Abstract
The Mycobacterium tuberculosis genome harbors a striking number (>40) of toxin-antitoxin systems. Among them are at least seven MazF orthologs, designated MazF-mt1 through MazF-mt7, four of which have been demonstrated to function as mRNA interferases that selectively target mRNA for cleavage at distinct consensus sequences. As is characteristic of all toxin-antitoxin systems, each of the mazF-mt toxin genes is organized in an operon downstream of putative antitoxin genes. However, only one of the seven putative upstream antitoxins (designated MazE-mt1 through MazE-mt7) has significant sequence similarity to Escherichia coli MazE, the cognate antitoxin for E. coli MazF. Interestingly, the M. tuberculosis genome contains two independent operons encoding E. coli MazE orthologs, but they are not paired with mazF-mt-like genes. Instead, the genes encoding these two MazE orthologs are each paired with proteins containing a PIN domain, indicating that they may be members of the very large VapBC toxin-antitoxin family. We tested a spectrum of pair-wise combinations of cognate and noncognate Mtb toxin-antitoxins using in vivo toxicity and rescue experiments along with in vitro interaction experiments. Surprisingly, we uncovered several examples of noncognate toxin-antitoxin association, even among different families (e.g. MazF toxins and VapB antitoxins). These results challenge the “one toxin for one antitoxin” dogma and suggest that M. tuberculosis may enlist a sophisticated toxin-antitoxin network to alter its physiology in response to environmental cues.
Keywords: mRNA, Protein Stability, Ribonuclease, Toxins, Translation, MazE, MazF, VapB, VapC, Tuberculosis
Introduction
Tuberculosis is a widespread disease in the developing world—at least one third of the world's population is infected with Mycobacterium tuberculosis (Mtb)4 (1, 2). This infection is extremely complex and is able to cause active tuberculosis or persist in a latent state that has enabled the extensive persistence of Mtb in the human population.
Toxin-antitoxin (TA) modules are specialized operons comprising adjacent antitoxin and toxin genes that are present in free-living bacteria (3–5). The toxin and its cognate antitoxin protein form a stable protein complex; however, the antitoxin is more labile than the toxin protein. When cellular conditions lead to a decrease in the amount of antitoxin, freed toxin is able to act on its intracellular target. Expression of Escherichia coli TA modules leads to cell death (6) that is preceded by a dormant state (7); the latter has been linked to persistence (8–11). Dormancy and persistence are properties associated with latent tuberculosis infection (12–14). However, the biochemical activities and physiological roles of the many Mtb TA systems are not yet understood. Therefore, it is unclear whether TA systems contribute to tuberculosis latency.
The ability of TA systems to mediate a reversible state of growth arrest was discovered during the study of the E. coli MazF toxin (derived from the mazEF TA module) and other TA systems (15). MazF is a ssRNA- and sequence-specific endoribonuclease that cuts before or after the first A at ACA sequences in mRNA (16, 17). MazF expression in E. coli leads to a type of suspended animation called “quasi-dormancy”, where cell growth is arrested but the cells retain the capacity for full metabolic activity (7). However, this state was found to exist only for a short window of time after MazF induction and appears to facilitate survival during periods of stress (18, 19).
Although E. coli possesses a single mazEF TA module, there are at least seven MazF counterparts in M. tuberculosis (4), of which at least four of the seven MazF-mt toxins function as mRNA interferases (i.e. sequence-specific endoribonucleases that exclusively target mRNA) (20, 21). All four of the MazF toxins characterized to date (MazF-mt1, -mt3, -mt6, and -mt7) have different sequence specificities, recognizing three- or five-base consensus sequences. MazF-mt1 preferentially cleaves mRNA between U and A in UAC triplet sequences (5′-U↓AC-3′), whereas MazF-mt6 preferentially cleaves U-rich regions with the degenerate consensus sequence of 5′-(U/C)U↓(A/U)C(U/C)-3′ (21). MazF-mt3 cleaves RNA at 5′-UU↓CCU-3′ and 5′-CU↓CCU-3′, whereas MazF-mt7 cleaves at 5′-U↓CGCU-3′ (20). Therefore, MazF family members in Mtb exhibit a range of cleavage specificities that are proposed to alter protein expression through differential mRNA degradation, some result in extensive mRNA cleavage whereas others target selected mRNAs for cleavage (20).
In this work, we used two Mtb MazF family members, MazF-mt1 and MazF-mt3, to dissect regulation of toxin activity in this pathogen. Surprisingly, we discovered that the MazF-mt3 protein (and to a lesser extent, MazF-mt1) could physically interact with other “noncognate” antitoxins (MazF toxins-VapB antitoxins) in addition to the protein product of its upstream gene (its cognate antitoxin). Likewise, we also documented noncognate interactions between VapC toxins and a MazE antitoxins. These interactions were physiologically significant because the noncognate antitoxins were also able to reduce toxicity when co-expressed in an E. coli host. Taken together, these results bring to light the possibility of significant cross-talk among Mtb TA system families, resulting in a network of toxins whose activities may modulate translation (or other essential cellular processes) in response to environmental cues.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
The E. coli strains BL21(DE3) (Novagen) and BW25113 (22) were used for recombinant protein expression and in vivo toxicity and rescue experiments, respectively. The VapB-mt24 (Rv0599c), VapB-mt25 (Rv2595), MazE-mt1 (Rv2801A), and MazE-mt3 (Rv1991A) open reading frames were PCR-amplified from M. tuberculosis H37Rv genomic DNA using primers containing 5′-NdeI-BamHI-3′ ends and cloned into the corresponding sites of the IPTG-inducible plasmid pINIII (23, 24) to create pIN-VapB-mt24, pIN-VapB-mt25, pIN-MazE-mt1, and pIN-MazE-mt3. Plasmids pIN-VapB-mt24, pIN-VapB-mt25, pIN-MazE-mt1, and pIN-MazE-mt3 were then digested with NdeI and BamHI and ligated into the corresponding sites of pET28a (Novagen) to create pET28a-VapB-mt24, pET28a-VapB-mt25, pET28a-MazE-mt1, and pET28a-MazE-mt3. The VapC-mt24 (Rv0598c), VapC-mt25 (Rv2596), MazF-mt1 (Rv2801c), and MazF-mt3 (Rv1991c) open reading frames were PCR-amplified from M. tuberculosis H37Rv genomic DNA using primers with 5′-NdeI-BamHI-3′ ends and ligated into the corresponding sites of pET21c (Novagen) to create pET21c-VapC-mt24, pET21c-VapC-mt25, pET21c-MazF-mt1, and pET21c-MazF-mt3.
The pBAD33 plasmid was used to facilitate tight, arabinose-regulated toxin expression (22). pET21c-MazF-mt1 and pET21c-MazF-mt3 were cut with XbaI and HindIII and ligated into the corresponding sites of pBAD33 to create pBAD-MazF-mt1 and pBAD-MazF-mt3.
pET21c-MazF-mt1, pET21c-MazF-mt3, pET21c-VapC-mt24, and pET21c-VapC-mt25 were each digested with NdeI and BamHI, and the toxin-containing DNA fragment was ligated into the corresponding sites in the pCold-PST vector (25) to create the respective protein S-tagged toxins. Plasmid pCold-TF-MazE-mt1 was constructed after digestion of pET28a-MazE-mt1 with NdeI and BamHI and ligated into the corresponding sites in the pCold-TF vector (Takara-Bio, Inc.) to express MazE-mt1 fused to trigger factor (TF), a ribosome-associated chaperone protein that facilitates co-translational folding of nascent polypeptides. Thus, the TF tag facilitated the expression of soluble MazE-mt1 by promoting correct protein folding. The accuracy of all DNA fragments synthesized by PCR was confirmed by DNA sequence analysis.
Purification of Protein S-tagged Toxins
PST-MazF-mt1, PST-MazF-mt3, PST-VapC-mt24, and PST-VapC-mt25 proteins tagged at the N terminus were purified from the BL21(DE3) strain carrying pCold-PST-MazF-mt1, pCold-PST-MazF-mt3, pCold-PST-VapC-mt24, or pCold-PST-VapC-mt25, respectively, using myxospores as described previously (25). Briefly, expression of the protein S-tagged proteins was induced at an A600 of 0.6 by adding IPTG to a final concentration of 1 mm and continuing to grow the cells at 15 °C for 16 h. The cells were then harvested, lysed by sonication, and incubated with myxospores in 1 mm CaCl2, 50 mm Tris-HCl (pH 8.0), 50 mm KCl, 5% glycerol at 4 °C for 1 h to facilitate binding of the protein S-tagged proteins to the myxospores.
Purification of MazE-mt1
TF-MazE-mt1 protein tagged at the N terminus was purified from the BL21(DE3) strain carrying pCold-TF-MazE-mt1 using Ni-NTA resin (Qiagen). The N-terminal TF tag was removed by digestion with thrombin (Sigma). MazE-mt1 was further purified by ion-exchange chromatography using Q-Sepharose Fast Flow and SP-Sepharose Fast Flow FPLC column chromatography (GE Healthcare).
RESULTS
Genome Arrangement of MazF-mt Toxin Genes and Their Putative Antitoxins
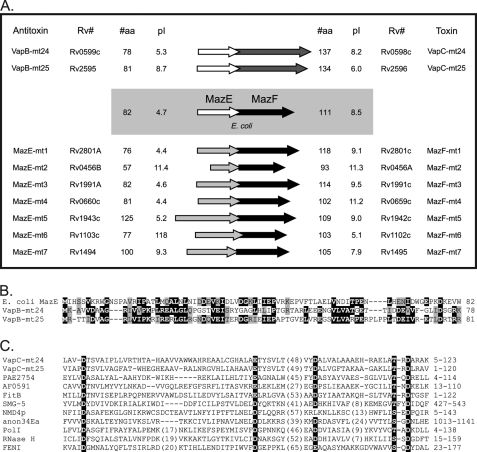
As with the E. coli MazE-MazF TA system (where MazE is the antitoxin for MazF and is positioned upstream of the toxin), the genes encoding each M. tuberculosis MazF toxin are co-localized with an upstream gene in an apparent operon (4, 21). We have designated the putative antitoxins upstream of MazF-mt1 through MazF-mt7 as MazE-mt1 through MazE-mt7, respectively (Fig. 1A). It is not known why the Mtb genome harbors so many MazF counterparts. Mtb MazF toxins are clearly orthologs of E. coli MazF because ∼20–45% of their amino acids are either identical or similar upon alignment (+1 or greater using the blosum62 matrix) (21). However, the putative MazE-mt antitoxins share <20% similarity with E. coli MazE (except for MazE-mt6; 26% similarity). Instead, we found that M. tuberculosis contained two other apparent TA modules encoding antitoxin proteins with 27 and 37% similarity to E. coli MazE (Fig. 1B). Curiously, these two antitoxins were upstream of genes encoding two apparent VapC toxins, which we designated VapC-mt24 and VapC-mt25 (Fig. 1C). These two VapC genes were not among the 23 Mtb VapC family members identified previously (4). However, as with all other VapC family members, they contain a PIN (PilT N terminus) domain. PIN domains were originally identified in a bacterial protein involved in pili synthesis. All PIN domain proteins in Eubacteria and Archaea are ∼140 amino acids in length and have four highly conserved acidic amino acids plus a fifth residue that is either a serine or threonine that form the Mg2+- or Mn2+-binding active site, highlighted in Fig. 1C (26). Eukaryotic proteins contain PIN domains within larger proteins. Although the precise function of the PIN domain has not yet been determined, it has been suggested that it is associated with nuclease activity based on the properties and structural features of PIN domain proteins in Eubacteria, Archaebacteria, and eukaryotes (26).
FIGURE 1.
Two genes encoding Mtb MazE orthologs are in operons with VapC toxin genes. A, arrows representing a gene in an operon. Arrow length is roughly proportional to relative gene length. Arrows in white represent the genes for MazE orthologs, dark grey arrows represent the genes for VapC orthologs, light grey arrows represent the genes for MazE-mt1 to -mt7, and black arrows represent the genes for MazF orthologs. The protein length, pI values, and Rv numbers are shown. B, alignment of E. coli MazE with Mtb VapB-mt24 and VapB-mt25. Identical amino acids are highlighted in black. Similar amino acids are highlighted in grey. Numbers on the right indicate the number of amino acids in the corresponding protein. C, alignment of VapC-mt24 and -25 with other PIN domain-containing proteins. The four conserved acidic residues (D or E) and a fifth invariant hydroxyl residue (S or T) comprising the catalytic site of PIN domain proteins are highlighted in black. The next three sequences are those of PIN domain proteins whose structures have been determined: PAE2754, hypothetical protein in Pyrobaculum aerophilum; AF0591, PIN domain protein in Archaeoglobus fulgidus; FitB, Neisseria gonorrhoeae (44). Caenorhabditis elegans SMG-5, Saccharomyces cerevisiae NMD4p, and Drosophila melanogaster anon34Ea are each involved in nonsense-mediated mRNA decay. Chlamydia pneumoniae DNA polymerase I (PolI), bacteriophage T4 RNase H and Methanococcus jannaschii Flap endonuclease-1 (FENI) possess 5′ to 3′ exonuclease catalytic domains (45). The numbers on the right indicate the amino acid numbers in the respective full-length protein used for the alignment.
As with MazF, it is not known why Mtb possesses such an abundance of VapBC modules. However, by convention all antitoxin genes upstream of Mtb VapC toxins are called VapB. Therefore, even though the genes upstream of VapC-mt24 and VapC-mt25 are more similar to E. coli MazE than any other gene in Mtb, we will refer to them as VapB-mt24 and VapB-mt25 in accordance with established nomenclature.
Analysis of Antitoxin Function by in Vivo Rescue of Toxicity
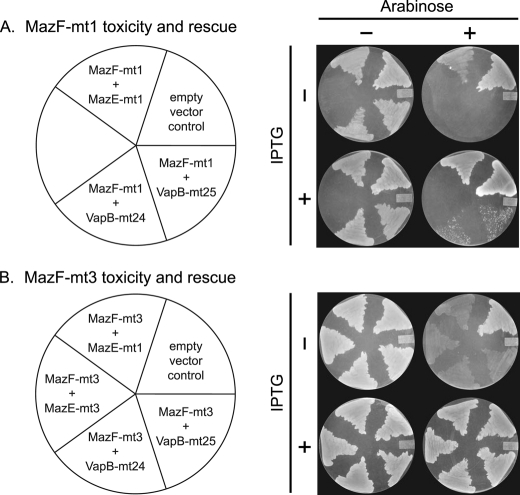
We tested whether the genes upstream of mazF-mt1, mazF-mt3, vapC-mt24, and vapC-mt25 functioned as antitoxins by performing in vivo toxicity and rescue experiments in E. coli. We were able to use E. coli as the host because we demonstrated previously that both MazF-mt1 and MazF-mt3 are toxic in this background (21). BW25113 E. coli cells containing an arabinose-inducible plasmid expressing MazF-mt1 or MazF-mt3 were transformed with a second plasmid that enabled IPTG-inducible co-expression of one of four different antitoxins (pIN-MazE-mt1, pIN-MazE-mt3, pIN-VapB-mt24, or pIN-VapB-mt25). We used these strains to perform plate toxicity and rescue experiments with the various toxin-antitoxin combinations.
For MazF-mt1 (Fig. 2A), no growth was observed when we used 0.02% arabinose to induce toxin expression. In comparison, a control strain that contained only pBAD and pIN vectors in BW25113 cells grew normally. However, when the putative cognate antitoxin MazE-mt1 was co-induced with the MazF-mt1 toxin on plates containing 1 mm IPTG plus 0.02% arabinose, cell growth was completely restored. Therefore, although not similar to E. coli MazE, MazE-mt1 functioned as an antitoxin for MazF-mt1. When the analogous experiment was performed with MazF-mt1 and the VapB-mt24 or VapB-mt25 noncognate antitoxins, growth was observed, but the colony sizes were small relative to rescue with the cognate antitoxin. We were unable to assess the effect of co-induction of MazF-mt1 with MazE-mt3 using this approach. Inexplicably, we were unable to recover BW25113 transformants containing both pBAD-MazF-mt1 and pIN-MazE-mt3 plasmids. Overall, these in vivo rescue experiments demonstrated that the toxic effect of MazF-mt1 could be fully neutralized by its cognate antitoxin MazE-mt1 and partially (because normal growth was not reconstituted) by either of the two noncognate VapB-mt24 and VapB-mt25 antitoxins.
FIGURE 2.
Antitoxin rescue of toxicity in vivo. Key for plasmids present in the BW25113 strains is shown on the left. The empty vector control strain contains pBAD33 and pINIII plasmids. Toxins in pBAD plasmids were induced with 0.02% arabinose, whereas antitoxins in pIN plasmids were induced with 1 mm IPTG.
Similar plate toxicity and rescue experiments were also carried out upon induction of the MazF-mt3 toxin using 0.02% arabinose. Consistent with published observations using high arabinose (0.2%) to induce the toxin, MazF-mt3 expression did not completely inhibit growth on plates (21) (Fig. 2B). However, a scorable phenotype, very weak growth on plates, was reproducibly observed, enabling us to assess rescue. BW25113 cells containing pBAD-MazF-mt3 were co-transformed with one of the following plasmids: pIN-MazE-mt1, pIN-MazE-mt3, pIN-VapB-mt24, or pIN-VapB-mt25. When we induced any one of the four antitoxins (cognate or noncognate) with the MazF-mt3 toxin, normal growth was observed (Fig. 2B). These results indicated that the toxic effect of MazF-mt3 could be reversed by co-expression with its cognate antitoxin, MazE-mt3, or with either noncognate antitoxin MazE-mt1, VapB-mt24 or VapB-mt25.
Analysis of Antitoxin Function Using in Vitro Interaction Studies
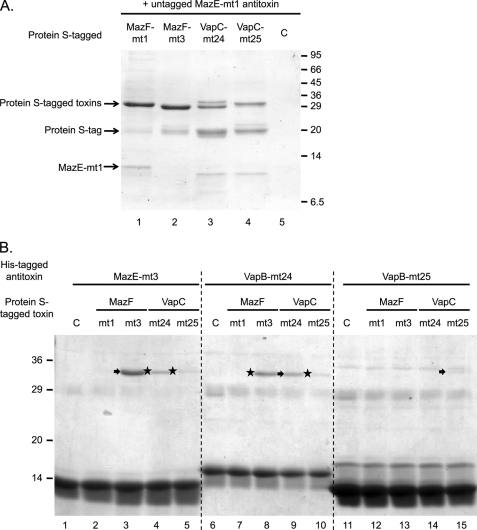
We next tested whether physical interactions between the same toxin and antitoxin combinations shown in Fig. 2 could substantiate our in vivo rescue results using pulldown experiments with either His-tagged proteins or protein S-tagged proteins (protein S is a major spore coat protein from Myxococcus xanthus).
Expression of Mtb TA toxins in E. coli is typically challenging. We have recently demonstrated that addition of the N-terminal protein S-tag (comprising two tandem N-terminal domains of protein S) enhances expression levels and protein solubility (25) when they are expressed at 15 °C from a vector derived from the pCold vectors developed in our laboratory (27). The addition of the 20-kDa protein S-tag also helps us to distinguish the toxin from the antitoxin on stained protein gels easily because all toxins and antitoxins have low molecular masses (∼10 kDa) whose mobilities often overlap. Protein S binds tightly to the surface of myxospores in the presence of Ca2+ (28, 29). Thus, a recombinant protein fused to protein S can be affinity-purified using myxospores.
We first demonstrated that untagged MazE-mt1 was able to bind protein S-tagged MazF-mt1 protein after myxospore affinity purification (Fig. 3A, lane 1); this interaction was consistent with our results in Fig. 2A. However, using the same analysis, we did not detect interactions between MazE-mt1 and the three other noncognate toxins (Fig. 3A, lanes 2–4).
FIGURE 3.
Protein interactions between cognate and noncognate toxin-antitoxin pairs. A, extracts derived from cells expressing protein S-tagged MazF-mt1, MazF-mt3, VapC-mt24, or VapC-mt25 were incubated with equivalent volumes of myxospores in the presence of 1 mm CaCl2 at 4 °C for 1 h, followed by the addition of 5 μg of purified MazE-mt1 protein and another 4 °C 1-h incubation. After washing the myxospores several times, Laemmli loading buffer was added to the myxospores, and the supernatant was heated to 95 °C for 5 min and loaded onto a 12% SDS-PAGE (29:1) gel. The control (lane C) represents proteins nonspecifically bound to myxospores. It is unclear why the protein S-tagged VapC-mt24 in lane 3 ran as a doublet. The position of the untagged MazE-mt1 is noted along with free protein S-tag (a percentage of the protein S is released from the fusion protein upon purification). The positions of the molecular mass markers are indicated on the right. The identity of the low molecular mass band (below MazE-mt1) in lanes 3 and 4 is not known; it may represent a protein that nonspecifically binds to VapC-mt24 and VapC-mt25 or a protein S degradation product. B, 5 μg of (His)6MazE-mt3, (His)6VapB-mt24, and (His)6VapB-mt25 was first incubated with Ni-NTA resin. The His-tagged antitoxins bound to the Ni-NTA resin were then incubated with 5 μg of purified protein S-tagged toxins. After washing several times, Laemmli loading buffer was added to the Ni-NTA resin, and the supernatant was loaded onto a 12% SDS-PAGE (29:1) gel. Lanes 1–5, His-tagged MazE-mt3 was incubated with buffer, protein S-tagged MazF-mt1, MazF-mt3, VapC-mt24, or VapC-mt25, respectively. Lanes 6–10, His-tagged VapB-mt24 was incubated with buffer, protein S-tagged MazF-mt1, MazF-mt3, VapC-mt24, or VapC-mt25, respectively. Lanes 11–15, His-tagged VapB-mt25 was incubated with buffer, protein S-tagged MazF-mt1, MazF-mt3, VapC-mt24, or VapC-mt25. Arrows highlight cognate toxin-antitoxin interactions; stars on the left highlight noncognate toxin-antitoxin interactions; lanes with a C contain buffer instead of a protein S-tagged toxin. The positions of the molecular mass markers are indicated on the left.
Next, we tested whether three individual His-tagged antitoxin proteins, MazE-mt3, VapB-mt24, and VapB-mt25, could interact with cognate or noncognate protein S-tagged toxins by Ni-NTA affinity chromatography (Fig. 3B). This was performed by first incubating the purified His-tagged antitoxin with Ni-NTA resin followed by addition of the purified protein S-tagged toxin (without a His-tag); after further incubation and washing to remove any noninteracting protein, the proteins retained on the Ni-NTA resin were visualized by SDS-PAGE followed by Coomassie staining. As expected, we observed interactions between the cognate toxin and antitoxin pairs (Fig. 3B, arrows to the left of the band in lane 3 (MazE-mt3 antitoxin with MazF-mt3 toxin), lane 9 (VapB-mt24 antitoxin with VapC-mt24 toxin), and lane 15 (VapB-mt25 antitoxin with VapC-mt25 toxin)). Notably, we also detected four noncognate toxin-antitoxin interactions (Fig. 3B, star to the left of the band in lane 4 (MazE-mt3 with VapC-mt24), lane 5 (MazE-mt3 with VapC-mt25), lane 8 (VapB-mt24 with MazF-mt3), and lane 10 (VapB-mt24 with VapC-mt25)).
We did not detect an interaction between PST-MazF-mt3 and (His)6VapB-mt25 (Fig. 3B, lane 13) even though expression of VapB-mt25 can neutralize the toxicity of MazF-mt3 in E. coli (Fig. 2B). The reason for this is unclear; however, it could be due to the instability of the protein complex in vitro or steric hindrance stemming from the presence of the protein S-tags and/or His-tags. Fig. 4 summarizes the data obtained in this study. In general, it appears that the in vivo rescue experiments enabled the detection of interactions that may not be as stable under nonphysiological conditions. Overall, our results clearly demonstrate that noncognate toxins and antitoxins are able to associate both in vivo and in vitro.
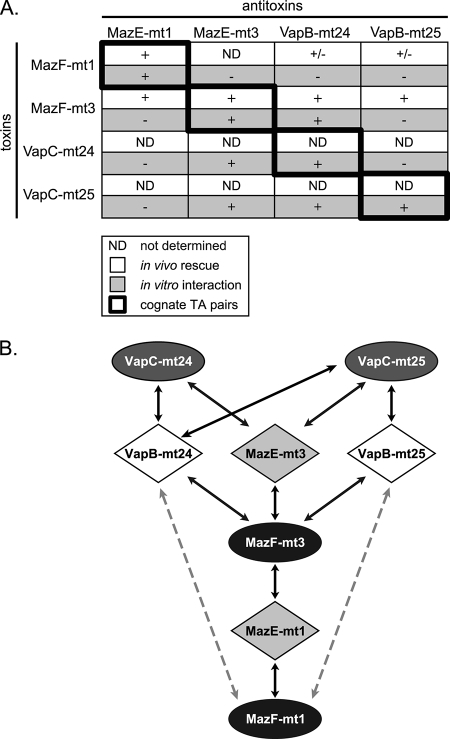
FIGURE 4.
Summary of toxin-antitoxin rescue and interaction data. A, key shown on bottom left. + for in vivo experiments denotes full rescue of toxicity on plates whereas ± denotes partial rescue of growth inhibition; for the in vitro interaction experiments a + denotes the identification of a detectable interaction (no indication of strong or weak interactions are noted). B, illustration of the data summarized in A. Colors correspond to those used in Fig. 1; toxins are represented by ovals and antitoxins by diamonds. Arrows at both ends indicate interaction between the proteins and/or inhibition of toxicity. Dotted gray line indicates a weaker interaction based on the partial rescue of toxicity.
MazF-mt3 and VapB-mt24 Are Up-regulated in Mtb Cells Exposed to Hypoxic Conditions
We mined published gene-profiling studies for supporting evidence that one or more of the in vitro and/or in vivo interactions that we discovered were physiologically relevant in Mtb. We were able to consistently identify only six of the eight Mtb protein-coding sequences related to our study (excluding MazE-mt1/Rv2801A and MazE-mt3/Rv1991A). These two antitoxin genes were among 82 newly identified protein-coding sequences (30) absent from the original annotation of the Mtb genome (31); consequently, they are not listed in many published microarray data sets.
Interestingly, we found that the steady-state levels of MazF-mt3 and VapB-mt24 mRNAs were elevated in Mtb cells subjected to gradual oxygen limitation (32) in a sealed, stirred culture according to the Wayne model (33, 34) (Table 1). This slow depletion enables Mtb cells to adapt and survive anaerobic conditions, thus modeling the transition of Mtb cells from active growth to the nonreplicating persistent (NRP) state characteristic of granulomas. Physiologically, NRP is divided into two stages: NRP1 and NRP2. Mtb cells enter NRP1 when the oxygen concentration reaches 1% of normal saturation (microaerophilic conditions), resulting in slow growth. Progression to NRP2 occurs when the oxygen concentration reaches 0.06% of normal saturation (anaerobic conditions) and growth ceases. The NRP state has many parallels to the dormant state caused by the action of TA toxins in E. coli.
TABLE 1.
Toxin and antitoxin mRNA expression levels from Muttucumaru et al. (32)
Gene profiling data derived from Mtb cells subjected to the Wayne model.
| Gene | Rv no. | NRP1 | NRP2 |
|---|---|---|---|
| mazE-mt1 | Rv2801A | NDa | ND |
| mazF-mt1 | Rv2801c | 0.86b | 1.09 |
| mazE-mt3 | Rv1991A | ND | ND |
| mazF-mt3 | Rv1991c | 1.60 | 1.62 |
| vapB-mt24 | Rv0599c | 1.54 | 1.44 |
| vapC-mt24 | Rv0598c | 0.52 | 0.88 |
| vapB-mt25 | Rv2595 | 0.54 | 0.57 |
| vapC-mt25 | Rv2596 | 0.55 | 0.59 |
a ND, not determined; this Rv number was not present in the dataset.
b Average fold changes in NRP1 and NRP2 compared to aerobic mid-log growth.
The ratio of transcript abundance in NRP1 or NRP2 cells relative to that for aerobically grown cells is shown in Table 1 (32). Only the noncognate MazF-mt3 and VapB-mt24 pair exhibited a clear increase in steady-state mRNA levels (ratios of ∼1.5 for each); this level was essentially the same at both NRP1 and NRP2 stages. Interestingly, this is the same pair for which we demonstrated both an in vitro and in vivo interaction (Fig. 4A). We were unable to determine whether the transcripts corresponding to the cognate TA pair MazF-mt3/MazE-mt3 were also elevated because the Rv number for MazE-mt3 was not listed in this dataset.
Activation of toxins is sometimes thought to occur by a combination of the degradation of the antitoxin by a protease activated by host cell stress followed by an increase in transcription of the TA operon. This would result in a net higher concentration of toxin in the cell compared with operons whose transcription rate did not increase. In the TA field, these data implicate the TA pair showing a relative increase in steady-state transcript abundance in having a hand in toxin-mediated growth modulation.
DISCUSSION
A general scheme for how TA systems are regulated has emerged from methodical studies of E. coli and bacteriophage TA systems, especially for the mazEF (6, 35, 36) and phd-doc (37–43) operons. The adjacent gene pairs encoding the toxin and its cognate antitoxin are contained within specialized operons. The toxin and antitoxin form a stable protein complex; however, the antitoxin is unstable relative to the toxin protein because it is susceptible to cleavage by one of the cellular proteases. The operon is autoregulated; both the antitoxin alone and the TA complex repress transcription upon binding to the palindrome upstream of the module. Antitoxin instability and operon autoregulation are key features of this very dynamic system. When stress conditions lead to activation of cellular proteases, the levels of antitoxin decrease, leading to a concomitant decrease in the concentration of both repressors (antitoxin only and the TA complex) of transcription. Therefore, protease degradation of the antitoxin now leads to diminished levels of antitoxin but a relative increase in module transcription because the antitoxin repressors are in short supply. These tandem events result in an excess of toxin. Consequently, any free toxin will act on its target (e.g. ACA sequences in mRNA for E. coli MazF) leading to transient growth arrest or eventual cell death if antitoxin synthesis does not resume within a window of time (18, 19).
The physiological consequences of this dynamic feature of TA systems have not been studied in cells such as Mtb that contain multiple family members. As with E. coli, Mtb possesses multiple proteases (including the ClpXP protease) (31) that likely influence antitoxin stability. However, its genome does not contain genes encoding an apparent lon protease or ClpA ATP-dependent subunit to associate with the ClpP proteolytic subunit (31). The role of Mtb proteases in regulating toxin activity has not yet been investigated. Also, the physiological triggers of antitoxin degradation (which precedes toxin activation) are not known.
Because the E. coli genome does not possess multiple toxins within a single family, as Mtb does, the general assumption in the field has been that each distinct toxin can pair with only one antitoxin. In this work, we presented data that challenges the one toxin-one antitoxin paradigm. It is not clear whether the networking of toxins and antitoxins exists in other bacteria or is unique to Mtb. The existing data suggest that the concerted action of these MazF and VapC toxins may facilitate adaptation to the environmental conditions encountered during Mtb infection.
Our data serve as a foundation for future studies on these TA systems in their natural host. It will be important to gain a deeper understanding of how the concerted action of this broad spectrum of TA systems manifests in vivo with respect to Mtb growth rate regulation, latency, and pathogenicity.
Acknowledgments
We thank Eric Rubin and Jeff Murry for valuable discussions, Jason Schifano for input on the manuscript, and Nancy Connell and Robert Husson for M. tuberculosis H37Rv genomic DNA.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01GM081567 (to M. I.). This work was also supported by Takara-Bio, Inc. (to M. I. and N. A. W.) and by National Institutes of Health T32 Training Grant AI07403, Virus-Host Interactions in Eukaryotic Cells from the NIAID to J. D. S. (awarded to S. Pestka).
- Mtb
- Mycobacterium tuberculosis
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- NRP
- nonreplicating persistent
- TA
- toxin-antitoxin
- TF
- trigger factor.
REFERENCES
- 1.Boshoff H. I., Barry C. E., 3rd (2005) Nat. Rev. Microbiol. 3, 70–80 [DOI] [PubMed] [Google Scholar]
- 2.Flynn J. L., Chan J. (2005) Trends Microbiol. 13, 98–102 [DOI] [PubMed] [Google Scholar]
- 3.Buts L., Lah J., Dao-Thi M. H., Wyns L., Loris R. (2005) Trends Biochem. Sci. 30, 672–679 [DOI] [PubMed] [Google Scholar]
- 4.Pandey D. P., Gerdes K. (2005) Nucleic Acids Res. 33, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerdes K., Christensen S. K., Løbner-Olesen A. (2005) Nat. Rev. Microbiol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 6.Aizenman E., Engelberg-Kulka H., Glaser G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6059–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki M., Zhang J., Liu M., Woychik N. A., Inouye M. (2005) Mol. Cell 18, 253–261 [DOI] [PubMed] [Google Scholar]
- 8.Falla T. J., Chopra I. (1998) Antimicrob. Agents Chemother. 42, 3282–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keren I., Shah D., Spoering A., Kaldalu N., Lewis K. (2004) J. Bacteriol. 186, 8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korch S. B., Hill T. M. (2006) J. Bacteriol. 188, 3826–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vázquez-Laslop N., Lee H., Neyfakh A. A. (2006) J. Bacteriol. 188, 3494–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Höner zu Bentrup K., Russell D. G. (2001) Trends Microbiol. 9, 597–605 [DOI] [PubMed] [Google Scholar]
- 13.Manabe Y. C., Bishai W. R. (2000) Nat. Med. 6, 1327–1329 [DOI] [PubMed] [Google Scholar]
- 14.Stewart G. R., Robertson B. D., Young D. B. (2003) Nat. Rev. Microbiol. 1, 97–105 [DOI] [PubMed] [Google Scholar]
- 15.Pedersen K., Christensen S. K., Gerdes K. (2002) Mol. Microbiol. 45, 501–510 [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Zhang J., Hara H., Kato I., Inouye M. (2005) J. Biol. Chem. 280, 3143–3150 [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Zhang J., Hoeflich K. P., Ikura M., Qing G., Inouye M. (2003) Mol. Cell 12, 913–923 [DOI] [PubMed] [Google Scholar]
- 18.Amitai S., Yassin Y., Engelberg-Kulka H. (2004) J. Bacteriol. 186, 8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolodkin-Gal I., Hazan R., Gaathon A., Carmeli S., Engelberg-Kulka H. (2007) Science 318, 652–655 [DOI] [PubMed] [Google Scholar]
- 20.Zhu L., Phadtare S., Nariya H., Ouyang M., Husson R. N., Inouye M. (2008) Mol. Microbiol. 69, 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu L., Zhang Y., Teh J. S., Zhang J., Connell N., Rubin H., Inouye M. (2006) J. Biol. Chem. 281, 18638–18643 [DOI] [PubMed] [Google Scholar]
- 22.Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura K., Inouye M. (1982) EMBO J. 1, 771–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakano E. T., Rao M. M., Perucho M., Inouye M. (1987) J. Virol. 61, 302–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi H., Yoshida T., Inouye M. (2009) Appl. Environ. Microbiol. 75, 5356–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arcus V. L., Rainey P. B., Turner S. J. (2005) Trends Microbiol. 13, 360–365 [DOI] [PubMed] [Google Scholar]
- 27.Qing G., Ma L. C., Khorchid A., Swapna G. V., Mal T. K., Takayama M. M., Xia B., Phadtare S., Ke H., Acton T., Montelione G. T., Ikura M., Inouye M. (2004) Nat. Biotechnol. 22, 877–882 [DOI] [PubMed] [Google Scholar]
- 28.Inouye M., Inouye S., Zusman D. R. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inouye S., Harada W., Zusman D., Inouye M. (1981) J. Bacteriol. 148, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camus J. C., Pryor M. J., Médigue C., Cole S. T. (2002) Microbiology 148, 2967–2973 [DOI] [PubMed] [Google Scholar]
- 31.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 32.Muttucumaru D. G., Roberts G., Hinds J., Stabler R. A., Parish T. (2004) Tuberculosis 84, 239–246 [DOI] [PubMed] [Google Scholar]
- 33.Wayne L. G., Hayes L. G. (1996) Infect. Immun. 64, 2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wayne L. G., Sohaskey C. D. (2001) Annu. Rev. Microbiol. 55, 139–163 [DOI] [PubMed] [Google Scholar]
- 35.Marianovsky I., Aizenman E., Engelberg-Kulka H., Glaser G. (2001) J. Biol. Chem. 276, 5975–5984 [DOI] [PubMed] [Google Scholar]
- 36.Zhang J., Zhang Y., Inouye M. (2003) J. Biol. Chem. 278, 32300–32306 [DOI] [PubMed] [Google Scholar]
- 37.Gazit E., Sauer R. T. (1999) J. Biol. Chem. 274, 16813–16818 [DOI] [PubMed] [Google Scholar]
- 38.Gazit E., Sauer R. T. (1999) J. Biol. Chem. 274, 2652–2657 [DOI] [PubMed] [Google Scholar]
- 39.Lehnherr H., Yarmolinsky M. B. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3274–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magnuson R., Lehnherr H., Mukhopadhyay G., Yarmolinsky M. B. (1996) J. Biol. Chem. 271, 18705–18710 [DOI] [PubMed] [Google Scholar]
- 41.Magnuson R., Yarmolinsky M. B. (1998) J. Bacteriol. 180, 6342–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinley J. E., Magnuson R. D. (2005) J. Bacteriol. 187, 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith J. A., Magnuson R. D. (2004) J. Bacteriol. 186, 2692–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattison K., Wilbur J. S., So M., Brennan R. G. (2006) J. Biol. Chem. 281, 37942–37951 [DOI] [PubMed] [Google Scholar]
- 45.Arcus V. L., Bäckbro K., Roos A., Daniel E. L., Baker E. N. (2004) J. Biol. Chem. 279, 16471–16478 [DOI] [PubMed] [Google Scholar]