Abstract
Autotransporters (ATs) constitute an important family of virulence factors secreted by Gram-negative bacteria. Following their translocation across the inner membrane (IM), ATs temporarily reside in the periplasmic space after which they are secreted into the extracellular environment. Previous studies have shown that the AT hemoglobin protease (Hbp) of Escherichia coli requires a functional signal recognition particle pathway and Sec translocon for optimal targeting to and translocation across the IM. Here, we analyzed the mode of IM translocation of Hbp in more detail. Using site-directed photocross-linking, we found that the Hbp signal peptide is adjacent to YidC early during biogenesis. Notably, YidC is in part associated with the Sec translocon but has until now primarily been implicated in the biogenesis of IM proteins. In vivo, YidC appeared critical for the biogenesis of the ATs Hbp and EspC. For Hbp, depletion of YidC resulted in the formation of secretion-incompetent intermediates that were sensitive to degradation by the periplasmic protease DegP, indicating that YidC activity affects Hbp biogenesis at a late stage, after translocation across the IM. This is the first demonstration of a role for YidC in the biogenesis of an extracellular protein. We propose that YidC is required for maintenance of the translocation-competent state of certain ATs in the periplasm. The large periplasmic domain of YidC is not critical for this novel functionality as it can be deleted without affecting Hbp biogenesis.
Keywords: Bacteria, Membrane, Protein Cross-linking, Protein Export, Protein Translocation, Autotransporter, Escherichia coli, Signal Peptide, Type V Secretion, YidC
Introduction
In Gram-negative bacteria, proteins destined for secretion into the extracellular environment must traverse a complex cell envelope comprising the inner membrane (IM)2 and outer membrane (OM) separated by the periplasmic space. Several pathways have evolved that enable protein transfer across the cell envelope. The simplest and most widely distributed of these is the autotransporter (AT) or type Va secretion pathway that is dedicated to the secretion of large virulence factors (1). ATs are organized in three domains: (i) the signal peptide at the N terminus to allow targeting to and translocation across the IM via the protein-conducting channel SecYEG (Sec translocon), (ii) the secreted passenger domain that carries the actual effector function of AT, and (iii) the β-domain at the C terminus that adopts a β-barrel conformation in the OM and plays a crucial role in transfer of the passenger domain across the OM (2).
Most ATs are produced with a classical signal peptide that mediates targeting to the Sec translocon via the post-translational SecB pathway and is cleaved by the major signal peptidase upon translocation of the AT across the IM (1). A subset of ATs, including all members of the subfamily of serine protease autotransporters of Enterobacteriaceae (SPATEs), are synthesized with remarkably long signal peptides that contain a conserved N-terminal extension (1, 3). Although some studies suggested that the extended signal peptides promote targeting of ATs via a strictly post-translational SecB-mediated mechanism (4–6), the signal peptide of the SPATE hemoglobin protease (Hbp) was shown to engage the signal recognition particle (SRP) that functions in a co-translational targeting pathway (7, 8). A number of studies suggest that the extended AT signal peptides are important at stages beyond the initial targeting step, slowing down either the progression of the ATs through the Sec translocon or the release from the IM after translocation (4, 5, 9). Interestingly, the signal peptide of the SPATE EspP appeared to prevent misfolding of the AT in the periplasmic space into a conformation that is incompatible with OM translocation, possibly by transiently anchoring the AT passenger to the IM (9).
The observation that Hbp can make use of the SRP targeting pathway was remarkable considering that SRP directs targeting of primarily integral inner membrane proteins (IMPs) with rather hydrophobic signal sequences (10). YidC has been identified as another component that plays a role in the biogenesis of IMPs. Available evidence suggests that it is in part associated with the SecYEG machinery (11–14) and acts downstream of it to facilitate lateral transfer (15, 16) and assembly of transmembrane segments (TMs) (17). Furthermore, YidC was shown to function in the assembly of specific protein complexes in the IM (18) and is believed to be involved in the folding of polytopic IMPs into their native structure (19, 20). A recent study suggested a linked role for YidC and the IM protease FtsH in the quality control of IMPs (21). YidC also functions independently from the translocon, catalyzing the insertion of relatively small/simple IMPs including subunits of the cytochrome o oxidase and F1F0-ATPase (22). As such, the Sec-independent function of YidC is essential for cell viability (23). YidC is an IMP with six TMs and a large periplasmic domain P1 (24). Recent crystal structures of the P1 domain (25, 26) revealed a conserved elongated cleft that was suggested to accommodate an unfolded polypeptide chain as its natural ligand (26). However, structure-function studies showed that the N-terminal 90% of this domain can be deleted without affecting cell viability and insertion of tested Sec-dependent and -independent substrates (14, 27), leaving the role of domain P1 unclear.
To gain more insight into the mode of AT translocation across the IM, we studied the molecular interactions of Hbp during the early stages of membrane insertion using an in vitro translation and photocross-linking approach. Remarkably, we found that the Hbp signal peptide is in contact with YidC. When YidC was depleted in vivo, Hbp secretion was impaired as a result of the formation of secretion-incompetent Hbp intermediates in the periplasmic space. This is the first demonstration of a role for YidC in the biogenesis of an extracellular protein.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
The YidC conditional strain JS7131 has been described previously (28). To construct the strain JS7131ΔdegP, the degP41(ΔPstI)::kanR allele was moved from strain KS476 (29) into JS7131 by P1 transduction. Transductants were selected at 37 °C on LB agar supplemented with kanamycin (50 μg/ml), 0.2% l-arabinose, and 10 mm sodium citrate. The absence of DegP was further confirmed by immunoblotting using DegP antiserum.
Strains were routinely grown in LB medium at 30 °C. For YidC depletion experiments, cells were grown overnight in the presence of 0.2% glucose and 0.2% l-arabinose, washed in the same medium lacking l-arabinose, and subcultured to an OD660 of 0.05 in medium containing 0.2% glucose. l-arabinose (0.2%), or l-rhamnose (0.2%) was added, where appropriate, to induce the expression of YidC and YidC derivatives. If required, the medium was supplemented with streptomycin (50 μg/ml), chloramphinicol (20 μg/ml), kanamycin (30 μg/ml), spectinomycin (25 μg/ml), or ampicilin (100 μg/ml).
Reagents, Enzymes, and Sera
Restriction enzymes, T4-DNA ligase, and Lumi-Light Western blotting substrate were purchased from Roche Applied Science. PfuUltra DNA Polymerase was obtained from Stratagene. l-Arabinose and l-rhamnose were supplied by Merck. Coomassie Brilliant Blue G was from Janssen Chimica. All other chemicals were supplied by Sigma-Aldrich. The antisera against YidC and trigger factor (TF) were from our own collection. The antisera against PspA, DegP, and OmpA were kind gifts from J. Tommassen (Utrecht University), J. Beckwith (Harvard Medical School), and J. W. de Gier (Stockholm University), respectively.
Plasmid Construction
The plasmid pEH3-Hbp (30) was used for the expression of Hbp. To allow expression of EspC in an identical genetic context, the gene encoding EspC was amplified by PCR using plasmid pJLM174 (31) as a template and the primers pEH-XbaI-EspC-fw and EspC-BamHI-rv. The resulting fragment was cloned into XbaI/BamHI digested pEH3-Hbp, yielding pEH3-EspC. The plasmid pEH3-PhoE was created in a similar way. The gene encoding phoE was amplified using Escherichia coli MC4100 genomic DNA as template and the primers pEH-XbaI-PhoE-fw and PhoE-BamHI-rv. The PCR fragment was cloned into XbaI/BamHI digested pEH3-Hbp, resulting in pEH3-PhoE.
Plasmids pWSK-empty and pWSK-DegP were created as follows. First, pWSK29 (32) was digested with restriction enzymes BsaAI and PvuII, and the resulting 750-bp fragment, including lacZα, multiple cloning site, and T7 and T3 RNA polymerase promoters, was removed. The remaining 4684-bp vector fragment was blunt-end self-ligated, yielding pWSK-empty. To obtain pWSK-DegP, the degP gene and its regulatory elements were amplified from E. coli MC4100 genomic DNA by PCR using primers that annealed ∼480 bp upstream and ∼135 bp downstream of the degP gene, respectively. The primers used were Genom/degP[-480bp]-fw and Genom/degP[TAA+135bp]-rv. The resulting fragment carried two flanking SmaI sites that were used to clone the fragment into BsaAI/PvuII digested pWSK29 (see above) through blunt-end ligation.
To allow l-rhamnose induced expression of YidC and YidCΔP1, a pWSK29 derivative was created that carried the multiple cloning site, rhaT promoter and rhaR/rhaS regulatory genes of pRHA-113 (33). To this end, a PCR fragment was generated using pRHA-113 as a template and the primers DrdI-Rha-fw and SapI-Rha-rv. The resulting fragment was cloned into the DrdI/SapI sites of pWSK29, yielding pWSK/Rha. To generate pWSK/Rha-YidC, the XbaI/SmaI fragment of pCL-ecOxa1, carrying the YidC coding sequence (23), was subcloned into pWSK/Rha using the XbaI/EcoRV restriction sites. To construct pWSK/Rha-YidCΔP1, the nucleotide sequence encoding residues 27–320 of YidC was deleted using the Phusion site-directed mutagenesis kit (Finnzymes) with the primers YidCΔ27–320-fw and YidCΔ27–320-rv.
To construct the plasmid pCMM-116HbpTAG41, a PCR fragment was generated using pC4Meth-ssHbp (8) as a template and the primers Hbp-EcoRI-fw and pCMM116Hbp-BamHI-rv. The resulting fragment was cloned into pCMM (15) using the EcoRI/BamHI restriction sites, yielding pCMM-116Hbp. To allow in vitro photocross-linking, a single amber codon (TAG) was introduced at position 41 by nested PCR as described previously (16), using the mutagenesis primers ssHbp-TAG41-fw and ssHbp-TAG41-rv. The primers used in this study are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer | Primer sequence |
|---|---|
| pEH-XbaI-EspC-fw | 5′-taactttctagattacaaaacttaggagggtttttaccatgaataaaatatacgcattaaaata-3′ |
| EspC-BamHI-rv | 5′-gactggatcctcagaaagaataacggaagttag-3′ |
| pEH-XbaI-PhoE-fw | 5′-taactttctagattacaaaacttaggagggtttttaccatgaaaaagagcactctggc-3′ |
| PhoE-BamHI-rv | 5′-gatcggatccttaaaactgatacgtcatgcc-3′ |
| Genom/degP[-480bp]-fw | 5′-gtacgtacccggggtgtttagccatccagatgtc-3′ |
| Genom/degP[TAA+135bp]-rv | 5′-gtacgtacccggggcttagcataaggaagtacg-3′ |
| DrdI-Rha-fw | 5′-tgcatgcagactccaacgtcagtactcgataagcttaattaatctttctgcg-3′ |
| SapI-Rha-rv | 5′-tgcatgcaaagcggaagagcagtactgtgatggatatctggagaattcg-3′ |
| YidCΔ27–320-fw | 5′-gctccgcacctggatctgacc-3′ |
| YidCΔ27–320-rv | 5′-atcctgctcccaggcttgccag-3′ |
| Hbp-EcoRI-fw | 5′-gccggaattctaatatgaacagaatttattctcttc-3′ |
| pCMM116Hbp-BamHI-rv | 5′-gtcaggatccagctgccttatccagcgta-3′ |
| ssHbp-TAG41-fw | 5′-ccggttttattgtagatcccggta-3′ |
| ssHbp-TAG41-rv | 5′-taccgggatctacaataaaaccgg-3′ |
In Vitro Translation and Photocross-linking
Truncated mRNA was prepared from HindIII linearized pCMM-116HbpTAG41 as described previously (12). In vitro translation, photocross-linking, and sodium carbonate extraction were carried out as described (16, 34). Samples were analyzed directly by SDS-PAGE or immunoprecipitated first using 3-fold the amount used for direct analysis. Radiolabeled proteins were visualized using a Molecular Dynamics PhosphorImager 473.
RESULTS
Hbp Signal Peptide Interacts with YidC during Inner Membrane Translocation
The unusual signal peptides that are found in a large subset of ATs are thought to modulate the rate at which ATs are transferred into the periplasm. Recent work suggested that these peptides transit through the Sec translocon relatively slowly and/or transiently tether the AT to the IM (4, 9). Conceivably, these signal peptides direct atypical routing through the Sec translocon. In a previous study, we investigated the molecular environment of the signal peptide of Hbp during membrane insertion using an in vitro translation and cross-linking approach. Molecular interactions of nascent Hbp were fixed using the homo-bifunctional lysine-specific cross-linker disuccinimidyl suberate, demonstrating contacts with SecA and the translocon component SecY (8). Of note, previous work from our group on the biogenesis of the IMP FtsQ suggested that, although disuccinimidyl suberate can be used effectively to reveal contacts between substrate proteins and hydrophilic exposed regions of IMPs, it seems less suited to probe contacts with IMPs that take place strictly within the lipid bilayer (16, 34). This is probably due to relative paucity of lysine residues in the TMs of IMPs. Therefore, we considered it opportune to reanalyze the molecular contacts of the Hbp signal peptide during membrane insertion using an unbiased site-specific in vitro translation and photocross-linking approach (16). In this approach, radiolabeled translation intermediates of specific length (nascent chains) are produced from truncated mRNA in a cell- and membrane-free E. coli lysate. During translation, a carbene generating photoreactive cross-linking probe is introduced site-specifically using previously described (Tmd)Phe-tRNAsup technology (35). The highly reactive carbene is known to attack neighboring molecules irrespective of their chemical nature (35, 36).
A 116-amino acid-long Hbp nascent chain (116Hbp) was synthesized that is identical to the construct used in a previous study (8). At this length, the signal peptide is exposed outside the ribosome to provide optimal targeting information (Fig. 1A). Translation was carried out in the presence of purified inverted inner membrane vesicles derived from wild-type E. coli cells to allow membrane insertion of the Hbp 116-mer and generation of an Hbp translocation intermediate. To probe the molecular contacts of Hbp during membrane translocation, a photoreactive probe was introduced at position 41 (TAG41) of the signal peptide, in the middle of the hydrophobic core (Fig. 1A). After translation, one-half of the sample was irradiated with UV light to induce cross-linking, whereas the other half was kept in the dark to serve as a negative control. Irradiation of 116HbpTAG41 resulted in two prominent cross-linking adducts of ∼60–70 kDa (Fig. 1B).
FIGURE 1.
In vitro photocross-linking of YidC to the signal peptide of nascent Hbp. A, schematic representation of nascent 116HbpTAG41. The classical region of the Hbp signal peptide is indicated as a thick solid line with a white central area representing the hydrophobic core. A signal peptide extension that is conserved among a subset of ATs is indicated by a hatched box. The position of the amber codon that is suppressed with (Tmd)Phe-tRNAsup to allow incorporation of a photoreactive probe is indicated (TAG41). The nascent chain is depicted with 35 amino acids in the ribosome. B, in vitro translation of 116HbpTAG41 in the presence of inverted inner membrane vesicles. After translation, samples were irradiated with UV light to induce cross-linking or left in the dark as indicated. C, UV-irradiated samples described under B were subjected to extraction with sodium carbonate (Na2CO3). The resulting carbonate pellet and supernatant (sup.) fractions were analyzed by SDS-PAGE directly, or first immunoprecipitated (IP) using antiserum against YidC, SecY, TF, or fifty-four homologue as indicated. Adducts representing cross-linking to either SecY or fifty-four homologue are indicated with an caret. Molecular mass (kDa) markers are indicated at the left side of the panels.
To gain insight into the nature of the cross-linking adducts, the UV-irradiated sample was extracted with sodium carbonate to separate the membrane integrated from the peripheral membrane and soluble proteins (Fig. 1C). Approximately 50% of nascent Hbp was detected in the carbonate pellet (Fig. 1C, cf. lanes 1 and 4), indicating efficient targeting to the inverted inner membrane vesicles as described previously (8). Interestingly, the slower migrating adduct that appeared upon UV irradiation (Fig. 1B, lane 2) was detected in the carbonate pellet fraction (Fig. 1C, lane 1). Immunoprecipitation using an antiserum against the accessory translocon component YidC identified this protein as the cross-linked partner (Fig. 1C, lane 2). Consistently, the molecular mass of the adduct corresponded to the added molecular mass of the Hbp nascent chain (∼13 kDa) and YidC (∼62 kDa). As a control, immunoprecipitation using antiserum against SecY did not precipitate the prominent ∼70-kDa adduct, but a faint ∼40-kDa band representing minor cross-linking to SecY (Fig. 1C, lane 3). Similar to other cross-linking studies, a ∼25-kDa adduct was also precipitated using SecY antiserum, which has been suggested to represent a degradation product (34, 37).
In the carbonate supernatant fraction containing nontargeted material (Fig. 1C, lane 4), a cross-linking adduct similar in size to the YidC adduct was detected. Immunoprecipitation showed that this adduct represented cross-linking to TF (Fig. 1C, lane 5), a cytosolic chaperone with a general affinity for nascent polypeptide chains (38). The most prominent adduct, however, was precipitated using an antiserum against fifty-four homologue (Fig. 1C, lane 6), the protein component of the SRP, reflecting incomplete release of SRP from nascent Hbp under the conditions tested. This confirms earlier data in which TF and SRP were specifically cross-linked to 116Hbp using the lysine-specific bifunctional cross-linker disuccinimidyl suberate (7, 8). Furthermore, the present, site-specific data demonstrate that both TF and SRP are close to the hydrophobic core of the signal peptide at this stage in biogenesis.
Importantly, the cross-linking data now show that upon membrane insertion of 116Hbp, the Hbp signal peptide is in close vicinity of YidC. Taken together with data that show cross-linking of 116Hbp to SecA/SecY in the membrane and fifty-four homologue in the cytosol (Fig. 1C) (7, 8), the molecular contacts of Hbp during targeting and insertion are remarkably similar to those of the nascent IMPs FtsQ and Lep (16, 39, 40). Interestingly, cross-linking of nascent Hbp to YidC in the membrane appeared rather prominent and almost exclusive at endogenous YidC expression levels, suggesting that it represents a functional interaction.
YidC Is Involved in AT Biogenesis in Vivo
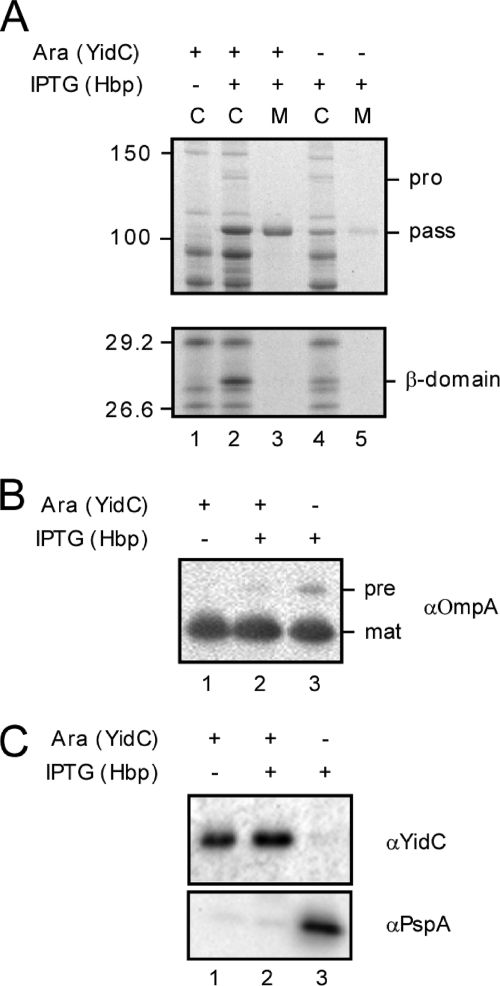
The in vitro cross-linking data suggest that YidC interacts with the Hbp signal peptide during passage of the IM. Next, we investigated whether YidC is involved in the biogenesis of Hbp in vivo. To this end, plasmid pEH3-Hbp, which carries the Hbp gene under the control of an IPTG-inducible lac promoter (30), was transformed into the YidC depletion strain JS7131. In this strain, the chromosomal yidC gene is disrupted and functionally substituted by an intact yidC gene that is under arabinose promoter control (28). Cells were grown either in the presence (control cells) or absence (YidC-depleted cells) of arabinose and supplemented with IPTG to induce expression of Hbp for 2 h. Subsequently, samples were withdrawn from the cultures and centrifuged to separate cells and medium. Both fractions were analyzed by SDS-PAGE and Coomassie staining to monitor expression and secretion of Hbp (Fig. 2).
FIGURE 2.
Depletion of YidC affects Hbp biogenesis in vivo. A, analysis of expression and secretion of Hbp in YidC depletion strain JS7131. Cells harboring pEH3-Hbp were grown in the presence or absence of l-arabinose to allow expression or depletion of YidC, respectively. After 5.5 h of growth, expression of Hbp was induced by addition of IPTG (1 mm), and growth was continued. Samples were collected 2 h after induction, separated in cells (C) and spent medium (M) and analyzed by SDS-PAGE and Coomassie staining. Cells were resuspended in SDS sample buffer directly, whereas medium samples were first TCA-precipitated. Cells loaded in lane 1 were collected after 5.5 h of growth, prior to addition of IPTG. The relevant areas of the same gel are shown. Molecular mass markers (kDa) are shown at the left side of the panels. The Hbp passenger (pass), proform (pro), and β-domain are indicated at the right side of the panels. B, immunoblot analysis of the OmpA content in 0.03 OD660 units of cells collected under A. The precursor (pre) and mature (mat) forms of OmpA are indicated at the right side of the panel. C, immunoblot analysis of the YidC and PspA content in 0.03 OD660 units of cells collected under A.
In nondepleted cells, Hbp is efficiently expressed, processed, and secreted as shown by the appearance of the ∼112-kDa passenger domain (30, 41) in the cells and medium as compared with cells that were not induced with IPTG (Fig. 2A, cf. lanes 1 and 2–3). Also, a ∼28-kDa product that represents the cleaved β-domain of Hbp (41) accumulated in the cells (Fig. 2A, lane 2). The identity of the passenger and β-domain was verified by immunoblotting using domain-specific antibodies (data not shown). Upon depletion of YidC, strongly reduced amounts of Hbp were observed in both the cells and medium fraction, suggesting that Hbp biogenesis was severely affected (Fig. 2A, lanes 2 and 3 and lanes 4 and 5). As a control, signal peptide processing of outer membrane A (OmpA) appeared virtually unaffected (Fig. 2B, lanes 2 and 3), demonstrating that indirect inactivation of the Sec translocon had not occurred under these conditions. Depletion of YidC was verified by analyzing the YidC content in the cell fraction by immunoblotting (Fig. 2C, lanes 2 and 3). In addition, up-regulation of the stress protein PspA was observed in these cells due to dissipation of the proton motive force (Fig. 2C, lane 3). This is caused by defects in the membrane assembly of the cytochrome o oxidase and F1F0-ATPase complexes, a direct consequence of YidC depletion (42).
Hbp belongs to the SPATE subfamily of ATs (1, 41). To extend our findings concerning the involvement of YidC, we wished to investigate whether they also apply to SPATE members other than Hbp. For this reason, we monitored the expression of the SPATE EspC (43) in the yidC conditional strain JS7131 in parallel to Hbp (supplemental Fig. S1). In the presence of arabinose, the ∼103-kDa EspC passenger domain was recovered from the culture medium indicating expression and secretion of EspC under these conditions (supplemental Fig. S1A, lane 8). In line with previous observations (43), the EspC passenger was virtually absent in whole cells as opposed to the Hbp passenger (supplemental Fig. S1A cf. lanes 3 and 7). In the absence of arabinose (see supplemental Fig. S1B, lane 3), significantly lower amounts of EspC passenger were detected (supplemental Fig. S1A, lanes 5 and 6 and lanes 7 and 8) comparable with what was observed for Hbp (supplemental Fig. S1A, lanes 1 and 2 and lanes 3 and 4), showing that the biogenesis of EspC and Hbp are similarly affected by depletion of YidC. To investigate whether the reduced levels of Hbp and EspC upon depletion of YidC reflect a functional dependence on YidC rather than a general defect in protein expression, we analyzed the effect of YidC depletion on the steady-state levels of an unrelated secretory protein (supplemental Fig. S2). To this end, the outer membrane protein PhoE was produced from the vector pEH3 (that was also used for Hbp and EspC expression) in strain JS7131 to similar levels as Hbp (supplemental Fig. S2A). Depletion of YidC did not alter the levels of PhoE significantly (supplemental Fig. S2A, lanes 5 and 6), suggesting that the effect on the biogenesis of Hbp and EspC were not due to more general secondary effects of YidC depletion. Taken together, the data point to a more general role for YidC in the biogenesis of SPATE-type ATs.
Depletion of YidC Leads to Accumulation of DegP-sensitive Hbp Intermediates in the Periplasmic Space
Depletion of YidC leads to reduced levels of Hbp. Pulse-chase analysis showed that synthesis of Hbp under these conditions is not affected (data not shown), suggesting that in the absence of YidC, Hbp is degraded. This raises the question in which cellular compartment the apparently unstable Hbp intermediates that are formed in the absence of YidC are degraded. We have recently shown that Hbp mutants that fail to be translocated across the OM are degraded by the periplasmic protease DegP (30). We further showed that expression of secretion incompetent Hbp derivatives in the absence of DegP resulted in the accumulation of unprocessed pro-Hbp in the periplasm (30). These findings prompted us to analyze whether degradation of Hbp upon YidC depletion takes place in the periplasm. For this purpose, a strain was engineered, JS7131ΔdegP, that allowed us to evaluate the effect of YidC depletion on Hbp biogenesis in the absence of DegP.
E. coli JS7131ΔdegP was transformed with pEH3-Hbp, and expression of Hbp was determined in the presence (control cells) or absence (YidC-depleted cells) of arabinose as before (Fig. 3). In contrast to the reduced levels of Hbp in standard JS7131 cells (Fig. 3A, lanes 1–4), YidC depletion in JS7131ΔdegP resulted in the accumulation of a ∼148-kDa product (Fig. 3A, asterisk) in the cells representing the unprocessed proform of Hbp (Fig. 3A, cf. lanes 7 and 5), as confirmed by immunoblotting using antisera against the Hbp passenger and β-domain (data not shown). Of note, the finding that proHbp was recovered specifically in the absence of DegP, a protease that is restricted to the periplasm, implies that proHbp accumulates in that compartment; i.e. after translocation across the IM. Furthermore, recovered proHbp material was not further processed and secreted into the medium, which suggests that these intermediates have attained a conformation that is incompatible with secretion. Efficient depletion of YidC, as well as the absence of DegP in JS7131ΔdegP cells, was verified by immunoblotting (Fig. 3B).
FIGURE 3.
Accumulation of DegP-sensitive Hbp intermediates in the periplasm upon depletion of YidC. A, analysis of expression and secretion of Hbp in strains JS7131 and JS7131ΔdegP grown in the presence or absence of l-arabinose to allow expression or depletion of YidC, respectively, as described in the legend to Fig. 2A. Cells (C) and medium (M) fractions are indicated at the top of the panel. Molecular mass markers (kDa) are shown at the left side of the panel. The Hbp passenger (pass) and proform (pro) are indicated at the right side of the panel. proHbp that accumulates upon depletion of YidC is indicated with an asterisk. B, immunoblot analysis of the YidC and DegP content of 0.03 OD660 units of cells collected under A.
To confirm that stabilization of proHbp in JS7131ΔdegP is the result of inactivation of the degP gene per se and not due to a secondary mutation on the chromosome, we investigated whether degradation of Hbp could be restored by providing a copy of the gene encoding DegP in trans (supplemental Fig. S3). For this purpose, strain JS7131ΔdegP harboring pEH3-Hbp was cotransformed with a compatible low copy number plasmid (pWSK29/DegP) carrying degP under control of its native promoter. Cells carrying both plasmids expressed DegP to levels similar to JS7131 cells that carried an intact chromosal copy of degP (supplemental Fig. S3B, lanes 1 and 3). In contrast, JS7131ΔdegP(pEH3-Hbp) cells, which carried empty pWSK29 or no extra plasmid at all, did not show DegP expression as expected (supplemental Fig. S3B, lanes 2 and 4). Furthermore, in contrast to the empty vector control, cells harboring the DegP expression plasmid survived growth at 42 °C (data not shown). Expression of Hbp in YidC-depleted JS7131ΔdegP carrying the empty vector resulted in accumulation of proHbp material (supplemental Fig. S3A, lane 5) as expected. Such accumulation of proHbp was not observed when pWSK29/DegP was present (Fig. S3A, lane 1). Instead, depletion of YidC again resulted in reduced amounts of Hbp in the cells and the medium (supplemental Fig. S3A, lanes 1 and 2 and 3 and 4), demonstrating that secretion incompetent Hbp intermediates formed under these conditions were degraded by plasmid-encoded DegP. The data suggest that stabilization of proHbp species in the periplasm of JS7131ΔdegP is a direct consequence of deletion of degP.
Taken together, the data demonstrate that upon depletion of YidC, secretion-incompetent and DegP-sensitive Hbp intermediates accumulate in the periplasmic space. Apparently, YidC is involved in biogenesis of Hbp at a late stage, after translocation across the IM.
Large Periplasmic Domain P1 of YidC Is Not Important for Biogenesis of Hbp
YidC is composed of six TMs with a large ∼35-kDa periplasmic domain P1 between TMs 1 and 2, corresponding to amino acid residues 24–342 (24). Thus far, the exact role of the domain P1 has remained unclear. Our in vivo data show that YidC is involved in late steps of Hbp biogenesis and suggest that it may be required to prevent misfolding of Hbp in the periplasmic space. Therefore, we considered the possibility that domain P1 of YidC plays a role in folding of Hbp.
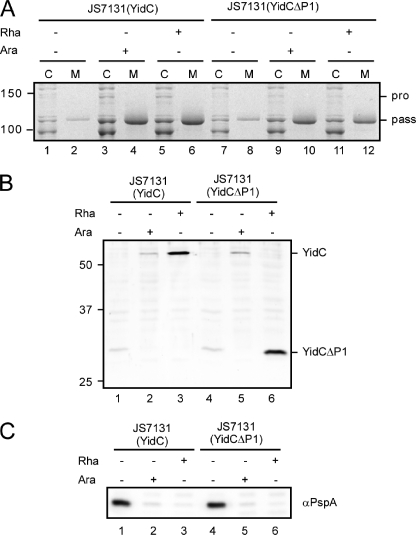
The construct YidCΔP1, which lacks residues 27–320 of YidC, was made. Importantly, this construct still contains the C-terminal region of the periplasmic domain that has been reported to be essential for YidC activity (14, 27). Indeed, when YidCΔP1 was placed on a low copy number vector under rhamnose promoter control and was introduced into JS7131(pEH3-Hbp), addition of rhamnose to the growth medium did restore the growth defect that was observed when cells were grown in the absence of arabinose to deplete YidC (data not shown). Furthermore, expression of YidCΔP1 (see Fig. 4B, lane 6) was shown to complement the PspA response that is associated with YidC depletion (Fig. 4C, cf. lanes 4 and 6). Wild-type YidC was properly depleted under these conditions as shown by immunoblotting (Fig. 4B, cf. lanes 5 and 6), validating our assay conditions. Taken together, YidCΔP1 can functionally complement YidC in growth and preclusion of the PspA response.
FIGURE 4.
The large periplasmic domain of YidC is dispensible for expression and secretion of Hbp. A, analysis of expression and secretion of Hbp in JS7131(pEH3-Hbp) upon complementation of YidC depletion with plasmid encoded YidCΔP1 (lanes 7–12) or wild-type YidC (lanes 1–6) as described in the legend to Fig. 2A. Cultures were supplemented with l-arabinose (Ara) to allow expression of YidC from the genome, or l-rhamnose (Rha) to allow expression of YidC and YidCΔP1 from pWSK/Rha, as indicated. Cells (C) and medium (M) fractions are indicated at the top of the panel. Molecular mass markers (kDa) are shown at the left side of the panel. The Hbp passenger (pass) and proform (pro) are indicated at the right side of the panel. B, immunoblot analysis of YidC and YidCΔP1 expression in 0.03 OD660 units of cells collected under A using antiserum against YidC. Molecular mass markers (kDa) are shown at the left side of the panel. YidC and YidCΔP1 are indicated at the right side of the panel. C, immunoblot analysis of the PspA content in 0.03 OD660 units of cells collected under A.
Upon complementation with YidCΔP1, the amount of Hbp in the cells and the culture medium were comparable with what was observed for cells grown in the presence of arabinose, expressing wild-type YidC from the chromosome (Fig. 4A, lanes 11 and 12 and 9 and 10), or cells complemented with wild-type YidC from a rhamnose-inducible plasmid (Fig. 4A, lanes 5 and 6). As a control, the levels of Hbp in the absence of both arabinose and rhamnose were significantly reduced (Fig. 4A, lanes 1 and 2 and 7 and 8). These results demonstrate that YidCΔP1 sustains proper biogenesis of Hbp, indicating that the periplasmic domain P1 of YidC is dispensable for the Hbp secretion process.
DISCUSSION
ATs of the SPATE subfamily are synthesized with an unusually long signal peptide to mediate targeting to and translocation across the bacterial IM. To investigate the mechanism of transfer across the IM, we probed the molecular interactions of the AT Hbp during IM translocation. In a previous study, we inferred that Hbp inserts into the membrane in or close to the Sec translocon, based on the observation that nascent Hbp was primarily cross-linked in vitro to SecY and SecA (8). In this study, a homo-bifunctional cross-linking reagent was used that is specific for lysine residues and less suited to probe interactions of lysine-deprived TMs and signal peptides that occur in the lipid bilayer (16). In the present study, we focused on the interactions of the unusual signal peptide of Hbp in the inner membrane using an unbiased site-specific in vitro photocross-linking approach. Surprisingly, the signal peptide in short nascent Hbp was efficiently cross-linked to YidC, which has been implicated in the biogenesis of IMPs both in a Sec-dependent and -independent process (the YidC-only pathway). In addition, Hbp was shown to require YidC for efficient secretion in vivo. In the absence of YidC, secretion-incompetent Hbp intermediates accumulated in the periplasmic space that appeared sensitive to degradation by the protease DegP. Based on these data and the previous observation that DegP degrades misfolded Hbp passenger derivatives that are unable to pass the outer membrane (30), we suggest that YidC is required to maintain or fold Hbp in the periplasm in a secretion-competent conformation.
Previously, we have shown that Hbp can make use of the SRP route for optimal targeting to the IM (7, 8). This is uncommon because, in bacteria, SRP primarily directs targeting of IMPs with relatively hydrophobic signal sequences (10). However, Hbp is not the only exception. SRP is also involved in the targeting of the periplasmic protein DsbA and the lipoproteins bacteriocin release protein and murein lipoprotein (44, 45). Interestingly, in both cases SRP dependence seemed correlated with an atypical use of the Sec translocon. Mutations in secY that specifically inhibit the insertion of IMPs, rather than the translocation of periplasmic and OM proteins, also affected processing of DsbA (45, 46). Furthermore, during IM translocation, bacteriocin release protein and murein lipoprotein were shown to interact with YidC (44). Finally, GFP fused to the cleavable hydrophobic FliP signal peptide appeared to rely on both SRP and YidC for IM translocation (47). Together, these observations hint at a correlation between the mode of targeting and the mode of translocation/insertion mediated by the Sec translocon.
Our current cross-link data (Fig. 1) show that the Hbp signal peptide interacts with YidC during IM translocation. Combined with observed cross-linking to SRP in the cytosol and SecA/SecY in the membrane (7, 8, this study), the molecular interactions of Hbp during targeting and insertion seem very similar to those of nascent IMPs (16, 39, 40). However, whereas bacteriocin release protein, murein lipoprotein, and FliP-GFP depend on YidC for their translocation across the IM (44, 47), Hbp is the first example of a secretory protein that relies on YidC at stages beyond the translocation step. Upon depletion of YidC, Hbp is degraded by the major periplasmic protease DegP (Fig. 3), suggesting that YidC is required to fold or maintain Hbp in a secretion-competent conformation in the periplasm. How does YidC influence the conformation of Hbp in the periplasm? YidC has already been implicated in the folding of polytopic IMPs mainly based on the impaired in vitro folding of LacY in YidC-depleted membrane vesicles (19). However, in this functionality, YidC is believed to assist in the assembly of TM segments into helical bundles. This view is supported by the observation that YidC is adjacent to multiple TM segments of nascent polytopic IMPs at a specific stage in biogenesis (17, 18, 20). Obviously, for Hbp that only contains a (cleavable) signal peptide YidC must play a different role. For the SPATE EspP, it was suggested that transient tethering to the IM is mediated by the signal peptide and necessary to prevent misfolding of the passenger in the periplasm (9). Given the efficient cross-linking of the Hbp signal peptide with YidC in inner membrane vesicles (Fig. 1), YidC might slow down the release of Hbp from the IM through a prolonged interaction with the Hbp signal peptide or by preventing access to the leader peptidase. If so, a critical role for the extended Hbp signal peptide is anticipated. However, preliminary data suggest that Hbp carrying a signal peptide of the SecB substrate PhoE (48) that cannot be cross-linked to YidC, is still degraded in the periplasm. Furthermore, pulse-chase experiments did not reveal accelerated processing of the Hbp signal peptide under conditions of YidC depletion (data not shown). Hence, an alternative role for YidC in the biogenesis of Hbp should be considered.
E. coli YidC comprises six TM segments and a large periplasmic domain P1 of which the function is not known. Interestingly, recent crystal structures of domain P1 revealed a cleft that might have unfolded polypeptide chains as its natural ligand (25, 26). Conceivably, P1 binds to regions of Hbp that emerge in the periplasm upon translocation through the Sec-YidC complex to slow down release and to assist folding or recruitment of chaperones such as Skp and SurA that rather delay folding and assist targeting to the outer membrane. However, such a scenario is difficult to reconcile with our observation that biogenesis and secretion of Hbp proceed unaffected when the P1 domain has been deleted from YidC (Fig. 4). Furthermore, limited proteolysis of surface exposed and secreted Hbp did not reveal conformational differences in Hbp passenger material secreted in the presence or absence of domain P1 (data not shown). Notably, YidC has been shown to associate with the SecDF-YajC accessory complex of the Sec translocon, and its overproduction suppresses the growth defect in the absence of SecDF-YajC (11, 14). Similar to YidC, SecD contains a large periplasmic domain that has been implicated in the release of preproteins from the translocon (49). Conceivably, the periplasmic domains of SecDF and YidC have (partially) overlapping functions, which might account for proper biogenesis of Hbp in the absence of the P1 domain. In any case, the conserved membrane domain of YidC is required and may be sufficient for the observed effects.
At present, we cannot exclude that YidC has a more indirect role in the biogenesis of ATs. Although a recent study showed that depletion of YidC has a global influence on the physiology of the cell (50), it should be stressed that the effect of YidC depletion on Hbp biogenesis appeared rather specific. For example, depletion of YidC strongly reduced the steady-state levels of Hbp and a second AT (EspC) but not the levels of an unrelated secretory protein (PhoE) that is translocated across the IM in a Sec-dependent but YidC-independent manner (Fig. 2 and supplemental Figs. S1 and S2). Furthermore, Hbp was efficiently translated in vivo under the depletion conditions used and also efficiently translocated across the IM via the Sec translocon. Also, in our experiments depletion of YidC did not cause up-regulation of DegP (Fig. 3B), an indicator of extracellular stress (51). Combined, these observations argue that the influence of YidC on Hbp biogenesis is not due to more general secondary effects. It remains possible that YidC recruits or modulates the activity of specific factors (e.g. chaperones) in the IM or periplasm that are required for folding and secretion of ATs.
The SPATEs Hbp and EspC are the first examples of ATs of which the biogenesis is influenced by YidC. Because ATs share a highly conserved domain organization and structure (2), it is anticipated that the biogenesis of other ATs also involves YidC. On the other hand, secretion and localization of the Shigella flexneri AT IcsA expressed in E. coli was not affected upon YidC depletion (52). Although this may relate to the expression of IcsA in a heterologous host organism, it is also possible that different ATs have solved the problems that come with the targeting and secretion of such large and complex proteins in different ways. The use of different signal peptides, targeting factors, and periplasmic chaperones indeed hint at disparate mechanisms of biogenesis.
Supplementary Material
Acknowledgments
We thank G. Koningstein for great technical assistance during the cross-linking analysis. We also thank P. van Ulsen and J. W. de Gier for useful comments on the manuscript. J. Tommassen, J. Beckwith, J. W. de Gier, J. Nataro, and S. Wagner are acknowledged for providing antisera, strains, and plasmids.
This work was supported by an “ALW Open Program” grant from the Netherlands Organization for Scientific Research (NWO) and a FEBS short term fellowship (to W. S. P. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- IM
- inner membrane
- Hbp
- hemoglobin protease
- OM
- outer membrane
- SRP
- signal recognition particle
- IMP
- inner membrane protein
- TM
- transmembrane segment
- TF
- trigger factor
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- fw
- forward
- rv
- reverse
- SPATE
- serine protease autotransporter of Enterobacteriaceae.
REFERENCES
- 1.Henderson I. R., Navarro-Garcia F., Desvaux M., Fernandez R. C., Ala'Aldeen D. (2004) Microbiol. Mol. Biol. Rev. 68, 692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dautin N., Bernstein H. D. (2007) Annu. Rev. Microbiol. 61, 89–112 [DOI] [PubMed] [Google Scholar]
- 3.Desvaux M., Cooper L. M., Filenko N. A., Scott-Tucker A., Turner S. M., Cole J. A., Henderson I. R. (2006) FEMS Microbiol. Lett. 264, 22–30 [DOI] [PubMed] [Google Scholar]
- 4.Chevalier N., Moser M., Koch H. G., Schimz K. L., Willery E., Locht C., Jacob-Dubuisson F., Müller M. (2004) J. Mol. Microbiol. Biotechnol. 8, 7–18 [DOI] [PubMed] [Google Scholar]
- 5.Desvaux M., Scott-Tucker A., Turner S. M., Cooper L. M., Huber D., Nataro J. P., Henderson I. R. (2007) Microbiology 153, 59–70 [DOI] [PubMed] [Google Scholar]
- 6.Peterson J. H., Szabady R. L., Bernstein H. D. (2006) J. Biol. Chem. 281, 9038–9048 [DOI] [PubMed] [Google Scholar]
- 7.Jong W. S., Luirink J. (2008) Biochem. Biophys. Res. Commun. 368, 522–527 [DOI] [PubMed] [Google Scholar]
- 8.Sijbrandi R., Urbanus M. L., ten Hagen-Jongman C. M., Bernstein H. D., Oudega B., Otto B. R., Luirink J. (2003) J. Biol. Chem. 278, 4654–4659 [DOI] [PubMed] [Google Scholar]
- 9.Szabady R. L., Peterson J. H., Skillman K. M., Bernstein H. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luirink J., Sinning I. (2004) Biochim. Biophys. Acta 1694, 17–35 [DOI] [PubMed] [Google Scholar]
- 11.Nouwen N., Driessen A. J. (2002) Mol. Microbiol. 44, 1397–1405 [DOI] [PubMed] [Google Scholar]
- 12.Scotti P. A., Valent Q. A., Manting E. H., Urbanus M. L., Driessen A. J., Oudega B., Luirink J. (1999) J. Biol. Chem. 274, 29883–29888 [DOI] [PubMed] [Google Scholar]
- 13.van Bloois E., Dekker H. L., Fröderberg L., Houben E. N., Urbanus M. L., de Koster C. G., de Gier J. W., Luirink J. (2008) FEBS Lett. 582, 1419–1424 [DOI] [PubMed] [Google Scholar]
- 14.Xie K., Kiefer D., Nagler G., Dalbey R. E., Kuhn A. (2006) Biochemistry 45, 13401–13408 [DOI] [PubMed] [Google Scholar]
- 15.Houben E. N., ten Hagen-Jongman C. M., Brunner J., Oudega B., Luirink J. (2004) EMBO Rep. 5, 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scotti P. A., Urbanus M. L., Brunner J., de Gier J. W., von Heijne G., van der Does C., Driessen A. J., Oudega B., Luirink J. (2000) EMBO J. 19, 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck K., Eisner G., Trescher D., Dalbey R. E., Brunner J., Müller M. (2001) EMBO Rep. 2, 709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pop O. I., Soprova Z., Koningstein G., Scheffers D. J., van Ulsen P., Wickström D., de Gier J. W., Luirink J. (2009) FEBS J. 276, 4891–4899 [DOI] [PubMed] [Google Scholar]
- 19.Nagamori S., Smirnova I. N., Kaback H. R. (2004) J. Cell Biol. 165, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner S., Pop O., Haan G. J., Baars L., Koningstein G., Klepsch M. M., Genevaux P., Luirink J., de Gier J. W. (2008) J. Biol. Chem. [DOI] [PubMed] [Google Scholar]
- 21.van Bloois E., ten Hagen-Jongman C. M., Luirink J. (2007) Biochem. Biophys. Res. Commun. 362, 727–733 [DOI] [PubMed] [Google Scholar]
- 22.Xie K., Dalbey R. E. (2008) Nat. Rev. Microbiol. 6, 234–244 [DOI] [PubMed] [Google Scholar]
- 23.van Bloois E., Nagamori S., Koningstein G., Ullers R. S., Preuss M., Oudega B., Harms N., Kaback H. R., Herrmann J. M., Luirink J. (2005) J. Biol. Chem. 280, 12996–13003 [DOI] [PubMed] [Google Scholar]
- 24.Sääf A., Monné M., de Gier J. W., von Heijne G. (1998) J. Biol. Chem. 273, 30415–30418 [DOI] [PubMed] [Google Scholar]
- 25.Oliver D. C., Paetzel M. (2008) J. Biol. Chem. 283, 5208–5216 [DOI] [PubMed] [Google Scholar]
- 26.Ravaud S., Stjepanovic G., Wild K., Sinning I. (2008) J. Biol. Chem. 283, 9350–9358 [DOI] [PubMed] [Google Scholar]
- 27.Jiang F., Chen M., Yi L., de Gier J. W., Kuhn A., Dalbey R. E. (2003) J. Biol. Chem. 278, 48965–48972 [DOI] [PubMed] [Google Scholar]
- 28.Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., Phillips G. J., Dalbey R. E. (2000) Nature 406, 637–641 [DOI] [PubMed] [Google Scholar]
- 29.Strauch K. L., Beckwith J. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 1576–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jong W. S., ten Hagen-Jongman C. M., den Blaauwen T., Slotboom D. J., Tame J. R., Wickström D., de Gier J. W., Otto B. R., Luirink J. (2007) Mol. Microbiol. 63, 1524–1536 [DOI] [PubMed] [Google Scholar]
- 31.Dutta P. R., Cappello R., Navarro-García F., Nataro J. P. (2002) Infect. Immun. 70, 7105–7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R. F., Kushner S. R. (1991) Gene 100, 195–199 [PubMed] [Google Scholar]
- 33.Giacalone M. J., Gentile A. M., Lovitt B. T., Berkley N. L., Gunderson C. W., Surber M. W. (2006) BioTechniques 40, 355–364 [DOI] [PubMed] [Google Scholar]
- 34.Urbanus M. L., Scotti P. A., Froderberg L., Saaf A., de Gier J. W., Brunner J., Samuelson J. C., Dalbey R. E., Oudega B., Luirink J. (2001) EMBO Rep. 2, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martoglio B., Dobberstein B. (1996) Trends Cell Biol. 6, 142–147 [DOI] [PubMed] [Google Scholar]
- 36.Brunner J. (1989) Methods Enzymol. 172, 628–687 [DOI] [PubMed] [Google Scholar]
- 37.Houben E. N., Urbanus M. L., Van Der Laan M., Ten Hagen-Jongman C. M., Driessen A. J., Brunner J., Oudega B., Luirink J. (2002) J. Biol. Chem. 277, 35880–35886 [DOI] [PubMed] [Google Scholar]
- 38.Ullers R. S., Houben E. N., Brunner J., Oudega B., Harms N., Luirink J. (2006) J. Biol. Chem. 281, 13999–14005 [DOI] [PubMed] [Google Scholar]
- 39.Houben E. N., Scotti P. A., Valent Q. A., Brunner J., de Gier J. L., Oudega B., Luirink J. (2000) FEBS Lett. 476, 229–233 [DOI] [PubMed] [Google Scholar]
- 40.Valent Q. A., de Gier J. W., von Heijne G., Kendall D. A., ten Hagen-Jongman C. M., Oudega B., Luirink J. (1997) Mol. Microbiol. 25, 53–64 [DOI] [PubMed] [Google Scholar]
- 41.Otto B. R., van Dooren S. J., Nuijens J. H., Luirink J., Oudega B. (1998) J. Exp. Med. 188, 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Laan M., Urbanus M. L., Ten Hagen-Jongman C. M., Nouwen N., Oudega B., Harms N., Driessen A. J., Luirink J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5801–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein M., Kenny B., Stein M. A., Finlay B. B. (1996) J. Bacteriol. 178, 6546–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fröderberg L., Houben E. N., Baars L., Luirink J., de Gier J. W. (2004) J. Biol. Chem. 279, 31026–31032 [DOI] [PubMed] [Google Scholar]
- 45.Shimohata N., Akiyama Y., Ito K. (2005) J. Bacteriol. 187, 3997–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimohata N., Nagamori S., Akiyama Y., Kaback H. R., Ito K. (2007) J. Cell Biol. 176, 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pradel N., Decorps A., Ye C., Santini C. L., Wu L. F. (2005) Biochimie 87, 191–196 [DOI] [PubMed] [Google Scholar]
- 48.Kusters R., de Vrije T., Breukink E., de Kruijff B. (1989) J. Biol. Chem. 264, 20827–20830 [PubMed] [Google Scholar]
- 49.Matsuyama S., Fujita Y., Mizushima S. (1993) EMBO J. 12, 265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang P., Kuhn A., Dalbey R. E. (2010) J. Bacteriol. 192, 2193–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz N., Silhavy T. J. (2005) Curr. Opin. Microbiol. 8, 122–126 [DOI] [PubMed] [Google Scholar]
- 52.Brandon L. D., Goehring N., Janakiraman A., Yan A. W., Wu T., Beckwith J., Goldberg M. B. (2003) Mol. Microbiol. 50, 45–60 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.