoblot membrane. Although this could be due to a relatively small amount of IKKγ being citrullinated, we do not exclude the alternative possibility that other proteins interacting with IKKγ might be citrullinated.
FIGURE 6.
Citrullination of IKKγ by PAD2. A, detection of citrullination in IKKγ immunoprecipitates (IP) from RAW 264.7 cells. Cells were incubated with 1 μg/ml LPS for 4 h after pretreatment with BAPTA/AM (30 μm) for 1 h, and immunoprecipitation was performed with an anti-IKKγ antibody (a-IKKγ). The immunoprecipitates were then immunoblotted (IB) with anti-citrulline antibody (a-citrulline). Proteincit, citrullinated proteins. B, in vitro citrullination assay. Purified IKKγ and PAD2 proteins were incubated in reaction buffer containing 10 mm Ca2+ for 2 h. The reaction mixture was then immunoblotted with the anti-citrulline antibody. Silver staining was done with sister mixture loaded on the gel (bottom panel). IKKγcit, citrullinated IKKγ.
Thus, in the second set of experiments, we performed the in vitro citrullination assay in which purified IKKγ and PAD2 proteins are incubated in the buffer containing Ca2+ (10 mm). Probed with the anti-citrulline antibody, the immunoblot showed citrullination of IKKγ (Fig. 6B). Citrullination did not occur without PAD2 or with IKKγ and PAD2 in Ca2+-free buffer.
DISCUSSION
Overview
The major findings from the present study are the following. 1) LPS induces Ca2+-dependent citrullination in the macrophage cells RAW 264.7; 2) PAD2 overexpression in macrophage cells reduces COX-2 production; 3) activated PAD2 suppresses IKKβ/γ activity; 4) PAD2 interacts with IKKγ in the presence of Ca2+; and 5) PAD2 can citrullinate IKKγ. These findings provide novel evidence that PAD2 can reduce inflammatory responses by suppressing IKKβ/γ and interacting with, and probably citrullinating, the regulatory protein IKKγ in macrophage cells.
Based on these findings, we propose a model for the effect of protein citrullination on cellular function in immune effector macrophages. LPS causes intracellular Ca2+ to rise (31), which then induces the conversion of inactive PAD2 to active PAD2 to mediate protein citrullination (supplemental material). Activated PAD2 then suppresses the IKK complex activity by binding to IKKγ. The binding of PAD2/IKKγ may lead to citrullination of IKKγ and/or IKKγ-interacting proteins. The essential part of the model is that PAD2 acts to down-regulate pro-inflammatory gene expression induced by LPS. Activation of PAD2 in inflammation might serve to self-limit inflammatory processes by modifying key LPS-sensing signaling molecules.
LPS Treatment Induces PAD Activity in RAW 264.7 Cells
Many reports show that citrullination occurs in a variety of proteins, including keratin (32), histones (10), vimentin (26), and myelin basic proteins (33). Citrullination also occurs in chemokines (34–36) such as CXCL8–12. Given the fact that the conversion of positively charged arginine into uncharged citrulline causes changes in protein folding, citrullination has been known to alter protein structure and function in cells (1, 2, 4). Citrullination has attained strong pathological and clinical interests because of its close association with autoimmune diseases such as rheumatoid arthritis (7). The goal of our study was to understand whether PAD2-mediated protein citrullination in macrophages is associated with inflammatory responses.
We found that protein citrullination is induced by LPS in RAW 264.7 cells (Fig. 1). Citrullination starts to occur within 1 h after LPS treatment and continues over time. LPS increases intracellular free Ca2+ levels by ∼150-fold within 4 min in RAW 264.7 cells (31). This increase is mediated by L-type Ca2+ channels (31). PAD is a Ca2+-dependent enzyme that requires Ca2+ for substrate binding. Thus, Ca2+ influx by LPS is sufficient to bring out PAD activity. Protein citrullination at early time points after LPS treatment is probably due to the conversion of inactive PAD to active PAD by transient increase in intracellular Ca2+. As for citrullination at later time points, PAD activity may be induced from pro-inflammatory proteins produced by LPS. Prostaglandin D2 produced by COX-2 acts to cells in autocrine and paracrine fashions (37). Prostaglandin D2 can induce PAD activity in RAW 264.7 cells (38). Prostaglandin D2 receptor (D2 receptor) activation also results in intracellular Ca2+ increase by Ca2+ influx (39).
PAD2 Reduces NF-κB Activity
The novel finding in our study is that PAD2 can reduce COX-2 production mediated by LPS in macrophage cells (Fig. 2, B and C). This reduction is due to the capability of PAD2 to suppress NF-κB activity (Fig. 2D). PAD2-mediated suppression of NF-κB is also observed in HEK 293 cells expressing PAD2 and IKKβ and IKKγ subunits (Fig. 3). IKKβ is a catalytic subunit that can phosphorylate IκBα, which binds to NF-κB and inhibits its function (25). Phosphorylation of IκBα results in ubiquitination. In contrast, IKKγ (also known as NEMO) is the regulatory subunit. IKKγ interacts preferentially with IKKβ and is required for the activation of the IKK complex (40). Our study shows that PAD2 markedly suppresses IKKβ alone as well as IKKβ/γ. The effect of PAD2 on IKKβ occurs regardless of Ca2+ in the medium, whereas the effect on IKKβ/γ appears to be dependent upon Ca2+. Thus, PAD2 appears to be activated to suppress IKKβ/γ activity. Liu et al. (41) reported that PAD4 overexpression in hematopoietic cells induces apoptosis. One might thus think that the reduced expression of COX-2 in our study would be due to apoptotic events caused by PAD2 overexpression in cells. However, in the previous report, the apoptotic event occurs >48 h after PAD4 overexpression, and we found that there is no difference in cell viability 24–32 h after PAD2 transfection (data not shown).
PAD2 Interacts with IKKγ
A number of proteins have been identified to interact with IKKγ. The essential function of these IKKγ-binding proteins is to modulate IKK activity. At least 16 IKKγ-binding proteins (30) are reported to enhance IKK complex activity, and at least eight IKKγ-interacting proteins are reported to suppress the complex (29). The mechanisms of their interactions with IKKγ vary depending upon the molecular and cellular nature of individual proteins. Our study shows that PAD2 is another member of the proteins interacting with IKKγ (Fig. 4). The significance of this interaction is its dependence on Ca2+. The interaction does not occur under normal conditions, in which intracellular Ca2+ is low. In response to LPS, however, intracellular Ca2+ levels increase to mediate the conversion of inactive PAD to active PAD2, and activated PAD2 then interacts with IKKγ. Therefore, the interaction is an induced event that occurs in response to inflammatory stimuli.
What would be the consequence of PAD2/IKKγ interaction? IKKγ plays important roles in regulating IKK complex activation. Proteins interacting with IKKγ activate or down-regulate NF-κB activity (29, 30). Obviously, PAD2/IKKγ interaction appears to down-regulate IKK complex activity. The interaction may serve as an inhibitory pathway to reduce the LPS-mediated signaling cascade. PAD2 decreases COX-2 expression by 50% (Fig. 2C) and NF-κB activity by 47% (Fig. 2D), both of which are substantial changes. These lead us to reevaluate the pathological role of protein citrullination in inflammation. Citrullination causes changes in protein folding, which lead to abnormal protein structure and function (3–5). Therefore, citrullination induced by LPS is clearly disadvantageous to cells. On the other hand, the activation of PAD2 occurring in inflammation may be a process that limits inflammation. PAD2 may play an anti-inflammatory role by reducing IKKγ activity and/or modifying key molecules responsible for LPS-induced inflammation (Fig. 5). In this sense, we note that Prostaglandin D2, which can induce citrullination (38), functions as an anti-inflammatory molecule in lung inflammation (42).
Mechanism of PAD2/IKKγ Interaction
Given the finding that the mutation of a putative Ca2+ coordination site Gln-358 completely abolishes the interaction (Fig. 4C), we conclude that PAD2/IKKγ interaction requires Ca2+. This result provides valuable information on the molecular mechanism of the interaction. Structural analysis of PAD4 (4) suggests that the binding of Ca2+ to the acidic concave surface in the C-terminal domain is critical for substrate specificity and binding. The binding of Ca2+ to PAD4 leads to conformational changes around the substrate-binding site, which then allows PAD4 to interact with arginine residues in other proteins. PAD4 has many residues for Ca2+ coordination, and one of these residues is Gln-349 (corresponding to Gln-358 in PAD2). Replacing Gln-349 with an Ala abolishes citrullination activity of PAD4. Thus, in our study, Q358A probably inhibits PAD2 from binding Ca2+ and subsequently fails to induce conformational change required for IKKγ binding. Nonetheless, this interpretation makes it difficult to explain the interaction between IKKγ and PAD2(1–385), which does not contain the site for substrate binding. The N-terminal domain of PAD4 is proposed to be responsible for a Ca2+-dependent regulation or protein-protein interaction (4). Thus, the interaction between IKKγ and PAD2 not only requires the C-terminal substrate-enzyme interaction, but it may also involve N-terminal protein-protein interaction.
Summary
Our study shows that protein citrullination is induced by LPS in macrophages and that citrullination can suppress up-regulation of pro-inflammatory gene expression. Activated PAD2 can interact with IKKγ and may citrullinate IKK subunits. PAD2-mediated suppression of IKK complex may be a novel mechanism that regulates inflammatory response. The exact mechanism underlying PAD2-mediated suppression needs further investigation.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants PO1 HL66196 (to B. W. C., M. J., and T. S. B.), R01-GM078502 (to I. C.), and HL81332 (to L. B. W.) and HL088263 (to L. B. W.). This work was also supported by an American Heart Association Established Investigator Award (L. B. W.).

The on-line version of this article (available at http://www.jbc.org) contains two supplemental figures.
- PAD
- peptidylarginine deiminase
- IKK
- IκB kinase
- NEMO
- NF-κB essential modulator
- BAPTA/AM
- 1,2-bis (o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetra (acetoxy-methyl) ester.
REFERENCES
- 1.Rogers G. E. (1962) Nature 194, 1149–1151 [DOI] [PubMed] [Google Scholar]
- 2.Vossenaar E. R., Zendman A. J., van Venrooij W. J., Pruijn G. J. (2003) Bioessays 25, 1106–1118 [DOI] [PubMed] [Google Scholar]
- 3.Tarcsa E., Marekov L. N., Mei G., Melino G., Lee S. C., Steinert P. M. (1996) J. Biol. Chem. 271, 30709–30716 [DOI] [PubMed] [Google Scholar]
- 4.Arita K., Hashimoto H., Shimizu T., Nakashima K., Yamada M., Sato M. (2004) Nat. Struct. Mol. Biol. 11, 777–783 [DOI] [PubMed] [Google Scholar]
- 5.Arita K., Shimizu T., Hashimoto H., Hidaka Y., Yamada M., Sato M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5291–5296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.György B., Tóth E., Tarcsa E., Falus A., Buzás E. I. (2006) Int. J. Biochem. Cell Biol. 38, 1662–1677 [DOI] [PubMed] [Google Scholar]
- 7.Foulquier C., Sebbag M., Clavel C., Chapuy-Regaud S., Al Badine R., Méchin M. C., Vincent C., Nachat R., Yamada M., Takahara H., Simon M., Guerrin M., Serre G. (2007) Arthritis Rheum. 56, 3541–3553 [DOI] [PubMed] [Google Scholar]
- 8.Wood D. D., Bilbao J. M., O'Connors P., Moscarello M. A. (1996) Ann. Neurol. 40, 18–24 [DOI] [PubMed] [Google Scholar]
- 9.Ishida-Yamamoto A., Senshu T., Takahashi H., Akiyama K., Nomura K., Iizuka H. (2000) J. Invest Dermatol. 114, 701–705 [DOI] [PubMed] [Google Scholar]
- 10.Neeli I., Khan S. N., Radic M. (2008) J. Immunol. 180, 1895–1902 [DOI] [PubMed] [Google Scholar]
- 11.Cuthbert G. L., Daujat S., Snowden A. W., Erdjument-Bromage H., Hagiwara T., Yamada M., Schneider R., Gregory P. D., Tempst P., Bannister A. J., Kouzarides T. (2004) Cell 118, 545–553 [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Wysocka J., Sayegh J., Lee Y. H., Perlin J. R., Leonelli L., Sonbuchner L. S., McDonald C. H., Cook R. G., Dou Y., Roeder R. G., Clarke S., Stallcup M. R., Allis C. D., Coonrod S. A. (2004) Science 306, 279–283 [DOI] [PubMed] [Google Scholar]
- 13.Makrygiannakis D., Hermansson M., Ulfgren A. K., Nicholas A. P., Zendman A. J., Eklund A., Grunewald J., Skold C. M., Klareskog L., Catrina A. I. (2008) Ann. Rheum. Dis. 67, 1488–1492 [DOI] [PubMed] [Google Scholar]
- 14.Vossenaar E. R., Radstake T. R., van der Heijden A., van Mansum M. A., Dieteren C., de Rooij D. J., Barrera P., Zendman A. J., van Venrooij W. J. (2004) Ann. Rheum. Dis. 63, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe K., Akiyama K., Hikichi K., Ohtsuka R., Okuyama A., Senshu T. (1988) Biochim. Biophys. Acta 966, 375–383 [DOI] [PubMed] [Google Scholar]
- 16.Asaga H., Nakashima K., Senshu T., Ishigami A., Yamada M. (2001) J. Leukoc. Biol. 70, 46–51 [PubMed] [Google Scholar]
- 17.Nakashima K., Hagiwara T., Ishigami A., Nagata S., Asaga H., Kuramoto M., Senshu T., Yamada M. (1999) J. Biol. Chem. 274, 27786–27792 [DOI] [PubMed] [Google Scholar]
- 18.Nakashima K., Hagiwara T., Yamada M. (2002) J. Biol. Chem. 277, 49562–49568 [DOI] [PubMed] [Google Scholar]
- 19.Beutler B. (2002) Curr. Top. Microbiol. Immunol. 270, 109–120 [DOI] [PubMed] [Google Scholar]
- 20.Beutler B., Rietschel E. T. (2003) Nat. Rev. Immunol. 3, 169–176 [DOI] [PubMed] [Google Scholar]
- 21.Dubois R. N., Abramson S. B., Crofford L., Gupta R. A., Simon L. S., Van De Putte L. B., Lipsky P. E. (1998) FASEB J. 12, 1063–1073 [PubMed] [Google Scholar]
- 22.Joo M., Kwon M., Sadikot R. T., Kingsley P. J., Marnett L. J., Blackwell T. S., Peebles R. S., Jr., Urade Y., Christman J. W. (2007) J. Immunol. 179, 2565–2575 [DOI] [PubMed] [Google Scholar]
- 23.Tanaka Y., Takizawa M., Igimi S., Amano F. (2004) Biol. Pharm. Bull. 27, 985–991 [DOI] [PubMed] [Google Scholar]
- 24.Kawai T., Akira S. (2007) Trends Mol. Med. 13, 460–469 [DOI] [PubMed] [Google Scholar]
- 25.Medzhitov R., Janeway C., Jr. (2000) N. Engl. J. Med. 343, 338–344 [DOI] [PubMed] [Google Scholar]
- 26.Vossenaar E. R., Després N., Lapointe E., van der Heijden A., Lora M., Senshu T., van Venrooij W. J., Ménard H. A. (2004) Arthritis Res. Ther. 6, R142–R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirai H., Blundell T. L., Mizuguchi K. (2001) Trends Biochem. Sci. 26, 465–468 [DOI] [PubMed] [Google Scholar]
- 28.Berchtold C. M., Wu Z. H., Huang T. T., Miyamoto S. (2007) Mol. Cell Biol. 27, 497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shifera A. S. (2010) Biochem. Biophys. Res. Commun. 396, 585–589 [DOI] [PubMed] [Google Scholar]
- 30.Shifera A. S. (2010) J. Cell Physiol. 223, 558–561 [DOI] [PubMed] [Google Scholar]
- 31.Kim Y., Moon J. S., Lee K. S., Park S. Y., Cheong J., Kang H. S., Lee H. Y., Kim H. D. (2004) Biochem. Biophys. Res. Commun. 314, 695–703 [DOI] [PubMed] [Google Scholar]
- 32.Senshu T., Akiyama K., Ishigami A., Nomura K. (1999) J. Dermatol. Sci. 21, 113–126 [DOI] [PubMed] [Google Scholar]
- 33.Moscarello M. A., Wood D. D., Ackerley C., Boulias C. (1994) J. Clin. Invest. 94, 146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proost P., Loos T., Mortier A., Schutyser E., Gouwy M., Noppen S., Dillen C., Ronsse I., Conings R., Struyf S., Opdenakker G., Maudgal P. C., Van Damme J. (2008) J. Exp. Med. 205, 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loos T., Mortier A., Gouwy M., Ronsse I., Put W., Lenaerts J. P., Van Damme J., Proost P. (2008) Blood 112, 2648–2656 [DOI] [PubMed] [Google Scholar]
- 36.Struyf S., Noppen S., Loos T., Mortier A., Gouwy M., Verbeke H., Huskens D., Luangsay S., Parmentier M., Geboes K., Schols D., Van Damme J., Proost P. (2009) J. Immunol. 182, 666–674 [DOI] [PubMed] [Google Scholar]
- 37.Ben-Baruch A., Michiel D. F., Oppenheim J. J. (1995) J. Biol. Chem. 270, 11703–11706 [DOI] [PubMed] [Google Scholar]
- 38.Lee H. J., Young G., Kwon M., Du R., Abdolrasulnia R., Christman B. W., Joo M. (2006) Proc. Amer. Thrac. Soc. 3, A232 [Google Scholar]
- 39.Boie Y., Sawyer N., Slipetz D. M., Metters K. M., Abramovitz M. (1995) J. Biol. Chem. 270, 18910–18916 [DOI] [PubMed] [Google Scholar]
- 40.Rothwarf D. M., Zandi E., Natoli G., Karin M. (1998) Nature 395, 297–300 [DOI] [PubMed] [Google Scholar]
- 41.Liu G. Y., Liao Y. F., Chang W. H., Liu C. C., Hsieh M. C., Hsu P. C., Tsay G. J., Hung H. C. (2006) Apoptosis 11, 183–196 [DOI] [PubMed] [Google Scholar]
- 42.Rajakariar R., Hilliard M., Lawrence T., Trivedi S., Colville-Nash P., Bellingan G., Fitzgerald D., Yaqoob M. M., Gilroy D. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20979–20984 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.