Abstract
The Syk protein-tyrosine kinase is phosphorylated on multiple tyrosines after the aggregation of the B cell antigen receptor. However, metabolic labeling experiments indicate that Syk is inducibly phosphorylated to an even greater extent on serine after receptor ligation. A combination of phosphopeptide mapping and mass spectrometric analyses indicates that serine 291 is a major site of phosphorylation. Serine 291 lies within a 23-amino acid insert located within the linker B region that distinguishes Syk from SykB and Zap-70. The phosphorylation of serine-291 by protein kinase C enhances the ability of Syk to couple the antigen receptor to the activation of the transcription factors NFAT and Elk-1. Protein interaction studies indicate a role for the phosphorylated linker insert in promoting an interaction between Syk and the chaperone protein, prohibitin.
Keywords: Lymphocyte, Protein Kinase C (PKC), Serine Threonine Protein Kinase, Signal Transduction, Tyrosine-protein Kinase (Tyrosine Kinase), Prohibitin, Syk
Introduction
The Syk protein-tyrosine kinase is an essential component of the protein machinery that couples receptors bearing immunoreceptor tyrosine-based activation motifs to intracellular signal transduction pathways in hematopoietic cells (for a recent review, see Ref. 1). Syk is recruited via its N-terminal pair of Src homology 2 domains to a pair of tyrosines located in the cytoplasmic immunoreceptor tyrosine-based activation motifs of immune recognition receptors that become phosphorylated after ligand-induced aggregation. For example, clustering of the B cell receptor for antigen (BCR)4 leads to the phosphorylation of the accessory molecules CD79a and CD79b, resulting in the recruitment of Syk to the clustered BCR complex. Receptor binding leads to the activation of Syk and to its phosphorylation on tyrosines that regulate its activity and interactions with downstream effectors that bear Src homology 2 or other phosphotyrosine binding domains. This in turn couples the BCR to the activation of, among others, the phospholipase γ/PKC, PI3K/Akt, and Ras/Raf/Erk signaling pathways. In the absence of Syk, few if any signals are sent after BCR clustering (2–4).
Syk contains a stretch of 23 amino acids known as the “linker insert” that lies within the linker B region that separates the tandem pair of Src homology 2 domains from the C-terminal catalytic domain. The presence of this insert distinguishes Syk from SykB, a shorter alternatively spliced isoform, and from ZAP-70, the Syk paralog found in T cells and natural killer cells (5, 6). The role of the linker insert in modulating the properties of Syk is poorly understood, but two functions have been proposed. In one, the linker insert harbors a signal sequence that allows the kinase to transit into the nucleus (7). In another, the insert enhances the ability of the kinase to interact with immunoreceptor tyrosine-based activation motif-bearing receptors (8). Within the linker insert (TWSPGGIISRIKSYSFPKPGHKK) are several potential sites of phosphorylation. The tyrosine (Tyr-290 in the murine Syk sequence) is a site of autophosphorylation in vitro (9) but to our knowledge is not known to be phosphorylated in response to receptor engagement. One study that analyzed phosphopeptides derived from proteins isolated from lung cancer cells did identify Tyr-290 as a site of phosphorylation (10). However, the replacement of Tyr-290 with phenylalanine is without effect on the ability of Syk to reconstitute signaling from either FcϵRI or the T cell antigen receptor (8). In this study we examined in more detail the role of phosphorylation within the linker insert in modulating the participation of Syk in immune cell signaling. Although the substitution of Tyr-290 with phenylalanine did not affect the ability of Syk to mediate BCR-stimulated signaling, it did alter the subcellular localization of the kinase by creating within the linker insert a nuclear export signal. We did find that Syk is phosphorylated within the linker insert after BCR engagement, but it was Ser-291 that was the major site of phosphorylation. Phosphorylation at this site, catalyzed by protein kinase C (PKC), enhanced the ability of Syk to couple the BCR to signaling pathways leading to the activation of NFAT and Elk1. Protein and peptide interaction studies indicated that the phosphorylated linker insert enhanced the ability of Syk to interact with the chaperone protein, prohibitin-1 (PHB1).
EXPERIMENTAL PROCEDURES
Cells and Cell Lines
DG75, U937, and DT40 cells were cultured in RPMI 1640 medium supplemented with 7.5% fetal calf serum, 50 μm 2-mercaptoethanol, 1 mm sodium pyruvate, 100 IU/ml penicillin G, and 100 μg/ml streptomycin. The culture media for DT40 cells also contained 5% chicken serum. PKD-deficient DT40 cells were a kind gift of Dr. Sharon Matthews (University of Dundee). B cells were enriched from murine spleens as described (11). Plasmids for the expression of murine Syk tagged at the C terminus with either a Myc-epitope or enhanced green fluorescent protein (EGFP) were as described previously (12, 13). Expression plasmids for Syk mutants with Ser-291 replaced by alanine or aspartic acid were generated by site-directed mutagenesis using either the Transformer (Clontech) or the QuikChange (Stratagene) kits. For the generation of stably transfected cells, Syk-deficient DT40 cells (2) were co-transfected by electroporation (300 V, 330 microfarads; Cell-Porator, Invitrogen) with plasmids (10 μg) coding for the expression of the indicated Syk mutant along with 1 μg of pBabePuro, a vector encoding a puromycin resistance gene. Cells were selected in media supplemented with puromycin (0.5 μg/ml).
Cellular Activation Assays
Stable or transiently transfected DT40 cells were transfected with an NFAT-luciferase reporter plasmid (pNFAT Luc (Stratagene)) or the Elk-1-GAL4 and GAL4-luciferase plasmids supplied with the Pathfinder kit from Stratagene. Cells were harvested 24 h post-transfection and plated at a density of 1 × 106/ml in chicken serum-free media. Cells were stimulated with goat anti-chicken IgM (Rockland) in the amounts indicated or with a mixture of PMA (50 ng/ml) and ionomycin (1.0 μm) at 37 °C. Luciferase activity was measured 6 h later using the luciferase assay system (Promega). The luciferase activity is reported as a ratio of the luciferase activity observed under the experimental conditions divided by the activity measured in cells treated with PMA plus ionomycin, which bypasses the need for receptor engagement. Syk expression levels were determined by Western blotting (anti-Syk N-19, Santa Cruz Biotechnology).
For the measurement of Ras activity, a fusion protein of GST and the Ras binding domain of Raf-1 (GST-RBD) expressed in bacteria was adsorbed to glutathione-Sepharose. DT40 cells expressing various forms of Syk and activated as indicated were lysed in buffer containing 25 mm HEPES, pH 7.2, 150 mm NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 10% glycerol, 10 mm NaF, 10 mm MgCl2, 1 mm EDTA, 1 mm sodium orthovanadate, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. Supernatants from a 5-min centrifugation at 8000 × g were incubated with immobilized GST-RBD for 30 min at 4 °C. Bound proteins were separated by SDS-PAGE and analyzed by Western blotting with antibodies to Ras (Ab-3, Calbiochem).
Metabolic Labeling and Phosphopeptide Mapping
DT40 cells (2 × 106 cells, 4 × 106/ml) were incubated in phosphate-free RPMI 1640 media for 2 h at 37 °C and then for an additional 3 h after the addition of 5 mCi of [32P]orthophosphate (PerkinElmer Life Sciences). Where indicated, cells were treated with the PKC inhibitor GÖ6976 (Calbiochem) dissolved in DMSO or with DMSO alone for 20 min and then stimulated with either anti-IgM (10 μg/ml) or PMA (100 ng/ml) for the times indicated. Cells were lysed in buffer containing 10 mm Tris/HCl, pH 7.2, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mm sodium orthovanadate, 5 mm p-nitrophenyl phosphate, 2 mm sodium fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin on ice for 15 min. The lysates were clarified by centrifugation at 18,000 × g for 5 min. The supernatants were mixed at 4 °C for 15 min with 2 μl of anti-EGFP rabbit sera (Clontech). Immune complexes were adsorbed to protein A-Sepharose, washed 4 times with lysis buffer, and then separated by SDS-PAGE. The proteins were then transferred to nitrocellulose membranes and detected by autoradiography. The band corresponding to phosphorylated Syk was excised from the membrane and digested with trypsin. The resulting phosphopeptides were separated by electrophoresis on alkaline 40% polyacrylamide gels (12) and detected by either autoradiography or with phosphorimaging.
For the analysis of phosphoamino acids, phosphopeptides were recovered from the polyacrylamide gels by extraction in water and hydrolyzed for 1 h at 110 °C in 6 n HCl. Phosphoamino acids were separated on Whatman cellulose thin-layer plates at pH 3.5 for 50 min at 1100 V in the presence of standards of phosphotyrosine, phosphothreonine, and phosphoserine and detected by autoradiography.
Fluorescence Microscopy
DT40 cells stably expressing Syk-EGFP or Syk-EGFP(S291A) were harvested and adhered to poly-l-lysine-coated coverslips. The cells were fixed with 3.7% formaldehyde in PBS for 10 min and then stained with 0.1 μg/ml 4′,6-diamidino-2-phenyl-indole (DAPI, Sigma) for 10 min. In some experiments cells were treated with PMA (100 ng/ml) for 90 min or leptomycin B (LMB) (50 ng/ml) for 2 h before fixation.
In Vitro Kinase Assays
For PKD kinase assays, Syk-deficient or PKD-deficient DT40 cells (5 × 107 cells/ml) were treated with 100 ng/ml PMA for 5 min at 37 °C and lysed by incubation in buffer containing 1% Nonidet P-40, 25 mm HEPES, pH 7.2, 150 mm NaCl, 5 mm EDTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm sodium vanadate, and 2 mm NaF for 15 min on ice. The lysates were cleared by centrifugation at 18,000 × g for 5 min. Anti-PKD (Santa Cruz Biotechnology, SC-935) immune complexes were isolated and incubated in a kinase reaction buffer containing 50 mm HEPES, pH 7.2, 10 mm NaF, 10 mm MgCl2, 5 μm ATP, 20 μCi of [γ-32P]ATP, and 25 μm concentrations of either Syntide (PLARTLSVAGLPGKK, MP Biomedicals) or biotin-Ser-291 peptide (biotin-GIISRIKSYSFPKPGHK) (Antagene). Phosphopeptides were isolated using P81 phosphocellulose paper and quantified by liquid scintillation spectrometry (14). Relative counts incorporated into peptide were corrected for background counts incorporated in the absence of PKD (i.e. immune complexes isolated from PKD-deficient cells).
For Syk kinase assays, DT40 cells stably expressing Syk-EGFP or Syk-EGFP(S291A) (1 × 106) were treated where indicated with 100 ng/ml PMA or DMSO vehicle control for 15 min at 37 °C. Cells were harvested and lysed as described above. Anti-GFP immune complexes were incubated in kinase reaction buffer containing 50 mm HEPES, pH 7.2, 10 mm NaF, 10 mm MnCl2, 0.20 mg/ml cytosolic domain of erythrocyte band 3 (cdb3), 10 μm ATP, and 30 μCi [γ-32P]ATP. The cdb3 was a generous gift of Dr. Philip Low (Purdue University). Reactions were terminated by the addition of SDS sample buffer. Conditions were established under which substrate phosphorylation was linear with respect to both time and enzyme concentration. The proteins were separated by SDS-PAGE, transferred to a PVDF membrane, and exposed to x-ray film. Protein bands corresponding to cdb3 were excised and analyzed by scintillation spectrometry.
Peptide Interaction Assays
For peptide pulldown assays, 0.5 μmol of one of two N-terminal biotinylated peptides (GIISRIKSYSFPKPGHK and GIISRIKSYpSFPKPGHK, Antagene) were added to lysates prepared from DG75 cells lysed in 1% Nonidet P-40, 25 mm HEPES, pH 7.2, 150 mm NaCl, 5 mm EDTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 100 mm sodium vanadate, and 500 mm NaF. After incubation at 4 °C for 1 h, peptides were recovered on streptavidin-agarose beads. Bound proteins were separated by SDS-PAGE and analyzed by Western blotting or by MS/MS (for detailed methods, see the supplemental Experimental Procedures). A cDNA coding for human PHB1 obtained from Origene was amplified by PCR and subcloned into the pGEX 4-T-2 plasmid for the bacterial expression of GST-PHB1. The bacterial cells were harvested and lysed by sonication. Lysates were incubated with the biotin-tagged peptides and recovered on streptavidin-agarose beads. Bound proteins were separated by SDS-PAGE and examined by Western blotting using a PHB1 (H-80 Santa Cruz) antibody.
Protein Interaction Assays
DT40 cells stably expressing Syk-EGFP or Syk-EGFP(S291A) and treated with anti-IgM (5 μg/ml), PMA (100 ng/ml), or DMSO carrier as indicated were lysed in 1% Nonidet P-40, 25 mm HEPES, pH 7.2, 150 mm NaCl, 5 mm EDTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm sodium vanadate, and 2 mm NaF for 15 min on ice. Lysates were adsorbed to GST or GST-14-3-3ζ or GST-14-3-3τ expressed in bacteria and bound to glutathione-Sepharose. Bound proteins were separated by SDS-PAGE and detected by Western blotting using the Syk antibody N-19 (Santa Cruz Biotechnology). Alternatively, lysates from DG75, U937, or resting spleen B cells pretreated with or without PMA (100 ng/ml) or piceatannol (20 μg/ml) were incubated with antibodies against PHB1 and then adsorbed onto protein A-Sepharose beads. Bound proteins were separated by SDS-PAGE and analyzed by Western blotting using antibodies against PHB1 and Syk.
RESULTS
Replacement of Tyr-290 with Phenylalanine
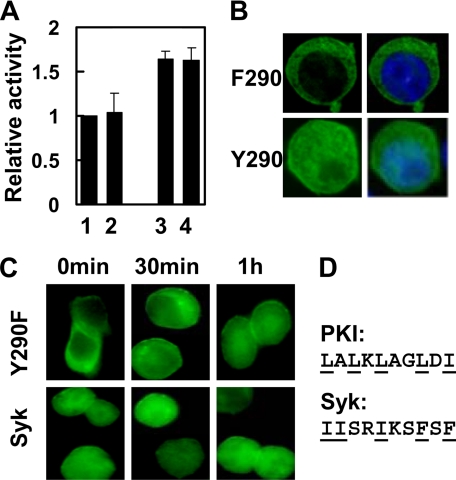
The replacement of Tyr-290 with phenylalanine was reported not to affect the ability of Syk to signal downstream of either FcϵRI or the T cell antigen receptor (8). To determine whether this was also true for the BCR, we expressed in Syk-deficient DT40 B cells Myc-tagged forms of wild-type Syk or a mutant in which Tyr-290 was replaced by phenylalanine (Syk(Y290F)). The ability of each to restore BCR-dependent signaling to Syk-deficient cells was measured by monitoring the activation of NFAT, a transcription factor regulated through changes in intracellular calcium. As shown in Fig. 1A, the replacement of Tyr-290 with phenylalanine had no obvious effect on the Syk ability to couple the BCR to the activation of NFAT as visualized using an NFAT-driven luciferase reporter plasmid. Similar results were seen when individual tyrosines within linker B were replaced with phenylalanines in combination with the Tyr-290 to phenylalanine substitution as shown in Fig. 1A for the Syk(Y317F) mutant. The Y290F substitution also failed to alter the signaling capability of Syk(Y342F), Syk(Y346F), or Syk(Y342F/Y346F) mutants (data not shown).
FIGURE 1.
Role of Tyr-290 in the location and activity of Syk. A, Syk-deficient DT40 cells were co-transfected with plasmids coding for either Myc-epitope tagged Syk (lane 1), Syk(Y290F) (lane 2), Syk(Y317F) (lane 3), or Syk(Y290F/Y317F) (lane 4) along with an NFAT-driven luciferase reporter plasmid stimulated with anti-IgM antibodies or a mixture of PMA and ionomycin and assayed for the expression of luciferase. The values reported indicate the activity produced by anti-IgM treatment divided by the activity produced in response to PMA + ionomycin to correct for differences in transfection efficiency and are normalized to a value of 1.0 for cells expressing wild-type Syk. Results represent the mean and S.E. from three experiments. B, Syk-deficient DT40 cells were transfected with plasmids coding for Syk-EGFP(Y290F) (F290) or Syk-EGFP(Y317F/Y342F/Y346F/Y358F) (Y290), fixed, stained with DAPI, and examined by fluorescence microscopy. Nuclear exclusion was observed in all cells expressing Syk-EGFP(Y290F). C, Syk-deficient cells expressing Syk-EGFP (Syk) or Syk-EGFP(Y290F) (Y290F) were treated with LMB for the times indicated and examined by fluorescence microscopy. D, alignment of the amino acid sequence of the NES of protein kinase I (PKI) with the linker insert from Syk(Y290F) is shown.
The linker insert has been reported to harbor a nuclear localization signal that regulates the trafficking of the kinase into the nucleus in breast epithelial cells (7). To determine whether the phosphorylation of Tyr-290 might play a role in regulating the transit of Syk into or out of the nucleus in B cells, we prepared plasmids for the expression of Syk or Syk(Y290F), both with EGFP tags at their C termini. The localization of each was then examined by fluorescence microscopy in transiently transfected, Syk-deficient DT40 cells. As shown previously, Syk-EGFP is localized throughout the cell in both the nucleus and cytoplasm (13). However, Syk-EGFP(Y290F) was restricted exclusively to the cytoplasm (Fig. 1B). To determine whether this effect was specific to a change at this particular tyrosine, we generated a mutant of Syk-EGFP in which all of the identified sites of tyrosine phosphorylation in linker B (Tyr-317, Tyr-342, Tyr-346, and Tyr-358) (9) with the exception of Tyr-290 were replaced by phenylalanines. When expressed in Syk-deficient DT40 cells, this mutant was evenly distributed throughout the cell (Fig. 1B) in a pattern similar to that of wild-type Syk (13). Thus, Tyr-290 was the only site of tyrosine phosphorylation in linker B whose replacement with phenylalanine resulted in the exclusion of Syk from the nucleus.
We reasoned that the substitution of Tyr-290 with phenylalanine could have inactivated a nuclear localization signal located within the linker insert. However, in our hands, removal of the entire linker insert does not block the transit of the kinase into the nucleus, and nuclear import instead requires a region near the C terminus of linker B (15). A second possibility that we considered was that the substitution of Tyr-290 had instead generated a nuclear export signal (NES) that restricted the location of Syk-EGFP(Y290F) to the cytoplasm. To examine this possibility, we treated cells expressing Syk-EGFP(Y290F) with LMB. LMB inhibits exportin1/Crm1, the karyopherin that recognizes cargo containing leucine-rich NES sequences. Treatment with LMB resulted in a relocalization of Syk-EGFP(Y290F) back into the nucleus, resulting in a final distribution pattern comparable with that of Syk-EGFP (Fig. 1C). As expected, the distribution of Syk-EGFP was not affected by LMB (Ref. 14 and Fig. 1C). This result suggested that the Y290F mutation had created an NES within the linker insert that resulted in the export of the kinase from the nucleus. A comparison of the sequence of the mutated linker insert with that of the well characterized NES of protein kinase inhibitor (16) revealed a similar distribution of hydrophobic amino acids, suggesting that this is a reasonable explanation (Fig. 1D). Because the Y290F substitution has no obvious effect on receptor-mediated signaling, the inability of Syk to reside within the nucleus does not appear to contribute to the BCR-mediated activation of NFAT.
Phosphorylation of Syk on Serine
The linker insert contains three serines that are potential sites of phosphorylation. Of these, Ser-291 is present within a sequence that matches closely the generalized consensus sequence of (R/K)1–3(X)0–2S/T(X)0–2(R/K)1–3 characteristic of substrates of PKC (17). To examine possible phosphorylation in this region, we incubated Syk-deficient DT40 cells stably expressing Syk-EGFP in media containing [32P]orthophosphate to label the intracellular pool of ATP. Cells were then left untreated or were treated with PMA, an activator of PKC. Syk-EGFP was recovered by immunoprecipitation from cell lysates and examined by autoradiography after separation by SDS-PAGE. The overall extent of phosphorylation of Syk-EGFP was increased after the treatment of cells with PMA (Fig. 2A).
FIGURE 2.
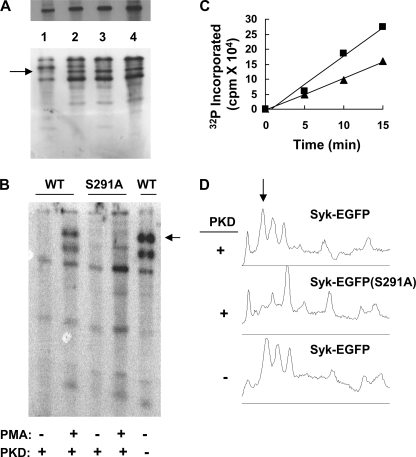
Phosphorylation of Syk on serine. A, Syk-deficient DT40 cells stably expressing Syk-EGFP were preincubated with [32P]orthophosphate and then treated with DMSO carrier alone (DMSO) or 100 ng/ml PMA (PMA). Anti-GFP immune complexes were isolated, separated by SDS-PAGE, and subjected to autoradiography. B, the 32P-labeled, EGFP-tagged Syk molecules shown in panel A were digested with trypsin. Phosphopeptides were separated by alkaline gel electrophoresis and detected by autoradiography. The major phosphopeptides are labeled 1–4. C, the tryptic phosphopeptides illustrated in panel B were excised and compared for extent of phosphorylation by scintillation spectrometry. D, phosphopeptides 1–4 were hydrolyzed and analyzed for phosphoamino acid content by thin-layer electrophoresis. The migration positions of phosphoserine (pSer), phosphothreonine (pThr), and phosphotyrosine (pTyr) are indicated.
To investigate possible phosphorylation within the linker insert, we first used a phosphopeptide mapping approach. Radiolabeled Syk-EGFP was transferred to a nitrocellulose membrane, excised, and treated with trypsin. The resulting phosphopeptides were then separated on an alkaline 40% polyacrylamide gel, which separates peptides on the basis of charge and size (12). Exposure of the gel to x-ray film revealed 4 major phosphopeptides (Fig. 2B). Individual phosphopeptides were recovered, and the relative extents of phosphate incorporation were quantified by scintillation counting (Fig. 2C). Treatment with PMA led to an increase in the amount of phosphate incorporated, primarily into peptide-2. This was of special interest as the relative migration position of peptide-2 on the 40% gel is the same as that of a phosphopeptide containing pTyr-290 as demonstrated previously (12). This suggested that peptide-2 could also be derived from the linker insert.
To determine which amino acid was phosphorylated, we recovered the phosphopeptides from the gels, hydrolyzed them in 6 n HCl, and separated the resulting phosphoamino acids by thin-layer electrophoresis. Phosphoserine was the only phosphoamino acid detected in all four phosphopeptides including peptide-2 (Fig. 2D). This suggested that either Ser-289 or Ser-291 had become phosphorylated in PMA-treated cells. To determine which of these was phosphorylated, we generated Syk-deficient DT40 cells that expressed Syk-EGFP(S289A), Syk-EGFP(S291A), or Syk-EGFP(S289A/S291A). The fusion proteins were recovered from the lysates of [32P]orthophosphate-treated cells and subjected to phosphopeptide mapping. Analysis of the resulting maps indicated that the replacement of Ser-291, but not Ser-289, selectively blocked the appearance of peptide-2 (Fig. 3A). Similarly, we compared phosphopeptide maps generated from Syk-EGFP and Syk-EGFP(S291A) isolated from metabolically labeled and PMA-treated cells. Again, the replacement of Ser-291 with alanine resulted in the loss of the PMA-enhanced peptide-2 (Fig. 3, B and D). As a final confirmation, we isolated Syk-EGFP from PMA-treated cells, digested the kinase with trypsin, and analyzed the resulting peptides by mass spectrometry. The major serine-phosphorylated peptide that was present in the protein digest corresponded to peptide-2 phosphorylated exclusively at position 291 (supplemental Fig. 1). Thus, after the treatment of cells with PMA, Syk clearly is phosphorylated in the linker insert region on Ser-291.
FIGURE 3.
Phosphorylation of Syk on Ser-291. A, anti-GFP immune complexes prepared from the lysates of Syk-deficient DT40 cells stably expressing Syk-EGFP (lane 1), Syk-EGFP(S289A) (lane 2), Syk-EGFP(S291A) (lane 3), or Syk-EGFP(S289A/S291A) (lane 4) and preincubated in [32P]orthophosphate were separated by SDS-PAGE and detected by autoradiography (upper panel). Tryptic phosphopeptides were generated from each Syk mutant, separated on alkaline gels, and detected by autoradiography (lower panel). The arrow marks the position of the phosphopeptide containing Ser-291 and Ser-289. B, anti-GFP immune complexes prepared from the lysates of Syk-deficient DT40 cells expressing Syk-EGFP or Syk-EGFP(S291A) or of PKD-deficient DT40 cells expressing Syk-EGFP that had been preincubated in [32P]orthophosphate and treated without (−) or with (+) PMA were separated by SDS-PAGE, isolated, and treated with trypsin. Tryptic phosphopeptides were separated on alkaline gels and detected using phosphorimaging. The arrow marks the position of the phosphopeptide containing Ser-291 and Ser-289. C, shown is the incorporation of 32P-phosphate into Syntide (■) or the Ser-291 peptide (▴) catalyzed by anti-PKD immune complexes isolated from Syk-deficient DT40 cells. Counts incorporated into peptide were corrected for counts incorporated using anti-PKD immune complexes isolated from PKD-deficient DT40 cells. D, the lanes in panel B representing phosphopeptides generated from Syk-EGFP isolated from PMA-treated cells were scanned using NIH image software. The arrow marks the position of the phosphopeptide containing Ser-291 and Ser-289.
Phosphorylation of Ser-291 by PKC
PKD/PKC-μ has been reported to associate with Syk and to phosphorylate it within linker B (18), making it an attractive candidate for the Ser-291 kinase. To examine if Ser-291 was a potential site of phosphorylation for PKD, we conducted an in vitro kinase assay using a peptide substrate with a sequence identical to that surrounding Ser-291. The phosphorylation of the Ser-291-containing peptide was compared with that of Syntide-2, a known peptide substrate for PKD (19). Both peptides were incubated in kinase reaction buffer containing [γ-32P]ATP, using as catalysts anti-PKD immune complexes prepared by incubating anti-PKD antibodies with lysates of either Syk-deficient or PKD-deficient DT40 cells. The anti-PKD immune complexes prepared from Syk-deficient cells, but not those from PKD-deficient cells, catalyzed the phosphorylation of both Syntide-2 and the Ser-291-containing peptide (Fig. 3C). The rate of phosphorylation of Syntide-2 was ∼1.7-fold higher than the rate of phosphorylation of the Ser-291-containing peptide. Thus, Ser-291 appeared to be a reasonable candidate as a substrate for PKD. We then asked if PKD was required for the phosphorylation of Syk in B cells. PKD-deficient DT40 cells that stably expressed Syk-EGFP were prepared, incubated in media containing [32P]orthophosphate, and then stimulated with PMA. Syk-EGFP was immunoprecipitated and subjected to phosphopeptide mapping (Fig. 3B). Cells lacking PKD did not demonstrate any obvious defect in the phosphorylation of peptide-2 (Fig. 3D). Thus, whereas we cannot rule out a role for PKD in the phosphorylation of Syk on Ser-291, it is clearly not essential, suggesting a role for other PKC family members.
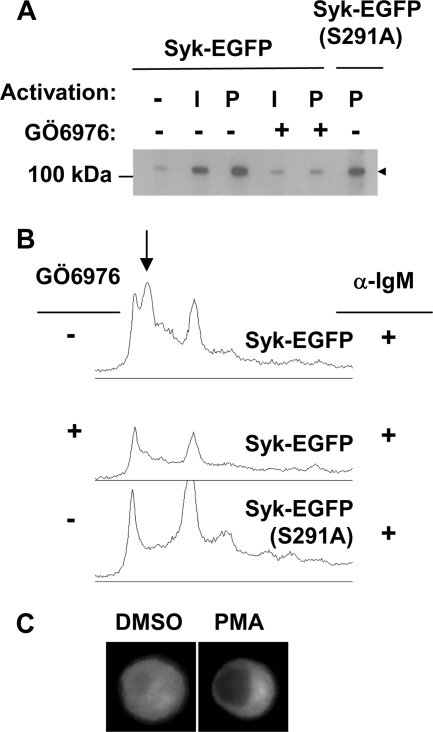
PKCs are activated in B cells after treatment with PMA or the cross-linking of surface IgM and can be inhibited by small molecule kinase inhibitors. To examine a role for one these enzymes, we preincubated Syk-EGFP-expressing DT40 B cells in media containing [32P]orthophosphate, pretreated the cells with or without the general PKC inhibitor GÖ6976 (20) for 10 min, and then stimulated them with either anti-IgM to cross-link the BCR or PMA. Syk-EGFP was immunoprecipitated from cell lysates and examined by autoradiography. The inclusion of the PKC inhibitor reduced both the anti-IgM and PMA-induced phosphorylation of Syk-EGFP (Fig. 4A). We analyzed by phosphopeptide mapping Syk-EGFP isolated from cells treated with anti-IgM in the presence or absence of the inhibitor and compared these to a map generated from Syk-EGFP(S291A). The treatment of cells with anti-IgM led to the phosphorylation of Syk on peptide-2, and this was blocked by the presence of the PKC inhibitor (Fig. 4B). These results are consistent with a role for a PKC family member in the phosphorylation of Syk on Ser-291 after BCR cross-linking.
FIGURE 4.
Phosphorylation of Ser-291 on Syk is inhibited by a PKC inhibitor. A, Syk-EGFP or Syk-EGFP(S291A) were immunoprecipitated from metabolically labeled DT40 cell lines that had been treated with either DMSO (−) or the PKC inhibitor GÖ6976 (+) and activated with goat anti-chicken IgM (I) or PMA (P). Proteins were separated by SDS-PAGE and detected by autoradiography. B, phosphopeptide maps were generated from Syk-EGFP or Syk-EGFP(S291A) isolated from anti-IgM-activated cells untreated (−) or treated (+) with GÖ6976. The arrow marks the position of the phosphopeptide containing Ser-291 and Ser-289. C, cells expressing Syk-EGFP(S291A) were treated with DMSO or PMA for 90 min and examined by fluorescence microscopy.
We then asked if the phosphorylation of Syk on Ser-291 affected its intracellular location. The treatment of B cells with PMA leads to the transient exclusion of Syk-EGFP from the nucleus (15). Consequently, we looked for a defect in this PMA-stimulated nuclear exclusion when Ser-291 was replaced by alanine. Syk-deficient DT40 cells expressing Syk-EGFP(S291A) were stimulated with 100 ng/ml PMA or DMSO vehicle control. Cells were fixed, stained with DAPI, and examined by fluorescence microscopy for the location of the fluorescently tagged kinase. In untreated cells, Syk-EGFP(S291A) was distributed evenly throughout the cytoplasm and nucleus, as was observed in cells expressing Syk-EGFP (Fig. 1C). Treatment of cells with PMA led to the exclusion of Syk-EGFP(S291A) from the nucleus (Fig. 4C), comparable with cells expressing Syk-EGFP (15). Thus, the phosphorylation of Syk on Ser-291 did not affect the intracellular location of the kinase or contribute in any significant way to the PMA-induced change in its localization.
Effect of Substitution of Ser-291 on Signaling
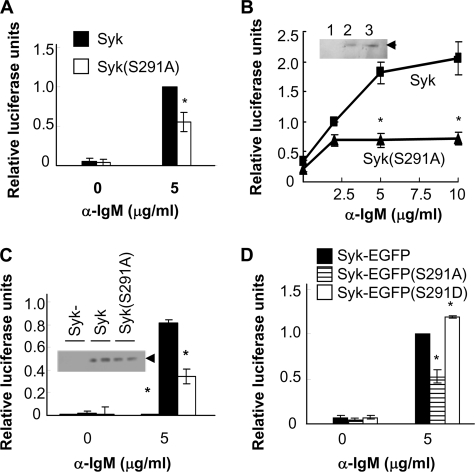
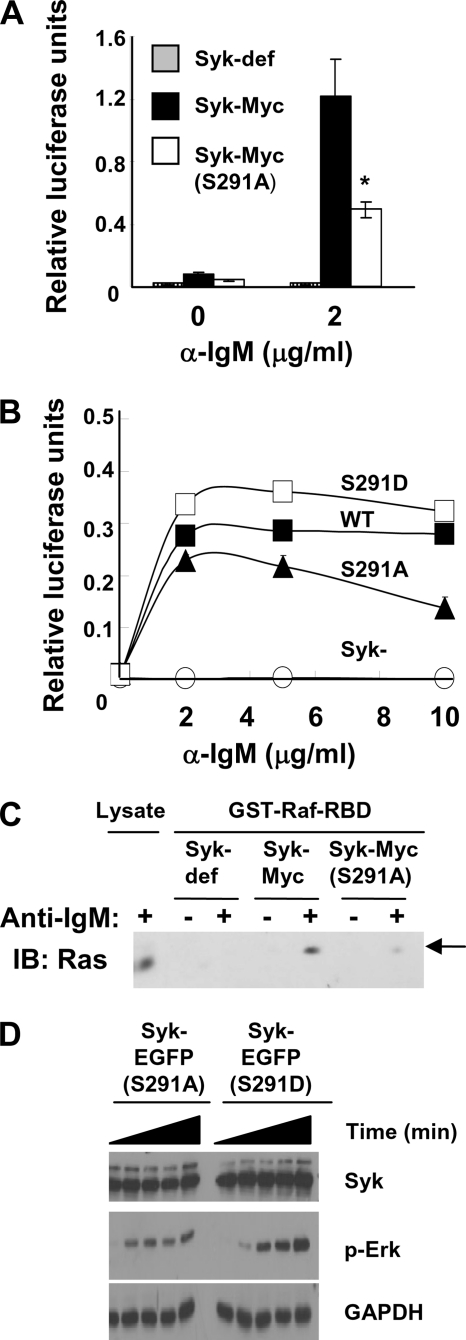
To determine whether the phosphorylation of Syk on Ser-291 influenced its ability to transduce signals from the BCR, we transiently transfected Syk-deficient DT40 cells with an NFAT-driven luciferase reporter plasmid and vectors coding for Myc-epitope-tagged forms of either Syk or Syk(S291A) and then stimulated them with anti-IgM. The replacement of Ser-291 with alanine decreased the ability of Syk to couple the BCR to the activation of NFAT as measured by the production of luciferase (Fig. 5A). This decreased signaling was apparent at multiple concentrations of activating anti-IgM antibody (Fig. 5B). A similar decrease was observed in cells transfected to express Syk-EGFP(S291A) as compared with Syk-EGFP (Fig. 5D). To confirm and extend this observation, we generated by antibiotic selection pools of stably transfected cells expressing either Myc-tagged Syk or Syk(S291A). Pooled cell lines expressed equivalent levels of each kinase (Fig. 5C) as well as equivalent levels of surface IgM as determined by FACS analysis (data not shown). The stably transfected pools of cells were transiently transfected with the NFAT-driven luciferase reporter plasmid, stimulated with anti-IgM, and assayed for luciferase expression. Again, cells expressing Syk(S291A) exhibited a decreased ability to activate NFAT in response to the clustering of surface BCR as compared with cells expressing Syk (Fig. 5C).
FIGURE 5.
Role of Ser-291 in the BCR-stimulated activation of NFAT. A, Syk-deficient DT40 cells were co-transfected with plasmids coding for either Myc-epitope tagged Syk or Syk(S291A) along with an NFAT-driven luciferase reporter plasmid, stimulated without or with anti-IgM antibodies or a mixture of PMA and ionomycin, and assayed for the expression of luciferase. B, Syk-deficient DT40 cells were transiently transfected as described in panel A. Cells were stimulated with varying concentrations of anti-IgM. The inset shows relative expression levels of Syk mutants as determined by Western blot of lysates from Syk-deficient cells (lane 1) or cells expressing Myc-Syk (lane 2) or Myc-Syk(S291A) (lane 3). C, Syk-deficient DT40 cells or stable pools of Syk-deficient cells expressing Myc-tagged Syk or Syk(S291A) were assayed for anti-IgM-induced expression of an NFAT-driven luciferase reporter plasmid as described above. Relative expression levels of Syk mutants were determined by Western blot (inset; each sample was run in duplicate in adjacent lanes). D, stable pools of Syk-deficient cells expressing Syk-EGFP, Syk-EGFP(S291A), or Syk-EGFP(S291D) were assayed for anti-IgM-induced expression of an NFAT-driven luciferase reporter plasmid as described above. The values reported indicate the activity produced by anti-IgM treatment divided by the activity produced in response to PMA + ionomycin to correct for differences in transfection efficiency and are normalized to a value of 1.0 for cells expressing wild-type Syk. Results represent the mean and S.E. for three experiments. *, p < 0.005.
We also generated a plasmid for the expression of a Syk mutant with Ser-291 replaced by aspartic acid. Syk-deficient DT40 cells or Syk-deficient DT40 cells stably expressing Syk-EGFP, Syk-EGFP(S291A), or Syk-EGFP(S291D) were generated and then transiently transfected with the NFAT-driven luciferase expression vector. Cells were left untreated or treated with anti-IgM, and the level of luciferase expression was measured. Although the cells expressing Syk-EGFP(S291A) were defective in the BCR-mediated activation of NFAT as compared with those expressing Syk-EGFP (Fig. 5D), the cells expressing Syk-EGFP(S291D) displayed an enhanced BCR-stimulated NFAT activity.
To determine whether other pathways also were affected by the replacement of Ser-291, we examined the Ras/Raf/Erk pathway. DT40 cells stably expressing Myc-tagged Syk or Syk(S291A) were transiently transfected with an Elk-1-driven luciferase reporter plasmid and then stimulated with anti-IgM. Elk-1 is a transcription factor that this activated after its phosphorylation by Erk. Cells expressing Syk(S291A) exhibited a reduced ability to couple the BCR to the activation of Elk-1 as compared with cells expressing wild-type Syk (Fig. 6A). Similarly, cells stably expressing Syk-EGFP(S291A) were less capable of supporting the activation of Elk-1 as compared with cells expressing either Syk-EGFP or Syk-EGFP(S291D) (Fig. 6B). The ability of anti-IgM to induce the activation of Ras, as measured by the binding of Ras-GTP to an immobilized fusion protein of GST linked to the Ras binding domain of Raf, was also decreased in cells expressing Syk(S291A) (Fig. 6C). Similarly, the activation of Erk was decreased in cells stably expressing Syk-EGFP(S291A) as compared with Syk-EGFP(S291D) (Fig. 6D).
FIGURE 6.
Role of Ser-291 in the BCR-stimulated activation of Elk-1. A, Syk-deficient DT40 cells were co-transfected with plasmids coding for either Myc epitope-tagged Syk or Syk(S291A) along with the Elk1-driven luciferase reporter plasmids, stimulated without or with anti-IgM antibodies or a mixture of PMA and ionomycin, and assayed for the expression of luciferase. The values reported indicate the activity produced by anti-IgM treatment divided by the activity produced in response to PMA + ionomycin to correct for differences in transfection efficiency and are normalized to a value of 1.0 for cells expressing wild-type Syk. Results represent the mean and S.E. for three experiments. *, p < 0.005. B, Syk-deficient DT40 cells (Syk-) or stable pools of Syk-deficient cells expressing Syk-EGFP (Syk), Syk-EGFP(S291A) (S291A) or Syk-EGFP(S291D) (S291D) were assayed for anti-IgM-induced expression of an Elk-1-driven luciferase reporter plasmid as described above. C, Syk-deficient DT40 cells or Syk-deficient DT40 cells stably expressing Myc-tagged Syk or Syk(S291A) were treated without (−) or with (+) anti-IgM (5 μg/ml) for 10 min. A cell lysate and proteins from lysates that bound to immobilized GST-RBD were examined by Western blotting (IB) using an antibody against Ras. Data are representative of three trials. D, Syk-deficient DT40 cells stably expressing Syk-EGFP(S291A) or Syk-EGFP(S291D) were treated without (−) or with (+) anti-IgM (5 μg/ml) for varying periods of time. Lysates were prepared and analyzed by Western blotting using antibodies against Syk, phosphorylated Erk (p-Erk), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Effect of Substitution of Ser-291 on the Association of Syk with 14-3-3
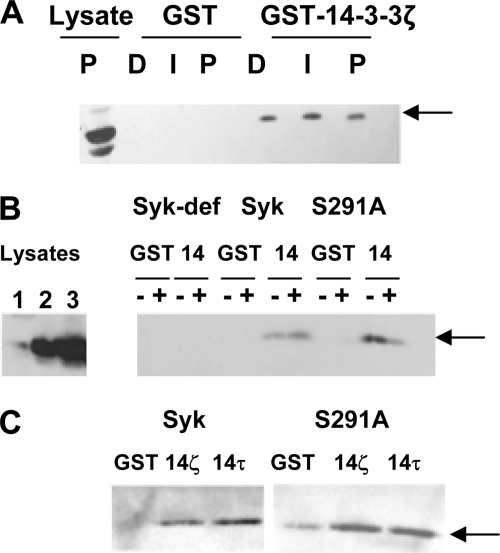
To examine possible mechanisms by which the phosphorylation of Ser-291 modulated Syk-mediated signaling, we first compared the intrinsic catalytic activities of Syk and Syk(S291A) in an in vitro kinase assay using the cytoplasmic domain of erythrocyte band 3 (cfb3) as a substrate. However, we were unable to detect any significant difference in the activity of Syk as compared with Syk(S291A) (data not shown). Similarly, the activity of Syk immunoprecipitated from PMA-treated cells was not significantly different from that of Syk isolated from untreated cells. Therefore, we examined other possible modes of regulation. The sequence surrounding Ser(P)-291 resembles a binding site for 14-3-3 proteins (21, 22), suggesting that phosphorylation of Ser-291 could promote an interaction. To explore this, we immunoprecipitated Syk-EGFP from DT40 cells stably expressing the fusion protein. Tryptic digests of the resulting immune complexes were analyzed by MS/MS to detect possible fragments generated from 14-3-3 isoforms. The analysis of peptides in the digest of Syk-associated proteins revealed two unique peptides derived from 14-3-3β/α, three from 14-3-3ϵ, and two from 14-3-3ζ (supplemental Table 1).
To determine whether an interaction between the two proteins might be mediated by the phosphorylation of Ser-291, we performed a GST pulldown assay using glutathione-Sepharose beads containing either immobilized GST or GST-14-3-3ζ. Beads with immobilized proteins were incubated with lysates from DT40 cells stably expressing Syk-EGFP that had been left untreated or were treated with either anti-IgM antibodies or PMA. Bound proteins were detected by Western blotting using antibodies against Syk. The immobilized 14-3-3ζ was able to recover Syk-EGFP from cell lysates, but binding was independent of the phosphorylation status of the kinase on Ser-291 as the treatment of cells with anti-IgM or PMA had no apparent effect on the interaction (Fig. 7, A and B). Similarly, no decrease was observed in the ability of the immobilized GST-14-3-3ζ to recover Syk-EGFP(S291A) from the lysates of cells treated with or without PMA. Similar results were observed in pulldown assays using immobilized GST-14-3-3τ (Fig. 7C) and in co-immunoprecipitation assays using antibodies against 14-3-3β (data not shown). Although we cannot rule out directly an interaction between Syk and 14-3-3 mediated by Ser-291, it is unlikely that phosphorylation on Ser-291 is essential for such an interaction. The actual mechanism by which Syk binds 14-3-3 proteins either directly or indirectly is at present unclear.
FIGURE 7.
Interaction of Syk with 14-3-3. A, DT40 cells stably expressing Syk-EGFP were treated with DMSO (D), 5 μg/ml anti-IgM (I), or 100 ng/ml PMA (P) for 30 min. Cell lysates were adsorbed to GST or GST-14-3-3ζ bound to glutathione-Sepharose. Bound Syk-EGFP was detected by Western blotting. B, DT40 cells lacking Syk (Syk-def) or stably expressing Syk-EGFP (Syk) or Syk-EGFP(S291A) (S291A) were treated with DMSO (−) or PMA (+) for 30 min. Lysates were adsorbed to immobilized GST (GST) or GST-14-3-3ζ (14). Syk-EGFP in the lysates of Syk-deficient cells (lane 1) or of cells expressing Syk-EGFP (lane 2) or Syk-EGFP(S291A) (lane 3) or bound to the resin was detected by Western blotting. The migration positions of Syk-EGFP proteins is indicated by the arrow. C, lysates from unstimulated cells described in panel B were adsorbed to resin containing GST-14-3-3ζ or GST-14-3-3τ. Bound Syk-EGFP was detected by Western blotting (arrow).
Effect of Substitution of Ser-291 on the Association of Syk with Prohibitin
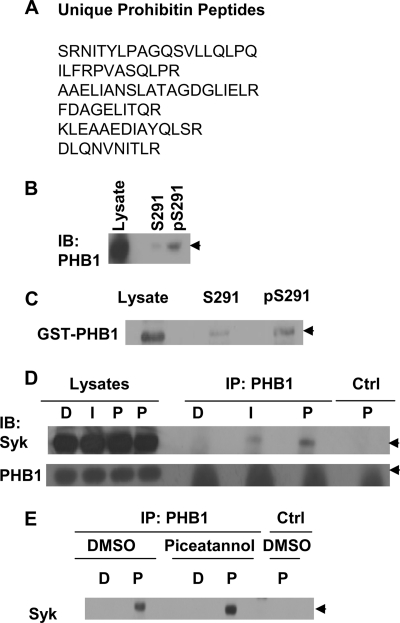
To examine the possibility that serine phosphorylation might influence Syk interactions with other proteins, we generated synthetic, N-terminal-biotinylated peptides corresponding in sequence to the region surrounding Ser-291 that contained either serine or phosphoserine. Peptides were added to detergent lysates prepared from human DG75 B cells and then recovered by adsorption onto streptavidin-agarose beads. After washing, bound proteins were eluted, digested with trypsin, and analyzed by MS/MS. Ten tryptic peptides derived from PHB1, six of which were unique, were identified exclusively in the sample that bound to the Ser(P)-291-containing peptide (Fig. 8A and supplemental Fig. 2, A-C). To verify this interaction, we examined proteins bound to each immobilized peptide by Western blotting with antibodies against PHB1. The results indicated that PHB1 preferentially bound to the immobilized peptide when it was phosphorylated (Fig. 8B). To determine whether this interaction was direct, we mixed each peptide with extracts of bacteria induced to express a GST-PHB1 fusion protein and then adsorbed these to streptavidin-agarose. Again, GST-PHB1 bound preferentially to the phosphorylated peptide (Fig. 8C).
FIGURE 8.
Interaction of Syk with PHB1. A, shown is the sequence of PHB1-derived peptides identified by mass spectrometric analysis of proteins binding to immobilized Ser(P)-291-derived peptide. B, lysates from DG75 cells were incubated with biotinylated peptides corresponding to the sequence surrounding Ser-291 with Ser-291-phosphorylated (pS291) or not (S291) and adsorbed onto streptavidin-agarose beads. Bound proteins were analyzed by Western blotting (IB) with an antibody against PHB1. The migration position of PHB1 is indicated by the arrowhead. C, lysates from Escherichia coli expressing GST-PHB1 were incubated with the Ser-291 or Ser(P)-291-containing peptides and adsorbed onto streptavidin-agarose. Bound GST-PHB1 (arrowhead) was detected by Western blotting. D, PHB1 was immunoprecipitated from lysates of DG75 cells pretreated with DMSO (D), 5 μg/ml anti-human IgM (I), or 100 ng/ml PMA (P). Syk and PHB1 were detected in the lysates and immune complexes by Western blotting. PHB1 migrates just above the immunoglobulin light chain. A mock immunoprecipitation performed in the absence of anti-PHB1 is also shown (Ctrl). E, DG75 cells were pretreated with DMSO or piceatannol (20 μg/ml) for 30 min and then treated with DMSO (D) or 100 ng/ml PMA (P) for 30 min. PHB1 was immunoprecipitated from cell lysates. The presence of Syk in the immune complexes was detected by Western blotting. A mock immunoprecipitation performed in the absence of anti-PHB1 is also shown (Ctrl).
A search of tryptic peptides present in the digest of the Syk-associated proteins described above that were analyzed by mass spectrometry also revealed two unique PHB1-derived peptides, suggesting that PHB1 can bind to the intact kinase (supplemental Table 1). To confirm this interaction, we immunoprecipitated PHB1 from lysates of human DG75 B cells that were left untreated or were treated with either anti-IgM or PMA. The resulting immune complexes were separated by SDS-PAGE and examined for the presence of Syk by Western blotting. Syk was found in the anti-PHB1 immune complexes isolated from cells activated through the BCR and to an even greater extent from cells treated with PMA (Fig. 8D). To determine whether this interaction required the catalytic activity of Syk, the experiment was repeated using cells pretreated with the Syk-selective inhibitor, piceatannol. Again, Syk could be identified in anti-PHB1 complexes when isolated from cells treated with PMA. However, piceatannol had no effect on the interaction (Fig. 8E), suggesting that the catalytic activity of the kinase was not required for its interaction with PHB1.
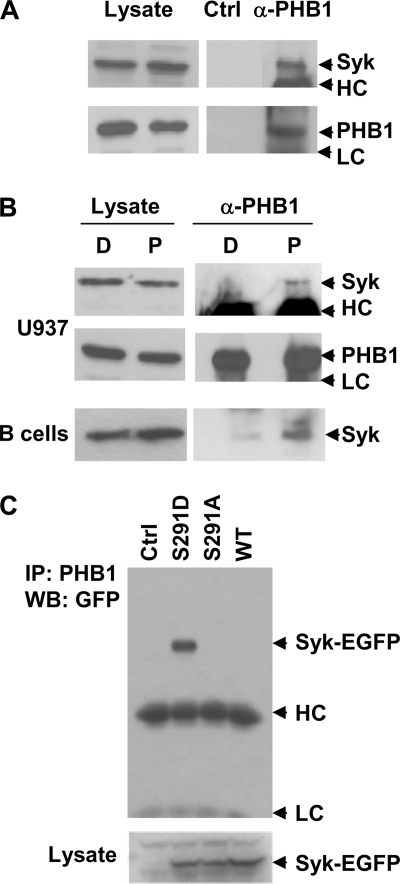
When immunoprecipitated from lysates of U937 monocytic cells, PHB1 associates with tyrosine-phosphorylated proteins of 72–75 kDa in a manner that is enhanced if cells are first treated with PMA (23). At least one component of this complex is the protein-serine/threonine kinase, Raf-1. Because the phosphorylation of Raf-1 on tyrosine is often difficult to detect (24), we reasoned that Syk, which is 72 kDa and readily phosphorylated on tyrosine, might be present in these anti-PHB1 immune complexes. To examine this, we immunoprecipitated PHB1 from detergent lysates of U937 cells that had been left untreated or treated with PMA. The resulting immune complexes were probed for the presence of Syk by Western blotting (Fig. 9, A and B). Indeed, Syk was found preferentially in the anti-PHB1 immune complexes isolated from the U937 cells that had been treated with PMA. Similar results were observed when PHB1 was immunoprecipitated from resting spleen B cells that were either untreated or treated with PMA (Fig. 9B). These results indicate that, in addition to Raf-1, Syk also associates with PHB1.
FIGURE 9.
Interaction of Syk and Syk(S291D) with PHB1. A, PHB1 was immunoprecipitated from lysates of U937 cells pretreated with 100 ng/ml PMA for 30 min. The presence of Syk (upper panels) or PHB1 (lower panels) in the immune complexes was detected by Western blotting. A mock immunoprecipitation performed in the absence of anti-PHB1 is also shown (Ctrl). Migration positions of immunoglobulin heavy (HC) and light (LC) chains are indicated. B, PHB1 was immunoprecipitated from lysates of U937 cells (U937) or resting mouse spleen B cells (B cells) pretreated with DMSO (D) or 100 ng/ml PMA (P) for 30 min. The presence of Syk (upper and lower panels) or PHB1 (middle panels) in the lysates and immune complexes was detected by Western blotting. Migration positions of immunoglobulin heavy (HC) and light (LC) chains are indicated. C, DG75 B cells were transiently transfected with plasmids for the expression of GFP (Ctrl), Syk-EGFP(S291D) (S291D), Syk-EGFP(S291A) (S291A), or Syk-EGFP (WT). The level of expression in cell lysates was compared by Western blotting (IB) with an anti-Syk antibody (lower panel). The presence of Syk fusion proteins in anti-PHB1 immune complexes was detected by Western blotting (WB) using an anti-GFP antibody (upper panel). The arrows mark the migration positions of Syk-EGFP fusion proteins.
The replacement of Ser-291 with aspartic acid enhanced the ability of Syk to couple the BCR to downstream signaling pathways. To determine whether this substitution altered the interaction between Syk and PHB1, we immunoprecipitated endogenous PHB1 from the lysates of untreated DG75 B cells that had been transiently transfected to express Syk-EGFP, Syk-EGFP(S291A), or Syk-EGFP(S291D). Western blotting analysis of the PHB1 immune complexes indicated that, of the three forms of Syk, only Syk-EGFP(S291D) strongly associated with PHB1 (Fig. 9C).
DISCUSSION
After the activation of B cells through the antigen receptor, Syk becomes phosphorylated at multiple sites. These include several tyrosines that have been characterized extensively whose modification alters the activity of the kinase and its interactions with other proteins (1). Our phosphopeptide mapping studies indicate that Syk also is phosphorylated on serine and that the extent of serine phosphorylation exceeds that of tyrosine phosphorylation. One of these serines, Ser-291 in the linker insert, is a major site that becomes phosphorylated after receptor engagement. The primary structure of the phosphorylation site matches the consensus sequence of a substrate for PKC. A role for PKC is consistent with Syk phosphorylation in response to PMA and the sensitivity of this phosphorylation to small molecule kinase inhibitors. PKCs are activated downstream of BCR engagement in a manner dependent on the presence of Syk and the subsequent phosphorylation and activation of phospholipase γ and the generation of diacylglycerol. Our studies indicate that, once activated, PKC can then catalyze the phosphorylation of Syk on Ser-291. PKD, which also is activated after receptor engagement (18, 25), might contribute to this phosphorylation but is clearly not required. Because the three-dimensional structure of full-length Syk has yet to be determined, the structure of the linker insert has not been characterized. In the crystal structure of Zap-70, which lacks a linker insert, the region of linker B in which the insert would be located is disordered and does not appear in the structure (26). This observation coupled with the large number of polar amino acids that are present in the insert suggests that it is likely to be exposed to the solvent, which is consistent with the fact that it is readily phosphorylated. Because this phosphorylation occurs in the presence of PMA even in the absence of receptor engagement, the association of Syk with the BCR is not required. In contrast, it is likely that the phosphorylation of Syk on tyrosine is restricted largely to the subset of the kinase that becomes physically associated with the antigen receptor. This difference likely accounts for the higher stoichiometry of serine, as compared with tyrosine phosphorylation. It is interesting that the sequence of the linker insert is highly conserved across species. This is in contrast to the regions that immediately precede and follow the insert, which bear several amino acid substitutions when comparing, for example, the murine and human sequences. This suggests a particularly important role for the insert itself in the some fundamental aspect of kinase function.
The inability of Syk(S291A) mutants to fully restore the BCR-mediated activation of NFAT, Ras, and Elk1 indicates a positive role for the phosphorylation of the linker insert in receptor-mediated signaling. There are reports in the literature that the treatment of cells with PMA leads to a partial activation of Syk as reflected by an increase in its phosphorylation on tyrosines within the activation loop (27). We were unable to detect any significant difference in the ability of Syk isolated by immunoprecipitation from untreated or PMA-treated cells to catalyze the in vitro phosphorylation of a protein substrate. Similarly, the activities of Syk and Syk(S291A) were not significantly different. However, mutant forms of Syk with activation loop tyrosines replaced with phenylalanines, although exhibiting signaling defects when expressed in cells, have similar catalytic activities when measured in vitro (28–31). Thus, it remains a possibility that the activity of Syk in an intact cell is enhanced by phosphorylation within the linker insert.
The linker insert region bears some resemblance to a bipartite nuclear localization signal and has been proposed to participate in the transit of the kinase into the nucleus (7). However, other sequences in the C-terminal region of linker B in Syk (15) and Zap-70, which transits into the nucleus in the absence of a linker insert (32), also clearly modulate nucleocytoplasmic shuttling. We were surprised to see that a Tyr-290 to phenylalanine mutation, which leaves the basic residues in the linker insert intact, leads to the exclusion of the kinase from the nucleus. The sensitivity of this exclusively cytoplasmic localization to LMB strongly implicates exportin 1 in the transport of the mutant from the nucleus. Thus, it appears likely that the replacement of tyrosine with the more hydrophobic phenylalanine created a nuclear export signal that was recognized by exportin 1. Although the treatment of DT40 cells expressing Syk-EGFP with PMA leads also to the exclusion of the kinase from the nucleus (15), this is apparently not due to the phosphorylation of Ser-291. This is consistent, however, with a previous study indicating that protein synthesis was required for the PMA-induced translocation of the kinase from the nucleus (15). Thus, the effect of phosphorylation by PKC on the signaling capabilities of Syk is unlikely to arise from an enhanced concentration of kinase in the nuclear or cytoplasmic compartment.
Our protein-protein interaction studies suggest another mode of regulation that is modulated by phosphorylation within the linker insert. Pulldown and co-immunoprecipitation assays using immobilized peptides and intact proteins indicate an interaction of Syk with PHB1. This interaction is stimulated by the phosphorylation of Syk on Ser-291 or enhanced by the replacement of Ser-291 with an acidic amino acid. PHB1 is an interesting protein that has been likened to 14-3-3 in its ability to interact with multiple binding partners (33). Interestingly, PHB1 along with its homolog PHB2 associates with surface IgM (antigen receptors) in B cells (34). Along with a variety of interesting binding partners that include Rb (retinoblastoma protein), E2F, and p53 (for review, see Ref. 35), PHB1 associates with at least two other protein kinases, Raf-1 and MLK2 (mixed lineage kinase 2) (23, 33, 36, 37) and PHB2 binds Akt (38). Raf-1 modulates the interactions of PHB1 with E2F, and PHB1, in turn, regulates the activation of Raf-1 (23, 33, 36). The expression of PHB1 is essential for the activation of the Raf/Mek/Erk pathway signaling pathway (33) through an enhancement in the recruitment of Raf-1 to the plasma membrane and displacement of 14-3-3 from an internal Raf-1 binding site (36). Based on these observations, it is interesting to speculate that an interaction with PHB1 also influences the association of Syk with the BCR complex at the plasma membrane to enhance its ability to couple the receptor to downstream signaling pathways.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA037372 (NCI; to R. L. G. and M. L. H.). This work was also supported by a National Science Foundation Career Award (to W. A. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. 1 and 2, and Table 1.
- BCR
- B cell receptor for antigen
- EGFP
- enhanced green fluorescent protein
- NFAT
- nuclear factor of activated T cells
- PHB1
- prohibitin-1
- PHB2
- prohibitin-2
- PKD
- protein kinase D
- PMA
- phorbol 12-myristate 13-acetate
- RBD
- Ras binding domain
- NES
- nuclear export signal
- LMB
- leptomycin B.
REFERENCES
- 1.Geahlen R. L. (2009) Biochim. Biophys. Acta 1793, 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takata M., Sabe H., Hata A., Inazu T., Homma Y., Nukada T., Yamamura H., Kurosaki. T. (1994) EMBO J. 13, 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner M., Mee P. J., Costello P. S., Williams O., Price A. A., Duddy L. P., Furlong M. T., Geahlen R. L., Tybulewicz V. L. J. (1995) Nature 378, 298–302 [DOI] [PubMed] [Google Scholar]
- 4.Cheng A. M., Rowley B., Pao W., Hayday A., Bolen J. B., Pawson T. (1995) Nature 378, 303–306 [DOI] [PubMed] [Google Scholar]
- 5.Yagi S., Suzuki K., Hasegawa A., Okumura K., Ra C. (1994) Biochem. Biophys. Res. Commun. 200, 28–34 [DOI] [PubMed] [Google Scholar]
- 6.Rowley R. B., Bolen J. B., Fargnoli J. (1995) J. Biol. Chem. 270, 12659–12664 [DOI] [PubMed] [Google Scholar]
- 7.Wang L., Duke L., Zhang P. S., Arlinghaus R. B., Symmans W. F., Sahin A., Mendez R., Dai J. L. (2003) Cancer Res. 63, 4724–4730 [PubMed] [Google Scholar]
- 8.Latour S., Zhang J., Siraganian R. P., Veillette A. (1998) EMBO J. 17, 2584–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furlong M. T., Mahrenholz A. M., Kim K. H., Ashendel C. L., Harrison M. L., Geahlen R. L. (1997) Biochim. Biophys. Acta 1355, 177–190 [DOI] [PubMed] [Google Scholar]
- 10.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 11.Severson C. D., Burg D. L., Lafrenz D. E., Feldbush T. L. (1987) Immunol. Lett. 15, 291–295 [DOI] [PubMed] [Google Scholar]
- 12.Keshvara L. M., Isaacson C. C., Yankee T. M., Sarac R., Harrison M. L., Geahlen R. L. (1998) J. Immunol. 161, 5276–5283 [PubMed] [Google Scholar]
- 13.Ma H., Yankee T. M., Hu J., Asai D. J., Harrison M. L., Geahlen R. L. (2001) J. Immunol. 166, 1507–1516 [DOI] [PubMed] [Google Scholar]
- 14.Glass D. B., Masaracchia R. A., Feramisco J. R., Kemp B. E. (1978) Anal. Biochem. 87, 566–575 [DOI] [PubMed] [Google Scholar]
- 15.Zhou F., Hu J., Ma H., Harrison M. L., Geahlen R. L. (2006) Mol. Cell. Biol. 26, 3478–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen W., Meinkoth J. L., Tsien R. Y., Taylor S. S. (1995) Cell 82, 463–473 [DOI] [PubMed] [Google Scholar]
- 17.Kennelly P. J., Krebs E. G. (1991) J. Biol. Chem. 266, 15555–15558 [PubMed] [Google Scholar]
- 18.Sidorenko S. P., Law C. L., Klaus S. J., Chandran K. A., Takata M., Kurosaki T., Clark E. A. (1996) Immunity 5, 353–363 [DOI] [PubMed] [Google Scholar]
- 19.Van Lint J. V., Sinnett-Smith J., Rozengurt E. (1995) J. Biol. Chem. 270, 1455–1461 [DOI] [PubMed] [Google Scholar]
- 20.Martiny-Baron G., Kazanietz M. G., Mischak H., Blumberg P. M., Kochs G., Hug H., Marmé D., Schächtele C. (1993) J. Biol. Chem. 268, 9194–9197 [PubMed] [Google Scholar]
- 21.Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 22.Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. (1996) Cell 84, 889–897 [DOI] [PubMed] [Google Scholar]
- 23.Wang S., Nath N., Fusaro G., Chellappan S. (1999) Mol. Cell. Biol. 19, 7447–7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia K., Lee R. S., Narsimhan R. P., Mukhopadhyay N. K., Neel B. G., Roberts T. M. (1999) Mol. Cell. Biol. 19, 4819–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews S. A., Rozengurt E., Cantrell D. (2000) J. Exp. Med. 191, 2075–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deindl S., Kadlecek T. A., Brdicka T., Cao X., Weiss A., Kuriyan J. (2007) Cell 129, 735–746 [DOI] [PubMed] [Google Scholar]
- 27.Hitomi T., Yanagi S., Inatome R., Ding J., Takano T., Yamamura H. (2001) Genes Cells 6, 475–485 [DOI] [PubMed] [Google Scholar]
- 28.Kurosaki T., Johnson S. A., Pao L., Sada K., Yamamura H., Cambier J. C. (1995) J. Exp. Med. 182, 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couture C., Williams S., Gauthier N., Tailor P., Mustelin T. (1997) Eur. J. Biochem. 246, 447–451 [DOI] [PubMed] [Google Scholar]
- 30.Zhang J., Kimura T., Siraganian R. P. (1998) J. Immunol. 161, 4366–4374 [PubMed] [Google Scholar]
- 31.Papp E., Tse J. K., Ho H., Wang S., Shaw D., Lee S., Barnett J., Swinney D. C., Bradshaw J. M. (2007) Biochemistry 46, 15103–15114 [DOI] [PubMed] [Google Scholar]
- 32.Sloan-Lancaster J., Zhang W., Presley J., Williams B. L., Abraham R. T., Lippincott-Schwartz J., Samelson L. E. (1997) J. Exp. Med. 186, 1713–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajalingam K., Wunder C., Brinkmann V., Churin Y., Hekman M., Sievers C., Rapp U. R., Rudel T. (2005) Nat. Cell Biol. 7, 837–843 [DOI] [PubMed] [Google Scholar]
- 34.Terashima M., Kim K. M., Adachi T., Nielsen P. J., Reth M., Köhler G., Lamers M. C. (1994) EMBO J. 13, 3782–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S., Faller D. V. (2008) Transl. Oncogenomics 3, 23–37 [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer A., Baljuls A., Reinders J., Nekhoroshkova E., Sibilski C., Metz R., Albert S., Rajalingam K., Hekman M., Rapp U. R. (2009) J. Biol. Chem. 284, 3183–3194 [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen R. K., Ji H., Eddes J. S., Moritz R. L., Reid G. E., Simpson R. J., Dorow D. S. (1997) Electrophoresis 18, 588–598 [DOI] [PubMed] [Google Scholar]
- 38.Sun L., Liu L., Yang X. J., Wu Z. (2004) J. Cell Sci. 117, 3021–3029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.