Abstract
Furaquinocin is a natural polyketide-isoprenoid hybrid (meroterpenoid) that exhibits antitumor activity and is produced by the Streptomyces sp. strain KO-3988. Bioinformatic analysis of furaquinocin biosynthesis has identified Fur7 as a possible prenyltransferase that attaches a geranyl group to an unidentified polyketide scaffold. Here, we report the identification of a physiological polyketide substrate for Fur7, as well as its reaction product and the biochemical characterization of Fur7. A Streptomyces albus transformant (S. albus/pWHM-Fur2_del7) harboring the furaquinocin biosynthetic gene cluster lacking the fur7 gene did not produce furaquinocin but synthesized the novel intermediate 2-methoxy-3-methyl-flaviolin. After expression and purification from Escherichia coli, the recombinant Fur7 enzyme catalyzed the transfer of a geranyl group to 2-methoxy-3-methyl-flaviolin to yield 6-prenyl-2-methoxy-3-methyl-flaviolin and 7-O-geranyl-2-methoxy-3-methyl-flaviolin in a 10:1 ratio. The reaction proceeded independently of divalent cations. When 6-prenyl-2-methoxy-3-methyl-flaviolin was added to the culture medium of S. albus/pWHM-Fur2_del7, furaquinocin production was restored. The promiscuous substrate specificity of Fur7 was demonstrated with respect to prenyl acceptor substrates and prenyl donor substrates. The steady-state kinetic constants of Fur7 with each prenyl acceptor substrate were also calculated.
Keywords: Bacterial Metabolism, Enzyme Kinetics, Enzymes, Isoprenoid, Metabolism, Streptomyces, Biosynthesis, Polyketide, Prenyltransferase
Introduction
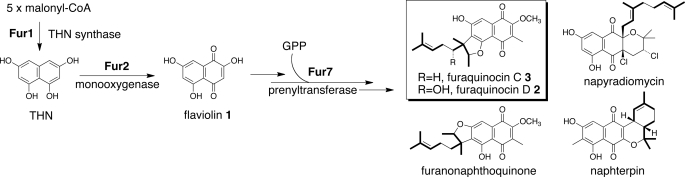
Several Streptomyces strains produce polyketide-isoprenoid hybrids (meroterpenoids), including furaquinocin (1, 2), naphterpin (3, 4), napyradiomycin (5), and furanonaphthoquinone (6). These meroterpenoids share a tetrahydroxynaphthalene (THN)-derived skeleton with an attached 10-carbon geranyl group (see Fig. 1). THN2 is generated through Claisen condensation reactions from five molecules of malonyl-CoA by the action of the ubiquitous Streptomyces type III polyketide synthase or THN synthase (7, 8). In fact, the THN synthase genes (e.g. fur1 for furaquinocin biosynthesis) are found in biosynthetic gene clusters for each meroterpenoid (9–12). The THN skeleton is presumed to be oxidized to 2,5,7-trihydroxynaphthalene-1,4-dione (flaviolin; 1) by a monooxygenase (13) (for example, fur2 for furaquinocin) and then further modified by methylation, prenylation, and cyclization before final biosynthetic elaboration of the meroterpenoids. However, no biosynthetic pathway for meroterpenoids has been elucidated biochemically.
FIGURE 1.
Biosynthesis of furaquinocin and other meroterpenoids produced by Streptomyces strains. The biosynthesis of furaquinocin involves the prenylation of an unidentified THN derivative using a GPP co-substrate.
Currently, four prenyltransferases that are presumed to conjugate a 10-carbon geranyl group have been identified in the biosynthetic gene clusters for furaquinocin (9), naphterpin (10), napyradiomycin (11), and furanonaphthoquinone (12). Of the prenyltransferases, two enzymes, NphB (10, 14) and Fnq26 (15), have been characterized. NphB is present in the naphterpin biosynthetic gene cluster from Streptomyces sp. strain CL190 and accepts a diverse collection of hydroxyl-containing aromatic substrates, such as flavonoids, to yield the corresponding prenylated products in the presence of divalent cations (14). The crystal structure of NphB has been solved: it is an architecturally new prenyltransferase, which consists of the characteristic five α-β-β-α secondary structure elements of the overall protein fold topology (10). Fnq26, cloned from the furanonaphthoquinone-producing S. cinnamonensis strain DSM 1042, shows 40% sequence identity to NphB. Independently of divalent cations, Fnq26 catalyzes the formation of a carbon-carbon bond between the C-3 of a geranyl group and the C-3 of 1 to yield 3-(3,7-dimethylocta-1,6-dien-3-yl)-2,5,7-trihydroxynaphthalene-1,4-dione (3-prenylflaviolin) (15). However, because 3-prenylflaviolin is not an intermediate of furanonaphthoquinone biosynthesis, 1 is not considered a genuine substrate of Fnq26. Therefore, no genuine substrates for the prenyltransferases that are responsible for meroterpenoid biosynthesis have been identified. In other words, the promiscuity of these prenyltransferases makes it difficult to identify their genuine physiological substrates.
All of the genes required for furaquinocin biosynthesis have been cloned, and heterologous production of furaquinocin has been demonstrated in Streptomyces lividans TK23 (9). Bioinformatic analysis of furaquinocin biosynthesis has identified Fur7 as a possible prenyltransferase that attaches a geranyl group to an unidentified polyketide skeleton (Fig. 1). Fur7 shows 35 and 64% sequence identities to NphB and Fnq26, respectively. Because the meroterpenoids share a common THN-derived skeleton, the prenyltransferases should play important roles in contributing to the structural diversity of meroterpenoids. However, neither the physiological substrates for the prenyltransferases nor their prenylated products have been identified. We report, for the first time, the identification of a physiological Fur7 prenyltransferase substrate and its prenylated products. We also demonstrate the conversion of one intermediate into the final furaquinocin, establishing its place in the chemical pathway. In addition, we demonstrate the promiscuity of Fur7 prenyltransferase with respect to prenyl acceptor substrates and prenyl donor substrates. The present study provides insight into the biosynthesis of meroterpenoids in various Streptomyces strains.
EXPERIMENTAL PROCEDURES
Plasmid, Strains, and Chemicals
The previously prepared plasmid pWHM-Fur2 (see Fig. 2A, GenBankTM accession no. AB212624) was used for the heterologous production of furaquinocin D (2) in S. lividans TK23 (9). Compound 2 was purified as described previously and used as a standard. Streptomyces albus was used as a heterologous host, and geranyl diphosphate (GPP) was synthesized as described previously (16).
FIGURE 2.
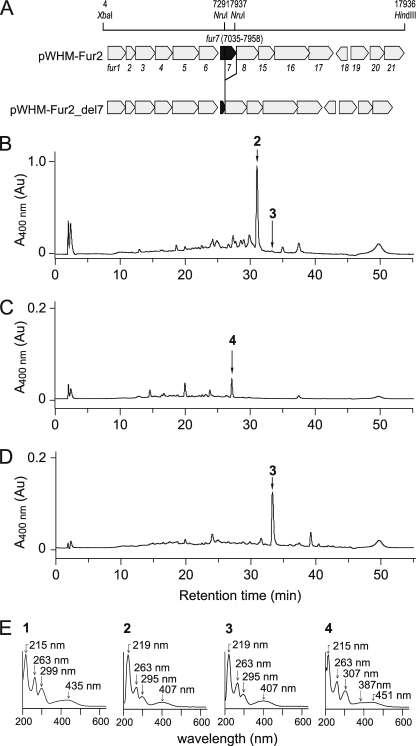
HPLC analysis of S. albus transformant broth. A, the structure of the furaquinocin biosynthetic gene cluster. pWHM-Fur2 contains all of the genes required for the biosynthesis of 2, whereas pWHM-Fur2_del7 lacks the fur7 gene. HPLC analysis of the products from the S. albus transformant broth. B, S. albus/pWHM-Fur2. C, S. albus/pWHM-Fur2_del7. D, S. albus/pWHM-Fur2_del7 supplemented with Fur7 and GPP). Small peaks at 10–25 min displayed UV-visible spectra that were distinctly different from that of 1. E, UV-visible spectra of 1 and 2 and the accumulated products 3 and 4. Au, absorbance units.
Structure Determination of Unidentified Products
The structures of unidentified products were analyzed by 1H NMR spectroscopy, 13C NMR spectroscopy, heteronuclear multiple-bond connectivity (HMBC) spectroscopy (600 MHz, JEOL ECA-600), and high resolution MS (JEOL JMS-T100LC). MS analysis was performed using electrospray ionization (ESI) in the negative ion mode.
Heterologous Expression of Furaquinocin Gene Cluster in S. albus
Transformation of S. albus was performed by the protoplast method (17). The S. albus/pWHM-Fur2 transformant was grown in 500-ml Erlenmeyer flasks containing 100 ml of NMMP (17) medium and 30 μg ml−1 thiostrepton. Fermentation was conducted for 4 days at 27 °C after 2 days of preculture at 30 °C in tryptone soya broth medium (Kanto Chemical) containing 30 μg ml−1 thiostrepton.
Construction of fur7 Deletion Plasmid
The plasmid pWHM-Fur2_del7, which contained the furaquinocin gene cluster lacking only the fur7 gene, was constructed by deleting the 646-bp NruI-NruI fragment located within the fur7 gene in pWHM-Fur2. The pWHM-Fur2 plasmid was digested with NruI and religated to construct pWHM-Fur2_del7 (Fig. 2A).
HPLC Analysis of Furaquinocins and Unidentified Biosynthetic Intermediates
Furaquinocins and the unidentified biosynthetic intermediates were analyzed on a HPLC system equipped with an MD-2010 Plus photodiode array (JASCO, Tokyo, Japan) and a PEGASIL ODS column (4.6 × 250 mm; column temperature, 25 °C; Senshu Scientific, Tokyo, Japan) under the following conditions: mobile phase A, acetonitrile + 0.1% acetate; mobile phase B, water + 0.1% acetate; 2–75% A over 30 min, 75–98% A over 5 min, 98% A for 10 min, and then 2% A for 10 min, at a flow rate of 1.0 ml min−1 (Fig. 2, B–D). The unidentified products were collected under these HPLC conditions and identified using a Hewlett Packard Series 1100 HPLC system linked to an Applied Biosystems API3000 LC/MS/MS spectrometer. The furaquinocin standards and the isolated products were loaded onto and separated with a PEGASIL ODS column (2.0 × 150 mm; temperature, 25 °C; Senshu Scientific) under the following conditions: mobile phase A, acetonitrile + 0.1% acetatel; mobile phase B = water + 0.1% acetate, 40–98% A over 18 min, 98% A for 2 min, and then 40% A for 10 min, at a flow rate of 0.2 ml min−1. MS analysis was performed by ESI in the negative ion mode.
Purification of Furaquinocin C (3)
The S. albus transformant harboring pWHM-Fur2_del7 was grown as described above for S. albus/pWHM-Fur2. After 1 day of culture, the Fur7 enzyme (at a final concentration of 0.1 mg ml−1) and GPP (at a final concentration of 0.1 mm) were added directly to the broth; the culture was then incubated for 2 days. The supernatant (1 liter) of the S. albus/pWHM-Fur2_del7 culture supplemented with the Fur7 enzyme and GPP was extracted twice with ethyl acetate. After being dried over Na2SO4, the organic layer was evaporated in vacuo. Next, the metabolites were purified by preparative HPLC with a PEGASIL ODS column (20 × 250 mm; Senshu Scientific) and an isocratic elution of 80% methanol at a flow rate of 8 ml min−1; the column eluate was monitored at 265 nm. Furaquinocin C (3; 1 mg) was obtained as a white solid and used as a standard.
NMR spectroscopy of 3 showed the following profile: 1H NMR (600 MHz, DMSO-d6) δ: 1.35 (s, 3H, Me-9′), 1.37 (d, J = 6.2 Hz, 3H, Me-1′), 1.40 (s, 3H, Me-10′), 1.53 (s, 3H, Me-8′), 1.65 (m, 2H, H-4′), 1.81 (m, 2H, H-5′), 1.86 (s, 3H, 3-Me), 3.86 (s, 3H, 2-O-Me), 4.40 (q, J = 6.2 Hz, 1H, H-2′), 4.95 (t, J = 6.8 Hz, 1H, H-6′), and 7.01 (s, 1H, H-8); and 13C NMR (150 MHz, DMSO-d6) δ: 9.7 (3-Me), 14.0 (C-1′), 17.9 (C-10′), 24.0 (C-9′), 25.9 (C-8′), 26.0 (C-5′), 35.1 (C-4′), 46.5 (C-3′), 60.8 (2-O-Me), 90.8 (C-2′), 108.3 (C-4a), 109.0 (C-8), 124.8 (C-6′), 127.2 (C-6), 131.1 (C-7′), 132.6 (C-3), 133.3 (C-8a), 156.8 (C-2, C-7), 159.6 (C-5), 180.8 (C-1), and 182.9 (C-4).
Purification of 2-methoxy-3-methylflaviolin 4
The S. albus transformant harboring pWHM-Fur2_del7 was grown as described above for S. albus/pWHM-Fur2. The supernatant (4 liters) of the S. albus/pWHM-Fur2_del7 culture was extracted twice with ethyl acetate. After being dried over Na2SO4, the organic layer was evaporated in vacuo. The metabolites were purified by preparative HPLC with a PEGASIL ODS column (20 × 250 mm; Senshu Scientific) and an isocratic elution of 60% methanol at a flow rate of 8 ml min−1; the column eluate was monitored at 265 nm. Solid, red 2-methoxy-3-methylflaviolin 4 (1 mg) was obtained. High resolution ESI-MS showed a peak of m/z 233.0404 (calculated for C12H9O5 (M-H)−, −4.6 mDa error).
NMR spectroscopy of 4 showed the following profile: 1H NMR (600 MHz, CD3OD-d4) δ: 1.99 (s, 3H, 3-Me), 4.03 (s, 3H, 2-O-Me), 6.45 (s, 1H, H-6), and 6.95 (s, 1H, H-8); and 13C NMR (150 MHz, CD3OD-d4) δ: 7.3 (3-Me), 60.9 (2-O-Me), 107.2 (C-6), 108.3 (C-4a), 107.8 (C-8), 131.1 (C-3), 133.5 (C-8a), 157.9 (C-2), 159.6 (C-5), 163.6 (C-7) 180.4 (C-1), and 189.5 (C-4).
Overexpression and Purification of Fur7
The previously prepared pWHM-Fur2 construct (9), which included fur7, was used as a PCR template for ligation into the E. coli expression vector pHIS8 (18). The gene (DDBJ/EMBL/GenBankTM accession no. AB187171) was amplified with the forward primer 5′-GGGCCATGGCCCGGTACGGACGATGTCGC-3′ (NcoI site underlined) and the reverse primer 5′-GGGGGATCCTCATGTCGACTCCTTGTCGCG-3′ (BamHI site underlined) to generate pHis8_fur7. E. coli BL21(DE3) cells transformed with the resulting plasmid pHis8_fur7 were cultivated in 1 liter of liquid TB medium containing 50 μg ml−1 kanamycin and grown at 37 °C to an A600 of 1.5. The cells were cooled on ice for 10 min and then induced with 0.5 mm isopropyl thiogalactoside for 20 h at 18 °C. The cells were harvested by centrifugation and resuspended in 100 ml of lysis buffer (50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 20% glycerol, 20 mm imidazole, and 1% Tween 20). The cell suspensions were sonicated with a Branson Sonifier 250 (Emerson Japan, Tokyo, Japan). To separate the cellular debris from the soluble protein, the lysate was centrifuged at 17,000 rpm at 4 °C for 20 min. Purification using nickel-nitrilotriacetic acid-agarose resin (GE Healthcare Life Sciences) was performed according to the manufacturer's instructions, and the protein was eluted with 250 mm imidazole in 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, and 20% glycerol. The His8-tag was removed by incubation with 20 units of thrombin for 16 h at 4 °C during dialysis against 50 mm Hepes (pH 7.5), and 500 mm NaCl. The dialyzed protein was passed over nickel-nitrilotriacetic acid-agarose and benzamidine-Sepharose (GE Healthcare) and concentrated by ultrafiltration with Vivaspin 20 (10,000 molecular weight cut-off; Sartrius Stedim Biotech). Approximately 50 mg of purified Fur7 were obtained.
Enzyme Assay with 2-Methoxy-3-methylflaviolin 4
The Fur7 assay was performed in 50 mm Hepes-NaOH (pH 7.5) containing 5 mm MgCl2, 1 mm 2-methoxy-3-methylflaviolin 4, 5 mm GPP, and 1 mg ml−1 Fur7. The reaction mixture was incubated at 30 °C for 2 or 16 h. After incubation, the reaction mixture was extracted with ethyl acetate. The extracts were evaporated, and the residue was redissolved in methanol. The reaction products were analyzed on an HPLC (JASCO, Tokyo, Japan) equipped with a MD-2010 plus photodiode array with a PEGASIL ODS column (2.0 × 150 mm, column temperature, 25 °C; Senshu Scientific) using an isocratic elution of 90% acetonitrile + 0.1% acetate at a flow rate of 0.2 ml min−1 for 15 min (Fig. 4A).
FIGURE 4.
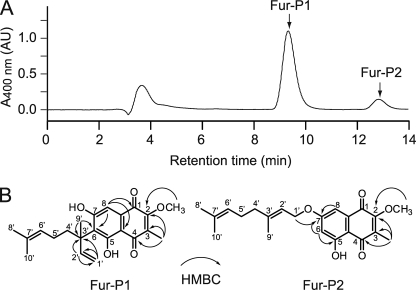
Prenylated products formed from 2-methoxy-3-methylflaviolin by the Fur7 prenyltransferase. A, HPLC analysis of the two prenylated products, Fur-P1 and Fur-P2, from the Fur7 reaction with 4 (1 mm) and GPP (5 mm). B, structures of the two products, Fur-P1 and Fur-P2, elucidated from NMR data. Carbon atoms are numbered. HMBC correlations are indicated. AU, absorbance units.
Large-scale Preparation of Geranylated Products
The large-scale production of geranylated products was carried out in a total volume of 10 ml. The Fur7 reactions were performed in 50 mm Hepes-NaOH (pH 7.5) containing 5 mm MgCl2, 2 mm of the aromatic substrate of interest, 5 mm GPP, and 1 mg ml−1 Fur7. The reaction mixture was incubated at 30 °C for 16 h and then extracted with 10 ml ethyl acetate three times. The combined extracts were evaporated, and the residue was redissolved in 1 ml of methanol. The prenylated products were purified by HPLC with a PEGASIL ODS column (20 × 250 mm, Senshu Scientific) and an isocratic elution of 90% acetonitrile + 0.1% acetate at a flow rate of 8 ml min−1. For 6-(3,7-dimethylocta-1,6-dien-3-yl)-5,7-dihydroxy-2-methoxy-3-methylnaphthalene-1,4-dione (6-prenyl-2-methoxy-3-methylflaviolin; Fur-P1), high resolution MS (ESI) identified a species with an m/z of 369.1733 (369.1702 calculated for C22H25O5 (M-H)).
NMR spectroscopy of Fur-P1 showed the following profile: 1H NMR (600 MHz, DMSO-d6) δ: 1.42 (s, 3H, Me-10′), 1.52 (s, 3H, Me-9′), 1.54 (s, 3H, Me-8′), 1.79 (m, 2H, H-4′), 1.86 (s, 3H, 3-Me), 1.94 (m, 2H, H-5′), 3.93 (s, 3H, 2-O-Me), 4.76 (d, JH = 10.3 Hz, 1H, H-1′), 4.82 (d, JH = 18.0 Hz, 1H, H-1′), 5.00 (t, JH = 6.8 Hz, 1H, H-6′), 6.26 (dd, JH = 10.3, 18.0 Hz, 1H, H-2′), and 6.91 (s, 1H, H-8); and 13C NMR (150 MHz, DMSO-d6) δ: 7.3 (3-Me), 17.9 (C-10′), 24.0 (C-9′), 25.9 (C-8′), 26.0 (C-5′), 35.1 (C-4′), 44.5 (C-3′), 60.0 (2-O-Me), 107.0 (C-8), 107.9 (C-4a, C-1′), 124.0 (C-6), 131.1 (C-6′), 133.1 (C-7′), 133.3 (C-8a), 133.5 (C-3), 149.2 (C-2′), 157.9 (C-2), 163.6 (C-7), 164.5 (C-5), 180.4 (C-1), and 189.5 (C-4).
For (E)-7-((3,7-dimethylocta-2,6-dien-1-yl)oxy)-5-hydroxy-2-methoxy-3-methylnaphthalene-1,4-dione (7-O-geranyl-2-methoxy-3-methylflaviolin; Fur-P2), NMR spectroscopy showed the following profile: 1H NMR (600 MHz, DMSO-d6) δ: 1.57 (s, 3H, Me-10′), 1.60 (s, 3H, Me-8′), 1.77 (s, 3H, Me-9′), 2.01 (s, 3H, 3-Me), 2.08 (m, 2H, H-4′), 2.11 (m, 2H, H-5′), 4.05 (s, 3H, 2-O-Me), 4.66 (d, JH = 6.8 Hz, 2H, H-1′), 5.05 (t, JH = 7.6 Hz, 1H, H-6′), 5.41 (t, JH = 6.2, 1H, H-2′), 6.64 (s, 1H, H-6), and 7.07 (s, 1H, H-8).
Conversion of Fur-P1 to Furaquinocin
The S. albus/pWHM-Fur2_del7 transformant was grown in 500-ml Erlenmeyer flasks containing 100 ml of NMMP medium and 30 μg ml−1 thiostrepton. Fermentation was conducted for 2 days at 27 °C after 2 days of preculture at 30 °C in tryptone soya broth medium containing 30 μg ml−1 thiostrepton, and 0.1 mg of Fur-P1 was then added to the 100 ml culture. After the addition of Fur-P1, the S. albus/pWHM-Fur2_del7 culture was further incubated for 15 h. The broth (25 ml) was extracted twice with 25 ml of ethyl acetate. The organic layer was collected and evaporated. The residue was dissolved in 500 μl methanol, and 40 μl of the resulting methanol solution was analyzed on the HPLC system under the conditions described above for furaquinocins (Fig. 5A). The product with the identical retention time to that of 3 was collected and analyzed by LC/MS/MS spectrometry under the conditions described above for furaquinocins (Fig. 5B).
FIGURE 5.
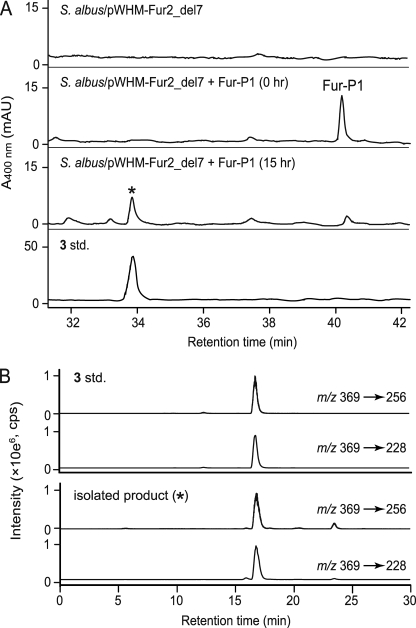
Conversion of Fur-P1 into furaquinocin C. A, HPLC analysis of the ethyl acetate extract from the S. albus/pWHM-Fur2_del7 culture supplemented with Fur-P1. Extracts were prepared at 0 and 15 h after the addition of Fur-P1. B, Identification of furaquinocin C (3) by HPLC-MS/MS. The standard (std.) 3 and the isolated product (*) were separated according to the method as described under “Experimental Procedures.” Compound 3 was identified using the combinations 369/259 and 369/228 m/z in the multiple reaction monitoring mode. mAU, milli-AU; cps, count per second.
Steady-state Kinetic Parameters
The steady-state kinetic parameters were determined with the Michaelis-Menten equation using SigmaPlot (version 10.0) and Enzyme Kinetic module 1.3 software packages (Systat Software, Point Richmond, CA). The concentration of GPP was 0.05–0.5 mm or 0.05–1.0 mm when the concentration was fixed at 0.5 mm Fur-P1, 5.0 mm 1,3-dihydroxy naphthalene (DHN), and 0.5 mm flaviolin (1). However, when the concentration of GPP was fixed at 2 mm, the concentration of Fur-P1 was 0.03–0.7 mm, the concentration of 1,3-DHN was 0.1–5 mm, and the concentration of 1 was 0.02–1 mm. In contrast, the concentration of 1 was 0.1–5 mm when the concentration of dimethylallyl diphosphate (DMAPP) was fixed at 4 mm; however, the concentration of DMAPP was 0.1–5 mm when the concentration of 1 was fixed at 2 mm. The incubation times were 10, 20, 30, and 40 min. All of the reactions were performed in 0.5 ml of 50 mm Hepes-NaOH (pH 7.5) containing 5 mm MgCl2, the respective aromatic substrates, GPP and 1 mg ml−1 Fur7. When 1 and GPP were used as the substrates, the concentration of Fur7 was 0.01 mg ml−1. The reaction was initiated by addition of Fur7 and then incubated at 30 °C. Aliquots (100 μl) were removed from the reaction mixture at the incubation times mentioned above. Each aliquot was extracted with two equal volumes of ethyl acetate. The organic layer was collected and evaporated. The residues were dissolved in 60 μl of methanol, and the formation of each product was quantified by HPLC using a standard curve. A plot of product formation versus time was used to determine the initial velocity. Because Fur7 was determined to be a monomer using a gel filtration experiment, kcat was calculated assuming one active site per Fur7 molecule.
Substrate Specificity
We tested 1,3-DHN, 1,6-DHN, 2,7-DHN, naringenin, apigenin, genistein, daidzein, olivetol, and resveratrol as aromatic substrates of Fur7. The Fur7 reactions and large-scale batch production of prenyl products were carried out as described above. The structures of the reaction products were elucidated from their 1H NMR, 13C NMR and HMBC spectroscopic data and high resolution MS data. MS analysis was performed using ESI in negative ion mode. The NMR data and high resolution MS data of the geranylated products are listed in the supplemental data.
RESULTS AND DISCUSSION
Heterologous Expression of Furaquinocin Gene Cluster in S. albus
The heterologous production of 2 has been demonstrated in S. lividans TK23 harboring pWHM-Fur2, which contains all of the genes required for the production of 2. The amount of 2 in the heterologous production experiment was sufficient for detection by HPLC. Therefore, we expected that S. lividans TK23 transformed with pWHM-Fur2_del7, which was made by deleting only the fur7 gene from pWHM-Fur2, should accumulate enough of the physiological polyketide substrate of the Fur7 prenyltransferase to be detected by HPLC. However, the transformant produced an unexpected red pigment, which impeded the purification of the possible substrate that accumulated in the transformant. Instead of S. lividans TK23, we decided to use S. albus as the host strain for heterologous production of the expected intermediate because S. albus produced less red pigment.
The pWHM-Fur2 and pWHM-Fur2_del7 constructs were introduced into S. albus to compare the products that accumulated in each transformant. These transformants were cultured, and the supernatants were extracted with ethyl acetate. HPLC analysis of the extracts identified 2 (retention time, 31 min) as the major product in the broth of S. albus/pWHM-Fur2 (Fig. 2B). The retention time, mass, and UV-visible spectrum were identical to those of an authentic sample of 2. As expected, S. albus/pWHM-Fur2_del7 did not produce 2 but did produce the unidentified product 4 (retention time, 27 min) (Fig. 2C). A characteristic UV-visible spectrum of the unidentified product closely resembled that of 1, suggesting that 4 is likely a flaviolin derivative (Fig. 2E). In contrast, small peaks appearing around 10–25 min displayed distinctly different UV-visible spectra from that of 1, indicating that the compounds resulting from the small peaks are unlikely to be involved in furaquinocin biosynthesis.
Next, to verify that the flaviolin derivative 4 is a possible physiological substrate of the Fur7 prenyltransferase, we added the recombinant Fur7 enzyme and GPP directly into the culture of S. albus/pWHM-Fur2_del7, with the expectation that furaquinocin production would be recapitulated. We assumed that the added Fur7 prenyltransferase would append a geranyl group to the possible substrate 4 to yield a geranylated product and that this product would be incorporated into the S. albus/pWHM-Fur2_del7 cells, which would finally convert it to 2. After adding the Fur7 enzyme and GPP, S. albus/pWHM-Fur2_del7 was cultured for 2 days, and the broth was extracted with ethyl acetate. As expected, HPLC analysis of the extract revealed that the flaviolin derivative 4 was completely consumed (Fig. 2D). Surprisingly, 2 was not detected in the HPLC analysis. Instead, an unanticipated compound 3 was detected at a retention time of 33 min (Fig. 2D). The UV-visible spectrum of 3 was identical to that of 2 (Fig. 2E), suggesting that compound 3 is likely a furaquinocin derivative. Thus, we purified compound 3 from the broth of S. albus/pWHM-Fur2_del7 supplemented with Fur7 and GPP. The NMR spectra of the purified compound were identical to those of the previously identified furaquinocin C, thereby allowing us to identify compound 3 as furaquinocin C (Fig. 1). A small amount of 3 was also detected in the culture broth of S. albus/pWHM-Fur2 (Fig. 2B).
Unexpectedly, the S. albus/pWHM-Fur2_del7 transformant produced 3 but not 2 under these culture conditions with Fur7 and GPP supplementation. Because 3 lacks a hydroxyl group at C-4′, the fur8 gene, which encodes the cytochrome P450 that is presumably involved in the hydroxylation of 3, may not have been translated in S. albus/pWHM-Fur2_del7. This loss of function in the fur8 gene product is most likely due to the disruption of the putative ribosome binding sequence (Shine-Dalgarno sequence) of the fur8 gene. In fact, the important sequence required for translation is located immediately after the NruI site that was used to disrupt the fur7 gene. Therefore, we confirmed that the flaviolin derivative 4 was a possible substrate of the Fur7 prenyltransferase.
Purification and Identification of Flaviolin Derivative 4
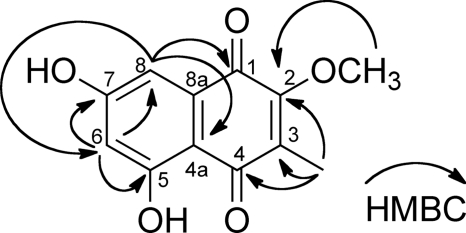
Because the flaviolin derivative 4 is a possible physiological substrate of the Fur7 prenyltransferase, we purified 4 from the broth of S. albus/pWHM-Fur2_del7. One milligram of 4 was purified from 4 liters of the S. albus/pWHM-Fur2_del7 culture using preparative HPLC. The 1H and 13C NMR spectra closely resembled those of 1, except for two signals (δH 1.99 (s), δC 7.3, and δH 4.03 (s), δC 60.9), which correspond to C-methyl and O-methyl groups, respectively. An extensive analysis of the HMBC NMR spectrum unequivocally determined that the structure of 4 was 5,7-dihydroxy-2-methoxy-3-methylnaphthalene-1,4-dione (2-methoxy-3-methylflaviolin; 4) (Fig. 3). Because the two methyl groups at O-2 and C-3 are already present in 4, as in 2 and because no flaviolin derivative other than 4 was detected in the S. albus/pWHM-Fur2_del7 culture, we presume that two methyltransferase reactions occur immediately before the Fur7 prenyltransferase reaction. The fur4 and fur6 gene products in the furaquinocin gene cluster likely encode methyltransferases because these genes show significant sequence similarity to other methyltransferases (9, 12).
FIGURE 3.
Structure of 2-methoxy-3-methylflaviolin. Carbon atoms are numbered. HMBC correlations are indicated.
Expression and Purification of Recombinant Fur7 Protein
To assess the enzymatic activity of Fur7, we overexpressed the fur7 gene in E. coli as an N-terminal His8-tagged protein and purified the recombinant protein to apparent homogeneity. The molecular mass of Fur7 was estimated to be 34 kDa by SDS-polyacrylamide gel electrophoresis and 34 kDa by gel filtration chromatography, suggesting that Fur7, similar to NphB, is likely a monomer.
Assay of Fur7 Prenyltransferase with 2-Methoxy-3-methylflaviolin 4
To assess the prenyltransferase activity of Fur7 using 4 as a substrate, we incubated the purified Fur7 protein with the substrate in the presence of GPP. HPLC analysis of the reaction mixture revealed the formation of two products, Fur-P1 and Fur-P2 (Fig. 4A). When 1 mm 4 was incubated with Fur7 prenyltransferase for 2 h, the ratio was 10:1 (Fig. 4A). Prolonged incubation (16 h) did not significantly alter this ratio.
These Fur7 reaction products were purified by preparative HPLC, and their structures were primarily elucidated by comparison of the 1H and 13C NMR spectra of Fur-P1 and Fur-P2 to that of substrate 4. The 1H NMR spectrum of Fur-P1 showed that the signals corresponded to a 3,7-dimethylocta-1,6-dien-3-yl moiety. The chemical shifts of H-1′ (4.76 ppm doublet and 4.82 ppm doublet) and H-2′ (6.26 ppm double doublet) unequivocally identified the structure. Notably, the signal of H-6 was absent from the 1H NMR spectrum of Fur-P1. Thus, we concluded that the structure of Fur-P1 was 6-(3,7-dimethylocta-1,6-dien-3-yl)-5,7-dihydroxy-2-methoxy-3-methylnaphthalene-1,4-dione (6-prenyl-2-methoxy-3-methylflaviolin) (Fig. 4B). We determined that Fur-P1 is most likely an intermediate in the last step of furaquinocin biosynthesis.
The 1H NMR spectrum of Fur-P2 showed signals corresponding to a 3,7-dimethylocta-2,6-dienyl (geranyl) moiety. The chemical shift of H-1′ of the geranyl moiety (4.66 ppm) indicated that the moiety was not attached to an aromatic carbon but to an oxygen atom. The HMBC spectrum showed a correlation between H-1′ and C-7. Thus, we concluded that the structure of Fur-P2 was (E)-7-((3,7-dimethylocta-2,6-dien-1-yl)oxy)-5-hydroxy-2-methoxy-3-methylnaphthalene-1,4-dione (7-O-geranyl-2-methoxy-3-methylflaviolin) (Fig. 4B).
Because it is conceivable that Fur-P2 is the primary enzymatic product, which then rearranges to Fur-P1 via a Claisen rearrangement, we incubated purified Fur-P2 with Fur7 prenyltransferase. However, no formation of Fur-P1 was detected by HPLC analysis even with overnight incubation, excluding the possibility that Fur-P1 was formed via the rearrangement of Fur-P2. Thus, we conclude that Fur7 catalyzes the prenyltransferase reaction of 4 to yield Fur-P1 and Fur-P2 simultaneously.
The addition of EDTA (1 mm) to deplete the assay mixture of divalent cations did not affect the enzyme activity, and the addition of 5 mm MgCl2·6H2O resulted in only a 30% increase in activity. The addition of 5 mm MnCl2·4H2O, CaCl2, or FeSO4·7H2O resulted in almost no change in enzyme activity. These results indicate that Fur7 is a divalent, cation-independent prenyltransferase. However, the addition of 5 mm CuCl2·2H2O resulted in the formation of a precipitate, and the enzyme lost nearly all activity.
Conversion of Fur-P1 to Furaquinocin
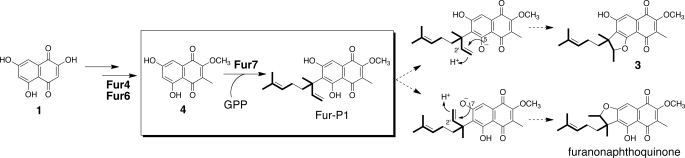
Next, to investigate whether Fur-P1 was an intermediate of furaquinocin, we evaluated whether Fur-P1 could be converted into furaquinocin. Fur-P1 was added into the S. albus/pWHM-Fur2_del7 culture, and the broth was extracted with ethyl acetate. After the addition of Fur-P1, the S. albus/pWHM-Fur2_del7 culture was incubated for 15 h before the extract was subjected to HPLC analysis. The results revealed that 3 was produced concomitantly with the nearly complete consumption of Fur-P1 (Fig. 5), which unambiguously demonstrates that Fur-P1, the Fur7 reaction product, was the intermediate of the late stage of furaquinocin biosynthesis. Presumably, Fur-P1 is subsequently converted into 3 through an acid/base-catalyzed cyclization catalyzed by an unidentified cyclase encoded in the furaquinocin biosynthetic gene cluster (Fig. 6). With the isolation of Fur-P1 as the possible substrate of the cyclase, it will be possible to identify the subsequent cyclase of furaquinocin biosynthesis in the gene cluster by biochemical analyses. Intermediate 3 is most likely finally converted to 2 by the action of cytochrome P450, which is encoded by the fur8 gene.
FIGURE 6.
Proposed biosynthetic pathway of furaquinocin. Compound 4 was identified as a genuine substrate for the Fur7 prenyltransferase in the present study. The formation of 4 appears to involve successive methylations of 1 by the Fur4 and Fur6 methyltransferases. Substrate 4 is attached to a 10-carbon geranyl group by the Fur7 prenyltransferase to yield Fur-P1. Fur-P1 is then presumably converted into 3 by an unidentified cyclase. Finally, 3 is most likely converted into 2 by the action of cytochrome P450, which is encoded by the fur8 gene. The present data were also used to construct the late stage of the furanonaphthoquine biosynthetic pathway.
Due to the structural similarity between furaquinocin and furanonaphthoquinone (12), Fur-P1 may be a common substrate for the cyclases involved in the biosynthesis of meroterpenoids. In furaquinocin biosynthesis, the cyclase presumably forms an ether bond between the 5-O and 2′-C atoms of Fur-P1 to yield 3. In furanonaphthoquine biosynthesis, however, the cyclase forms an ether bond between the 7-O and 2′-C atoms of Fur-P1 to yield furanonaphthoquinone (Fig. 6). Future work will detail the reaction mechanism and structural basis for the regiospecific cyclization of Fur-P1 by each cyclase involved in furaquinocin and furanonaphthoquine biosynthesis.
Fnq26 has been identified as a prenyltransferase for furanonaphthoquinone biosynthesis. The Fnq26 prenyltransferase shows 64% similarity to Fur7 and displays similar substrate specificity to that of Fur7 (described below). Therefore, Fnq26, like Fur7, would accept 4 as a substrate to yield Fur-P1.
Biochemical Characterization of Fur7 Prenyltransferase
We calculated the steady-state kinetic constants of Fur7 using 4 and GPP as substrates.
A typical hyperbolic curve of product formation over substrate concentration was obtained (supplemental Fig. S1) using different concentrations of 4 in the presence of 2 mm GPP, indicating that the reaction exhibits Michaelis-Menten kinetics. The apparent steady-state kinetic constants were estimated by quantitatively measuring the amount of the prenylated products using HPLC analysis (Km = 0.054 ± 0.005 mm and kcat (×103) = 0.66 ± 0.02 s−1). The calculated catalytic efficiency (kcat/Km) of Fur7 was 12 m−1 s−1 (Table 1); this value compares favorably with those of the well characterized NphB prenyltransferase (≈10 m−1 s−1) for naphterpin synthesis (14) and the Fnq26 prenyltransferase (≈10 m−1 s−1) for furanonaphthoquinone synthesis (15).
TABLE 1.
Steady-state kinetic parameters of Fur7
Values are expressed as the means ± S.D. of three independent experiments.
| Aromatic substrate | Prenyl donor |
Km |
Vmax | kcat × 103 | kcat/Km | |
|---|---|---|---|---|---|---|
| Aromatic substrate | Prenyl donor | |||||
| >mm | nmol/min/mg | s−1 | m−1s−1 | |||
| Fur-P1 | GPP | 0.054 ± 0.005 | 0.098 ± 0.024 | 1.1 ± 0.1 | 0.66 ± 0.02 | 12 |
| Flaviolin 1 | GPP | 0.048 ± 0.002 | 0.059 ± 0.010 | 47 ± 5 | 28 ± 3 | 580 |
| Flaviolin 1 | DMAPP | 0.35 ± 0.05 | 0.72 ± 0.06 | 1.4 ± 0.0 | 0.87 ± 0.03 | 2.5 |
| 1,3-DHN | GPP | 0.32 ± 0.02 | 0.063 ± 0.008 | 0.22 ± 0.01 | 0.13 ± 0.05 | 0.41 |
Using a constant concentration of 4 (0.5 mm) and varying concentrations of GPP, a typical hyperbolic curve of product formation over substrate concentration was obtained (Fig. S2), indicating that this reaction also followed Michaelis-Menten kinetics (KmGPP = 0.098 ± 0.024 mm).
Substrate Specificity of Fur7
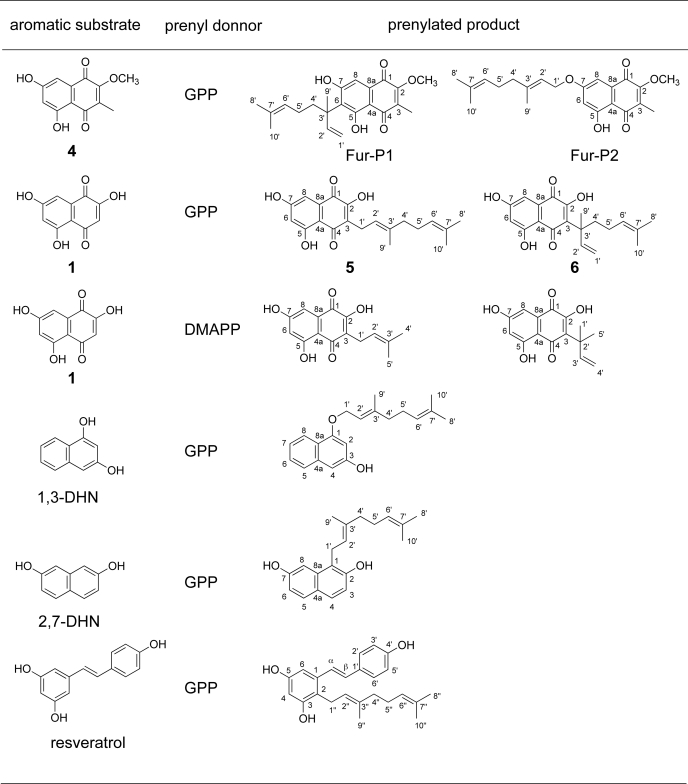
Next, we examined the substrate specificity of Fur7 because Streptomyces prenyltransferases display broad substrate specificities (14, 15, 19, 20) and are the expected tools for chemoenzymatic synthesis of prenylated compounds (21, 22). We focused on DHN, 1,6-DHN, 2,7-DHN, flaviolin 1, the two plant polyketides olivetol and resveratrol, and the (iso)flavonoid subclass, including naringenin, apigenin, genistein, and daidzein. 1,3-DHN, 2,7-DHN, 1, and resveratrol were geranylated, yielding one or two products (Fig. 7). However, none of the (iso)flavonoids tested yielded prenylated products.
FIGURE 7.
Prenylated products synthesized by Fur7. Carbon atoms are numbered.
The Fur7 reaction products contained a geranyl group in the ortho position with respect to the neighboring hydroxyl moiety. Fur7 also catalyzed the geranylation of a hydroxyl group, leading to the formation of an O-prenyl linkage in 1,3-DHN and 1. When 1 was incubated with GPP and Fur7, two products (5 and 6) were observed. The geranyl moieties of 5 and 6 were linked to C-3 of 1 via the C′-1 or C′-3 atoms, respectively. Interestingly, Fur7 attached the dimethylallyl moiety to C-3 of 1 via the C′-1 and C′-3 atoms (Fig. 7); the geranyl moiety was added to 1 in the same manner. Fur7 is the first prenyltransferase found to be capable of accepting both GPP and DMAPP as prenyl donor substrates.
The steady-state kinetic constants for Fur7 with each substrate and either GPP or DMAPP as substrates were calculated (supplemental Figs. S3–S8) and are listed in Table 1. Curiously, the kcat/Km of the Fur7-catalyzed prenylation of 1 (580 m−1 s−1) was significantly higher than that of the Fur7-catalyzed prenylation of 4 (12 m−1 s−1). However, because 1 was not detected in the S. albus/pWHM-Fur2_del7 culture (Fig. 1C), the Fur7-catalyzed prenylation of 1 should not occur during the biosynthetic elaboration of furaquinocin. Neither of the Fur7-catalyzed prenylated flaviolins 5 and 6 appears to be a biosynthetic intermediate of furaquinocin due to discrepancies in the prenylation position. In light of its relatively low Km value (0.054 mm), we conclude that 4 is a genuine physiological substrate of the Fur7 prenyltransferase that is responsible for furaquinocin biosynthesis. This conclusion is strongly supported by evidence that the Fur7 reaction product Fur-P1 was efficiently converted into furaquinocin by S. albus/pWHM-Fur2_del7 (Fig. 5A).
Conclusions
In the present study, we identified, for the first time, the physiological Fur7 prenyltransferase substrate 2-methoxy-3-methylflaviolin 4 and its reaction product Fur-P1; both compounds are novel biosynthetic intermediates of the furaquinocin biosynthetic pathway. In addition, we demonstrate that Fur7 displays promiscuous prenyltransferase activity against both prenyl donor substrates and prenyl acceptor substrates. The present data will contribute to the understanding of the biosynthesis of meroterpenoids produced by Streptomyces strains.
Supplementary Material
Acknowledgment
We are grateful to Tohru Dairi of Hokkaido University for providing pWHM-Fur2.
This work was supported in part by the “Development of Fundamental Technologies for Production of High Value Materials using Transgenic Plants” R&D project funded by the Ministry of Economy, Trade, and Industry and by a grant from the Naito Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental data and Figs. S1–S8.
- THN
- tetrahydroxynaphthalene
- GPP
- geranyl diphosphate
- HMBC
- heteronuclear multiple-bond connectivity
- ESI
- electrospray ionization
- DMAPP
- dimethylallyl diphosphate.
REFERENCES
- 1.Komiyama K., Funayama S., Anraku Y., Ishibashi M., Takahashi Y., Omura S. (1990) J. Antibiot. 43, 247–252 [DOI] [PubMed] [Google Scholar]
- 2.Ishibashi M., Funayama S., Anraku Y., Komiyama K., Omura S. (1991) J. Antibiot. 44, 390–395 [DOI] [PubMed] [Google Scholar]
- 3.Shin-ya K., Imai S., Furihata K., Hayakawa Y., Kato Y., Vanduyne G. D., Clardy J., Seto H. (1990) J. Antibiot. 43, 444–447 [DOI] [PubMed] [Google Scholar]
- 4.Shin-ya K., Furihata K., Hayakawa Y., Seto H. (1990) Tetrahedron Lett. 31, 6025–6026 [Google Scholar]
- 5.Shiomi K., Nakamura H., Iinuma H., Naganawa H., Isshiki K., Takeuchi T., Umezawa H., Iitaka Y. (1986) J. Antibiot. 39, 494–501 [DOI] [PubMed] [Google Scholar]
- 6.Sedmera P., Pospísil S., Novák J. (1991) J. Nat. Prod. 54, 870–872 [Google Scholar]
- 7.Funa N., Ohnishi Y., Fujii I., Shibuya M., Ebizuka Y., Horinouchi S. (1999) Nature 400, 897–899 [DOI] [PubMed] [Google Scholar]
- 8.Izumikawa M., Shipley P. R., Hopke J. N., O'Hare T., Xiang L., Noel J. P., Moore B. S. (2003) J. Ind. Microbiol. Biotechnol. 30, 510–515 [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki T., Hayashi Y., Kuzuyama T., Furihata K., Itoh N., Seto H., Dairi T. (2006) J. Bacteriol. 188, 1236–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuzuyama T., Noel J. P., Richard S. B. (2005) Nature 435, 983–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter J. M., Moffitt M. C., Zazopoulos E., McAlpine J. B., Dorrestein P. C., Moore B. S. (2007) J. Biol. Chem. 282, 16362–16368 [DOI] [PubMed] [Google Scholar]
- 12.Haagen Y., Glück K., Fay K., Kammerer B., Gust B., Heide L. (2006) Chembiochem 7, 2016–2027 [DOI] [PubMed] [Google Scholar]
- 13.Funa N., Funabashi M., Yoshimura E., Horinouchi S. (2005) J. Biol. Chem. 280, 14514–14523 [DOI] [PubMed] [Google Scholar]
- 14.Kumano T., Richard S. B., Noel J. P., Nishiyama M., Kuzuyama T. (2008) Bioorg. Med. Chem. 16, 8117–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haagen Y., Unsöld I., Westrich L., Gust B., Richard S. B., Noel J. P., Heide L. (2007) FEBS Lett. 581, 2889–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davisson V. J., Woodside A. B., Poulter C. D. (1985) Methods Enzymol. 110, 130–144 [DOI] [PubMed] [Google Scholar]
- 17.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (eds) (2000) Practical Streptomyces Genetics, The John Innes Foundation, Norwich, UK [Google Scholar]
- 18.Jez J. M., Ferrer J. L., Bowman M. E., Dixon R. A., Noel J. P. (2000) Biochemistry 39, 890–902 [DOI] [PubMed] [Google Scholar]
- 19.Ozaki T., Mishima S., Nishiyama M., Kuzuyama T. (2009) J. Antibiot. 62, 385–392 [DOI] [PubMed] [Google Scholar]
- 20.Takahashi S., Takagi H., Toyoda A., Uramoto M., Nogawa T., Ueki M., Sakaki Y., Osada H. (2010) J. Bacteriol. 192, 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Botta B., Delle Monache G., Menendez P., Boffi A. (2005) Trends Pharmacol. Sci. 26, 606–608 [DOI] [PubMed] [Google Scholar]
- 22.Tello M., Kuzuyama T., Heide L., Noel J. P., Richard S. B. (2008) Cell. Mol. Life Sci. 65, 1459–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.