Abstract
The mobilization and extracellular release of nuclear high mobility group box-1 (HMGB1) by ischemic cells activates inflammatory pathways following liver ischemia/reperfusion (I/R) injury. In immune cells such as macrophages, post-translational modification by acetylation appears to be critical for active HMGB1 release. Hyperacetylation shifts its equilibrium from a predominant nuclear location toward cytosolic accumulation and subsequent release. However, mechanisms governing its release by parenchymal cells such as hepatocytes are unknown. In this study, we found that serum HMGB1 released following liver I/R in vivo is acetylated, and that hepatocytes exposed to oxidative stress in vitro also released acetylated HMGB1. Histone deacetylases (HDACs) are a family of enzymes that remove acetyl groups and control the acetylation status of histones and various intracellular proteins. Levels of acetylated HMGB1 increased with a concomitant decrease in total nuclear HDAC activity, suggesting that suppression in HDAC activity contributes to the increase in acetylated HMGB1 release after oxidative stress in hepatocytes. We identified the isoforms HDAC1 and HDAC4 as critical in regulating acetylated HMGB1 release. Activation of HDAC1 was decreased in the nucleus of hepatocytes undergoing oxidative stress. In addition, HDAC1 knockdown with siRNA promoted HMGB1 translocation and release. Furthermore, we demonstrate that HDAC4 is shuttled from the nucleus to cytoplasm in response to oxidative stress, resulting in decreased HDAC activity in the nucleus. Together, these findings suggest that decreased nuclear HDAC1 and HDAC4 activities in hepatocytes following liver I/R is a mechanism that promotes the hyperacetylation and subsequent release of HMGB1.
Keywords: Hepatocyte, Histone Deacetylase, Ischemia, Liver Injury, Post-translational Modification, Danger Signaling, High Mobility Group Box 1
Introduction
High Mobility Group Box Protein 1 (HMGB1)3 is a ubiquitously expressed nuclear molecule that functions as a structural protein of chromatin (1). In addition to its nuclear role, HMGB1 also functions as an inflammatory cytokine when released from necrotic cells or actively secreted from stressed cells. Its proinflammatory properties were first highlighted in experiments showing that HMGB1 is actively secreted by activated macrophages, serving as a late mediator of lethality in sepsis (2). Whereas HMGB1 is involved in the late systemic inflammatory response to sepsis, our laboratory demonstrated that HMGB1 is a central and necessary mediator of organ damage following acute, sterile organ injury (3, 4). HMGB1 is rapidly mobilized and released by hepatocytes in the setting of hepatic ischemia and reperfusion injury. Extracellular HMGB1 functions as a damage-associated molecular pattern (DAMP) molecule and activates proinflammatory signaling pathways by activating pattern recognition receptors including Toll-like receptor 4 (TLR4) and the receptor for advanced glycation end-products (RAGE) (5, 6). Mounting evidence suggests HMGB1 may also function to facilitate the recognition of other immune co-activators such as LPS, DNA, and IL-1 through avid binding to these molecules (7–9).
Thorough understanding of the pathophysiology of liver I/R is vital, as it is commonly encountered clinically during elective liver surgical procedures, solid organ transplantation, trauma, and hypovolemic shock. The liver exhibits both direct cellular damage as the result of the ischemic insult as well as further dysfunction and damage resulting from activation of inflammatory pathways (10). We have previously shown that hepatocytes actively release HMGB1 in response to oxidative stress, suggesting that these parenchymal cells can provide danger signals to neighboring immune cells in the liver to promote inflammation and organ damage (11). Although HMGB1 can be passively released following necrosis, our findings demonstrate that oxidative stress in hepatocytes leads to early shuttling from the nucleus to cytoplasm, followed by its subsequent release in the absence of cell death. This suggests that HMGB1 mobilization in these cells is an active, regulated process. The pathways governing HMGB1 release from hepatocytes are unclear, although proximal events are known to involve TLR4 activation and calcium signaling through calcium/calmodulin-dependent protein kinases (CaMKs) (11). In other cell types, downstream events governing HMGB1 release have been linked to oxidation/reduction (12) and post-translational modifications that include phosphorylation (13) and acetylation (14). However, in liver I/R it is unknown if post-translational modifications of HMGB1 regulate its release from hepatocytes.
Histone deacetylases (HDACs) are a family of enzymes, which remove acetyl groups from lysine residues (15). While these enzymes were named according to their ability to modify histone proteins, they also play important roles in regulating the acetylation status of non-histone proteins (16, 17). Recent findings demonstrate that HMGB1 export from the nucleus and subsequent excretion in monocytes stimulated with bacterial lipopolysaccharide (LPS) involved hyperacetylation. Interestingly, HDAC inhibition also promoted HMGB1 hyperacetylation and extracellular release, suggesting a role for HDACs in regulating HMGB1 (14).
The purpose of this study was to examine the role of HDACs in mediating HMGB1 acetylation and release in hepatocytes following liver I/R. We show in an in vivo model that serum HMGB1 is acetylated, and that this process is dependent on a reduction in nuclear HDAC activity in response to oxidative stress. Thus, the fraction of hyperacetylated HMGB1 increases, promoting its translocation and release. We further investigate the role of the HDAC isoforms, HDAC1 and HDAC4, in regulating post-translational modifications of HMGB1 to influence its translocation and release from oxidative stressed hepatocytes.
EXPERIMENTAL PROCEDURES
Animals
All animals were maintained in a laminar-flow, pathogen-free atmosphere at the University of Pittsburgh. Animal protocols were approved by the Animal Care and Use Committee of the University of Pittsburgh, and the experiments were conducted in adherence to the National Institute of Health Guidelines for the Use of Laboratory Animals. Male C57BL/6 mice were purchased from Jackson Laboratories and were used at the age of 8–12 weeks and the weight of 25–35 g.
In Vivo Warm I/R Model
A previously described I/R protocol involving a nonlethal model of segmental (70%) hepatic warm ischemia was used (18). Sham animals underwent anesthesia, laparotomy, and exposure of the portal triad without hepatic ischemia. Baseline or untreated animals were given anesthesia and sacrificed without exposure of the portal triad.
Hepatocyte Isolation
Hepatocytes were isolated from mice by an in situ collagenase (type IV, Sigma) perfusion technique as described previously (19). Hepatocyte purity exceeded 98% as assessed by light microscopy, and viability was typically greater than 95% as determined by trypan blue exclusion assay.
Cell Culture and Treatment
Hepatocytes (3 × 106) were plated onto 6-cm gelatin-coated plastic tissue culture dishes. The initial culture medium was William's medium E containing 10% calf serum, 15 mm Hepes, 2 mm l-glutamine, and 100 units/ml of penicillin and streptomycin. Hepatocytes were allowed to attach to the plates overnight. The medium was then changed before addition of chemical inhibitors. For hypoxia experiments, the medium was replaced with hypoxic medium (equilibrated with 1% O2, 5% CO2, and 94% N2), and placed into a hypoxia chamber (Coy).
Isolation of Nuclear and Cytoplasmic Proteins
Frozen liver tissues were suspended in a buffer that contained 10 mm Tris, pH 7.5, 1.5 mm MgCl2, 10 mm KCl, and 0.1% Triton X-100 and lysed by homogenization. The samples were gently vortexed every 5 min for 30 min and then centrifuged at 7,500 rpm for 5 min. The supernatant that contained cytoplasmic and membrane protein was collected and stored at −80 °C. Nuclear proteins were extracted at 4 °C by gently resuspending the nuclei pellet in buffer that contained 20 mm Tris, pH 7.5, 20% glycerol, 1.5 mm MgCl2, 420 mm NaCl, 0.2 mm EDTA, and 0.1% Triton X-100. After 1 h with occasional vortexing, resuspended nuclear proteins were centrifuged at 13,000 rpm for 15 min at 4 °C, and the supernatant that contained nuclear proteins was collected. Protein concentration was determined with BCA protein assay reagent (Pierce).
SDS-PAGE and Western Blotting
20 μg of protein was diluted in SDS buffer and run on SDS-PAGE gels (8, 10, 12%). Samples were transferred to nitrocellulose overnight, and blocked with 5% BSA for 8 h. They were then incubated overnight with primary antibodies, which included anti-HMGB1 (1:1000; Abcam); anti-acetyl-lysine (1:1000, Cell Signaling), anti-HDAC1 (1:1000; Santa Cruz Biotechnology); anti-pHDAC1 (1:1000; Abcam); anti-HDAC4 (1:1000; Santa Cruz Biotechnology); anti-pHDAC4 (1:1000; Abcam), anti-histone H3, Ac-H3 Lys9, Ac-H3 Lys18 (1:1000; Cell Signaling). After washing, membranes were incubated with horseradish peroxidase-coupled rabbit or mouse secondary antibodies (1:5000 to 1:10,000) in 5% milk for 1 h and developed with the Super Signal West Pico chemiluminescent kit (Pierce).
HDAC Colorimetric Activity Assay
HDAC Activity Colorimetric Assay kit (Abcam, ab1432) was used according to the manufacturer's protocol. Briefly, 100 μg of nuclear protein was diluted in a final volume of 85 μl of ddH2O and placed in a round bottom 96-well plate. 10 μl of the 10× assay buffer was added to each well followed by 5 μl of the HDAC colorimetric substrate. The mixture was incubated at 37 °C for 3 h. 10 μl of Lysine Developer was then added, and the plate was incubated at 37 °C for 30 min. The sample was read in an ELISA plate reader at 405 nm.
Immunofluorescent Staining
Cells were cultured on coverslips and washed twice with cold PBS, fixed with 2% paraformaldehyde in PBS for 15 min, permeabilized with 0.1% Triton X-100 in PBS for 30 min at room temperature, and blocked for 1 h with 5% BSA in PBS. Then cells were incubated with the specific primary antibody for HMGB1 (Abcam, ab18256) in 1% BSA for 1 h, washed, and incubated with secondary antibody (AlexaFluor 488 goat anti-rabbit, Invitrogen). F-actin was stained with rhodamine phalloidin (Invitrogen). Cells were mounted with Vecta-Shield Mounting media with DAPI nuclear stain. Slides were viewed with Olympus Provis and Leica TSL-SL immunofluorescent microscopes.
Cell Transfection and RT-PCR
Murine hepatocytes at a concentration of 2.0 × 106 were seeded in 6-cm dishes. Hepatocytes were allowed to adhere overnight. Cells were washed and transfected for 6 h in Opti-Mem serum-free medium with siRNA for HDAC1 (Santa Cruz Biotechnology, sc-29344) or a scrambled, control siRNA (Santa Cruz Biotechnology, sc-37007) using Lipofectin transfection reagent (Invitrogen). Total levels of HDAC1 mRNA and protein were analyzed at 24 h and 40 h, respectively. For HDAC1 mRNA expression, total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RT-PCR was performed using the Titanium One-Step PCR kit (Clontech) with primers for HDAC1-HDAC9.
Co-immunoprecipitation
Immunoprecipitation was performed with 1 μg of antibodies against acetyl-lysine or HMGB1 in 200 μg whole lysate protein or 40 μl of serum diluted in IP buffer (50 mm Hepes, 0.5% Nonidet P-40, 150 mm NaCl, 10% glycerol, 1 mm EDTA). Normal rabbit IgG was used as a negative control. Samples were first precleared with a nonspecific IgG antibody. Precleared lysates were then incubated with antibody either to anti-acetyl-lysine or HMGB1 overnight, and then incubated for 2 h with protein A/G-agarose. Samples were washed four times with PBS and subjected to Western blot analysis.
HMGB1 Translocation Quantification
Hepatocytes were analyzed using a high content analysis platform on an Arrayscan VTi (Cellomics). Hepatocytes in 96-well plates were fixed and stained for HMGB1, and subsequently analyzed for nuclear and cytoplasmic staining intensities using Spotfire Decisionsite software. Hepatocytes that accumulated HMGB1 in the cytoplasm in each treatment group were quantified using a staining intensity threshold for cytoplasmic HMGB1.
RESULTS
HMGB1 Is Acetylated and Released in Warm Liver I/R Injury and Hypoxia
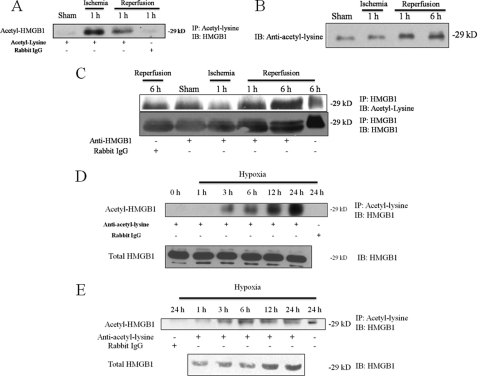
Acetylation is a post-translational modification that can influence protein localization and function. In monocytes stimulated with LPS, hyperacetylation of HMGB1 allows nuclear shuttling of the protein into the cytoplasm and subsequent release (14). To determine the acetylation status of HMGB1 after liver I/R injury, mice were subjected to hepatic I/R and levels of acetylated HMGB1 assessed by co-immunoprecipitation from liver whole cell lysates (Fig. 1A). One hour of ischemia without reperfusion induced a detectable difference in the amount of acetylated HMGB1, and it was also detectable at 1 h of reperfusion. These results were followed by the appearance of acetylated HMGB1 in the serum after reperfusion (Fig. 1, B and C). First we immunoblotted serum samples with anti-acetyl-lysine and observed a strong band appearing at 29 kDa, the molecular mass of HMGB1. Furthermore, the band intensity increased at 1 h and 6 h reperfusion. To identify acetylated HMGB1 in the serum, we co-immunoprecipitated with anti-HMGB1 and immunoblotted with anti-acetyl-lysine. Again, the strongest signals were observed at 1 h and 6 h of reperfusion. We should note, however, that we observed a low but detectable signal in both the control Rabbit IgG pull-down sample and sham-operated animals co-immunoprecipitated with anti-HMGB1. We believe this is due to HMGB1's nonspecific interaction IgG (20), as we observed this phenomenon only in serum samples. The blots were stripped and re-probed for HMGB1 to confirm pull-down, and we observed an identical pattern, suggesting that serum HMGB1 is acetylated after hepatic I/R injury.
FIGURE 1.
HMGB1 is acetylated and released in warm liver I/R injury and hypoxia. Co-immunoprecipitation analysis from mouse liver tissue lysates and serum following I/R. Mice were divided into sham or I/R groups. I/R groups underwent 60 min of warm ischemia followed by reperfusion. Blot shown is representative of three experiments with similar results. A, ischemic liver tissue lysates were immunoprecipitated with an anti-acetyl antibody and immunoblotted for HMGB1. Anti-rabbit IgG was used as a negative control. The blot shown is representative of three experiments with similar results. B, Western blot of mouse serum samples following I/R for anti-acetyl-lysine. C, co-immunoprecipitation of mouse serum following I/R. Samples were pulled down with anti-HMGB1 and immunoblotted with anti-acetyl-lysine. The blot was then stripped and re-probed for HMGB1. D, whole cell lysates of hepatocytes exposed to 1% hypoxia were immunoprecipitated with an anti-acetyl-lysine antibody and immunoblotted for HMGB1. The blot shown is representative of three experiments with similar results. E, co-immunoprecipitation of cell culture supernatants following stimulus with hypoxia (1% O2) for acetylated HMGB1.
To further investigate the role of acetylation in regulating HMGB1 release in vitro, we used primary cultured hepatocytes, which we have shown to be the main source of actively secreted HMGB1 in liver I/R injury. Hypoxia is believed to be the initiating event during I/R; thus, we assessed acetylated HMGB1 in cultured hepatocytes exposed to hypoxia. We found that 1% hypoxia also induced acetylation of HMGB1 in hepatocytes (Fig. 1, D and E). Using co-immunoprecipitation from hepatocyte whole cell lysates, we found a time-dependent increase in acetylated HMGB1 beginning as early as 3 h of hypoxia. A parallel pattern was observed in the cell culture supernatants (Fig. 1E). Combined, these findings demonstrate that liver I/R and hypoxic stress in hepatocytes result in the acetylation and release of HMGB1.
Nuclear HDAC Activity Decreases after Liver I/R and Hypoxia
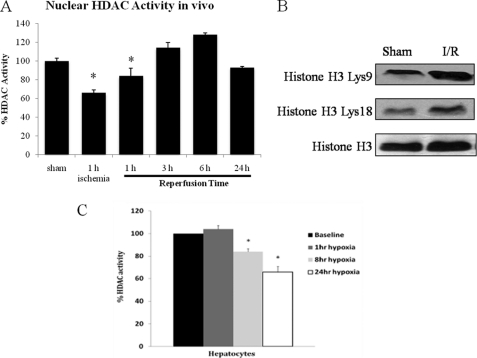
Because the level of acetylated HMGB1 increased with liver I/R and hypoxia, we used colorimetric assays to asses nuclear HDAC activity during I/R. We found that nuclear HDAC activity was significantly decreased in liver tissue from animals subjected to I/R compared with baseline animals during ischemia and early reperfusion (Fig. 2A). Furthermore, levels of acetylated histone H3 were also increased after I/R at Lys-9 and Lys-18 (Fig. 2B). These results suggest that a reduction in nuclear HDAC activity leads a net increase in the nuclear acetylation/deacetylation balance, which promotes HMGB1 hyperacetylation. In vitro, nuclear HDAC activity was also suppressed in hepatocytes by 1% hypoxia at 8 h and 24 h (Fig. 2C). These findings suggest that HDAC activity is suppressed in vivo during I/R and in vitro during hypoxia. This, in turn, contributes to hyperacetylation of HDAC substrates, including histones and HMGB1.
FIGURE 2.
Nuclear HDAC activity is decreased after liver I/R and hypoxia. A, nuclear protein was extracted from mice livers subjected to a time course of ischemia/reperfusion. HDAC activity was determined by colorimetric assay. *, p < 0.05. Assay shown is representative of three experiments with similar results. B, Western blot of acetylated (Lys-9 and Lys-18) and total H3 in sham and 1 h I/R mice. C, hepatocytes were exposed to hypoxia (1% O2) for 1, 4, 8, and 24 h and nuclear protein was analyzed for HDAC activity. *, p < 0.05. Assay shown is representative of three experiments with similar results.
Inhibition of HDACs Results in Nuclear-Cytosolic Translocation of HMGB1 and HMGB1 Release
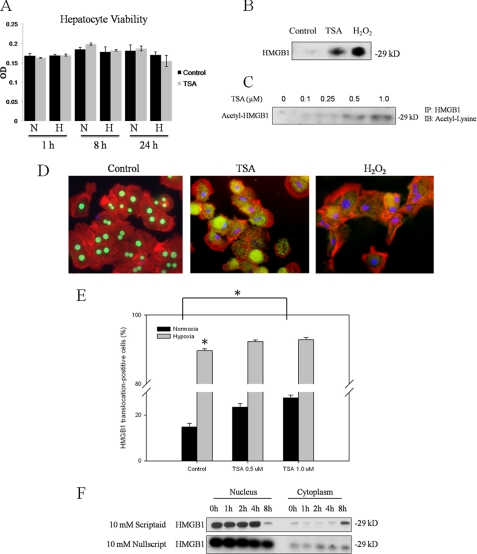
To further investigate the relationship between HDACs and HMGB1, mouse hepatocytes were treated with an HDAC inhibitor, trichostatin A (TSA). TSA has previously been examined in cultured rat hepatocytes (21, 22), with doses of up to 50 μm being well tolerated. We found a dose of 1 μm to be non-toxic to hepatocytes in normoxic or hypoxic culture conditions up to 24 h following treatment (Fig. 3A). Hepatocytes were treated with TSA for 24 h, and Western blot analysis of the cell culture supernatants for HMGB1 was performed. In contrast to the low amount of HMGB1 in the supernatant seen at baseline, TSA induced levels of HMGB1 release into the supernatant similar to that seen with H2O2 treatment (Fig. 3B). In addition, we found a dose-dependent relationship between TSA treatment and the amount of acetylated HMGB1 in cell culture supernatants, suggesting a relationship between HMGB1 acetylation and its extracellular release (Fig. 3C). Next, we treated hepatocytes with TSA for 8 h and performed immunofluorescent staining for HMGB1. Untreated hepatocytes exhibited strong nuclear localization of HMGB1, while both HDAC inhibition and stimulation with H2O2 induced nuclear-cytoplasmic translocation of HMGB1 at 8 h (Fig. 3D). We also used high content analysis to quantify the effect of HDAC inhibition on HMGB1 translocation in hepatocytes (Fig. 3E). As expected, treatment with 8 h hypoxia resulted in a dramatic increase in the fraction of viable hepatocytes in which HMGB1 translocated from the nucleus to the cytoplasm. Furthermore, TSA treatment in normoxia induced a dose-dependent, statistically significant increase in the fraction of HMGB1 translocation-positive cells. We also used another HDAC inhibitor, Scriptaid, and its inactive analog, Nullscript, to investigate the effect of HDAC inhibition on HMGB1 translocation. Crystal violet staining revealed a dose of 10 mm to be non-toxic to hepatocytes (data not shown). By Western blot analysis, the majority of HMGB1 was found in the nucleus at baseline. Addition of Scriptaid induced HMGB1 translocation from the nucleus to the cytosol at 8 h (Fig. 3F). In contrast, the Nullscript-treated hepatocytes maintained a constant nuclear-to-cytosolic ratio of HMGB1. These results show that HDAC inhibition promotes nuclear-to-cytoplasmic translocation and release of acetylated HMGB1 from hepatocytes, and they support a role for HDAC regulation of HMGB1.
FIGURE 3.
Inhibition of HDAC results in nuclear-cytosolic translocation of HMGB1 and HMGB1 release. A, hepatocyte cell viability was examined by Crystal Violet staining in normoxia and hypoxia following treatment with 1 μm TSA. B, Western blot analysis of rat hepatocyte cell supernatants for HMGB1 after TSA treatment or 500 μm H202 treatment. Blot shown is representative of three experiments with similar results. C, co-immunoprecipitation of cell supernatants following treatment with TSA. Cells were immunoprecipitated with anti-HMGB1 and immunoblotted for anti-acetyl-lysine. Blot shown is representative of three experiments with similar results. D, rat hepatocytes were treated with 1 μm TSA or 500 μm H202 and stained for HMGB1. Green, HMGB1; blue, nuclei; red, F-actin. Imaging shown is representative of three experiments with similar results. E, high content analysis for HMGB1 translocation in hepatocytes cell cultures. Cells were immunostained for HMGB1 and staining intensities were quantified in both nuclear and cytosplasmic compartments using a CellomicsTM Arrayscan® platform. “HMGB1 translocation-positive” and “HMGB1 translocation-negative” cell populations were identified using Spotfire Decisionsite Software. *, p < 0.05. F, Western blot analysis for HMGB1 of nuclear and cytoplasmic protein from primary rat hepatocytes treated with 10 mm of the HDAC inhibitor Scriptaid, or its inactive analog, Nullscript and subjected to hypoxia (1% O2) for various time points.
HDAC1 and HDAC4 Are Expressed in Mouse Hepatocytes
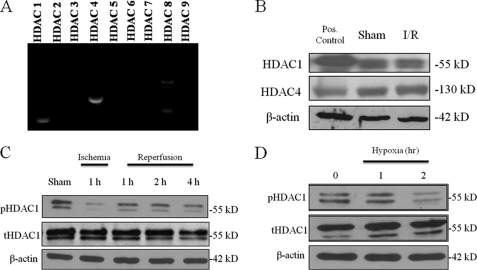
The above inhibitors of HDACs are not specific for a particular HDAC isoform, and the HDAC activity assays measure the activity of all HDACs. To first determine HDAC subtype(s) expressed most strongly in mouse hepatocytes, RT-PCR was performed on RNA isolated from hepatocyte cultures. The mostly highly expressed HDAC mRNAs were HDAC1 and 4 (Fig. 4A). Next, to assess whether HDAC1 and HDAC4 were altered in response to I/R injury, mice were subjected to liver I/R, and Western blots analysis was performed on protein from sham and 1 h I/R animals. Consistent with the in vitro results, HDAC1 and HDAC4 proteins were expressed in the mouse liver (Fig. 4B). In addition, protein levels of HDAC1 and HDAC4 were unchanged following I/R. Thus, we hypothesized that that the reduction in overall nuclear HDAC activity following I/R or oxidative stress was due to regulation of HDAC1 and HDAC4 rather than changes in their overall total protein levels.
FIGURE 4.
HDAC1 and HDAC4 are expressed in mouse hepatocytes. A, RT-PCR for HDAC 1–9 in rat hepatocytes. B, Western blot analysis of mouse liver nuclear protein extracts for HDAC 1 and HDAC4. Jurkat nuclear extracts were used as positive controls for HDAC1 and -4. The blot shown is representative of three experiments with similar results. C, Western blot analysis of nuclear extracts from ischemic liver tissue following I/R. Immunoblots were performed for phospho-HDAC1 and total HDAC1. The blot shown is representative of three experiments with similar results. D, mouse hepatocytes were subjected to hypoxia for and analyzed for pHDAC1 and tHDAC1 by Western blot analysis. The blot shown is representative of three experiments with similar results.
Next, we focused on deacetylase activity of HDAC1, as its activity is determined, in part, by its phosphorylation status (23, 24). We examined if liver I/R altered HDAC1 phosphorylation status. Compared with baseline, 1 h of ischemia resulted in a decrease in phosporylated HDAC1 (Fig. 4C). Upon reperfusion, levels of phosphorylated HDAC1 remained low but increased compared with ischemia. We then exposed hepatocytes to 1% hypoxia and Western blot analysis for phosphorylated HDAC1 in nuclear extracts was performed. By 2 h, levels of phosporylated HDAC1 decreased significantly. Thus, hypoxia appears to be a strong stimulus for the dephosphorylation of HDAC1 in hepatocytes.
Specific Inhibition of HDAC1 Promotes Translocation of Nuclear HMGB1 to the Cytoplasm and Increases HMGB1 Release
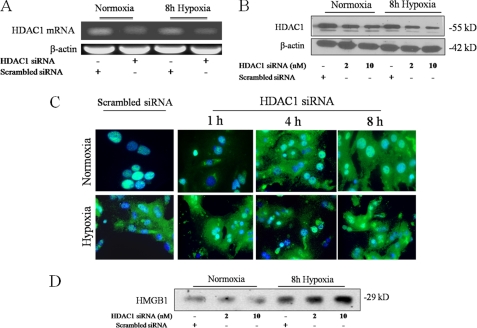
To more specifically investigate the role of HDAC1 in HMGB1 release, hepatocytes were transfected with HDAC1 siRNA and subjected to 8 h of hypoxia. Addition of HDAC1 siRNA, but not scrambled siRNA, decreased the levels of HDAC1 mRNA and protein in both normoxia and hypoxia (Fig. 5, A and B). We then assessed HMGB1 localization under baseline and hypoxia conditions in the HDAC1 siRNA treated cells. Addition of HDAC1 siRNA to normoxic cells mobilized HMGB1 out of the nucleus into the cytoplasm, and after hypoxia, even more HMGB1 translocated into the cytoplasm (Fig. 5C). Western blot analysis of supernatants from these hepatocytes supported these findings. Hypoxia alone caused an increase in HMGB1 release. This effect was augmented by HDAC1 knockdown, while the addition of scrambled siRNA had no effect (Fig. 5D). Our findings show HDAC1 has a potential role in altering HMGB1 localization and release.
FIGURE 5.
Specific inhibition of HDAC1 promotes translocation of nuclear HMGB1 to the cytoplasm and increases HMGB1 release. Mouse hepatocytes were transfected with scrambled siRNA or HDAC1 siRNA and subjected to RT-PCR 24 h post-transfection (A) or Western blot (B) analysis 40 h post-transfection. Blots shown are representative of three experiments with similar results. C, hepatocytes were transfected with 10 μm HDAC1 siRNA analyzed by immunofluorescent staining 40–48 h later. Cells were analyzed in normoxia or after 1, 4, and 8 h of hypoxia (1% O2) by immunostaining. Green, HMGB1; blue, nuclei. Imaging shown is representative of three experiments with similar results. D, Western blot for HMGB1 in cell supernatants of hepatocytes transfected with HDAC1 siRNA following 8 h of hypoxia. The blot shown is representative of three experiments with similar results.
HDAC4 Is Shuttled from the Nucleus to the Cytoplasm during I/R
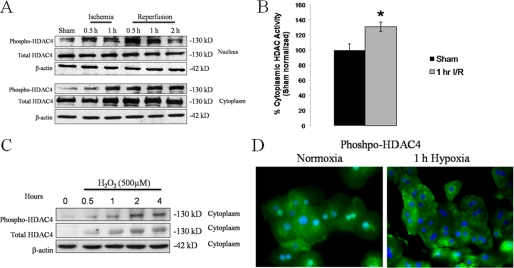
The nuclear activities of class II HDACs (e.g. HDAC4) are regulated in part by shuttling between the nucleus and cytoplasm. Phosphorylation of HDAC4 promotes its translocation to the cytoplasm, resulting in decreased HDAC activity in the nucleus (25, 26). We hypothesized that liver I/R is also a stimulus for HDAC4 phosphorylation and cytoplasmic shuttling. We performed Western blot analysis for phospho-HDAC4 on nuclear and cytoplasmic fractions of liver tissue from mice subjected to I/R (Fig. 6A). In sham-treated animals, a low basal level of phospho-HDAC4 was detected in both the nuclear and cytoplasmic fractions. Nuclear phospho-HDAC4 levels increased as early as 30 min reperfusion and peaked at 1 h. By 2 h reperfusion, the amount of nuclear phospho-HDAC4 began to decrease. This pattern was consistent with results observed in the cytoplasmic fraction. We observed a time-dependent increase in cytoplasmic phospho-HDAC4 from baseline sham levels. We also measured total cytoplasmic HDAC activity in these samples by colorimetric HDAC assay. Compared with sham-operated mice, total cytoplasmic HDAC activity increased following I/R (Fig. 6B). We interpret these results to indicate that the overall cytoplasmic HDAC activity increase is due, at least in part, to shuttling of active, phosphorylated HDAC4 into the cytoplasm. We also investigated HDAC4 shuttling in vitro in hepatocytes. In response to oxidative stress by H2O2 and hypoxia, increased levels of phospho-HDAC4 were observed in the cytoplasm. Western blot analysis for HDAC4 and phosopho-HDAC4 were performed on hepatocyte cytoplasmic fractions following H2O2 stimulation, where we observed a time-dependent increase in this compartment (Fig. 6C). Immunofluorescent staining for phospho-HDAC4 was also performed on hepatocytes stimulated with hypoxia for 1 h (Fig. 6D). In normoxic controls, phospho-HDAC4 staining was positive in both the nuclear and cytoplasmic compartments, while in hypoxic cells the nuclear phospho-HDAC4 signal was not observed. Collectively, these data demonstrate that nuclear HDAC4 is phosphorylated in vivo by I/R and in vitro by oxidative stress, where it is rapidly shuttled into the cytoplasm. This shuttling contributes to a decrease in total nuclear HDAC activity observed after liver I/R, which favors hyperacetylation and release of HMGB1.
FIGURE 6.
HDAC4 is shuttled from the nucleus to the cytoplasm during I/R. A, Western blot analysis of liver nuclear and cytoplasmic fractions for phospho-HDAC4 during I/R. B, total HDAC activity as measured by colorimetric assay at 1 h reperfusion. C, in vitro, hepatocytes were stimulated with 500 μm H202 and Western blot analysis was performed on cytoplasmic fractions for phospho-HDAC4 and total HDAC4. D, hepatocytes were subjected to hypoxia for 1 h and stained for phospho-HDAC4.
DISCUSSION
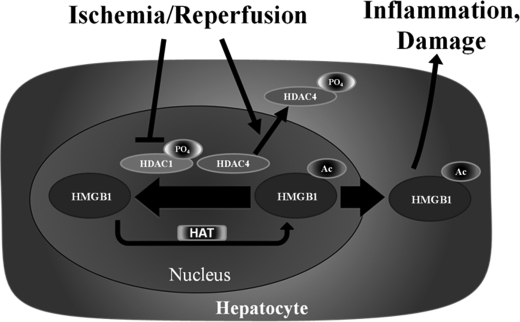
Damaged cells activate innate immunity and recruit inflammatory cells by releasing danger signals collectively known as DAMPs. One such molecule is HMGB1, which has been implicated as an early mediator of organ damage in I/R injury and hemorrhagic shock, as well as a late mediator of lethality in endotoxic shock (2, 3, 27). HMGB1 release occurs passively through necrosis (28), but it is also actively secreted by apoptotic (29) and oxidative-stressed (11) cells. In most cases the cells actively secreting HMGB1 appear to be immune cells such as macrophages, natural killer cells (30), and dendritic cells (31). However, it is becoming increasingly clear that non-immune parenchymal cells also participate in active HMGB1 secretion (32, 33). Our previous work demonstrated that the main source of actively secreted HMGB1 comes from parenchymal cells of the liver, hepatocytes. It has also been shown that neutralizing antibodies to HMGB1 ameliorate liver I/R injury, thereby suggesting a therapeutic benefit of blocking active HMGB1 release to minimize I/R-associated damage. This study was undertaken to determine the role of acetylation in ischemia-induced HMGB1 mobilization and release from hepatocytes following I/R. We provide evidence that ischemia reduces total nuclear HDAC activity by inactivation of HDAC1 and cytoplasmic shuttling of HDAC4. This, in turn, is associated with an increase in HMGB1 acetylation, translocation to the cytoplasm, and extracellular release (Fig. 7).
FIGURE 7.
I/R induces release of acetylated-HMGB1 from hepatocytes through HDAC1 deactivation and HDAC4 shuttling. The proposed model showing release of acetylated HMGB1 following I/R. Both de-activation of nuclear HDAC1 and cytosplasmic shuttling of HDAC4 contribute to a reduction in overall nuclear HDAC activity, tilting the acetylation/deacetylation balance of HMGB1 toward net acetylation and release.
It has not yet been shown that HMGB1 can be acetylated in an in vivo model of acute, sterile organ injury. Here, we show that liver I/R injury induced HMGB1 acetylation in hepatocytes as early as 1 h of ischemia and that acetylated HMGB1 was released into the serum. The ability of oxidative stress to induce acetylation of HMGB1 was also confirmed in an in vitro model, whereby 1% hypoxia induced HMGB1 acetylation. These data suggest that acetylation is a post-translational modification that directs HMGB1 movement across the nuclear membrane in non-hematopoietic cells under redox stress. Understanding mechanisms of HMGB1 release in redox-stressed cells is a crucial step in devising therapies to limit injury after I/R. A number of mechanisms have been described about the regulation of HMGB1 translocation and release in various cell types.
Acetylation, phosphorylation, and redox control of HMGB1 have all been shown to influence its subcellular location in response to various stimuli (12, 14, 34). Acetylated HMGB1 was first identified in 1979 (35) and has since been shown to be involved in regulating HMGB1 DNA binding properties along with its subcellular location, as hyperacetylation of HMGB1 shifts its equilibrium from a predominant nuclear location toward cytosolic accumulation. In vitro experiments showed that in macrophages, lysine residues of HMGB1 between 27 and 43 represent functional nuclear localizing signals (14). Phosphorylation of HMGB1 is another regulatory mechanism that influences its subcellular location(13). Recently, it was shown that HMGB1 phosphorylation by calcium/calmodulin protein kinase IV caused nuclear-to-cytoplasmic shuttling and release in LPS-stimulated macrophages (34). Also, phosphorylation mechanisms involving the classical Protein Kinase C have been described (36). Redox control mechanisms are less well known; however, it has been shown that Cys-106 of HMGB1 is required for its nuclear import (12). In all of these instances, post-translational modifications influence HMGB1 subcellular location by altering the availability or binding properties of its nuclear localization signals or nuclear export signals. Because these modifications occur at different amino acid residues on the HMGB1 protein, it is likely that a combination of these mechanisms work in parallel to regulate HMGB1 release.
Our observation that hepatocytes treated with HDAC inhibitors, TSA or Scriptaid, induced nuclear-cytosolic translocation and extracellular release of HMGB1 suggests that one or more HDAC subtypes control acetylation status of HMGB1. TSA is a pan-HDAC inhibitor with the ability to induce apoptosis in certain cell types, and recently it has been shown that HMGB1 can be released from late apoptotic cells (29, 37). Whereas it is a possibility that HMGB1 release occurred secondary to the induction of apoptosis in TSA-treated cells, we confirmed (data not shown) the finding that TSA does not induce apoptosis in primary cultured hepatocytes (38, 39), although this effect is observed in a number of other liver cancer cell lines (40, 41).
Histone deacetylase activity is dependent on a multitude of signaling pathways that control gene expression, growth, and response to stress. They are subjected to a number of post-translational modifications that have been well characterized (42). Phosphorylation is a key mechanism regulating virtually all members of the HDAC family. HDAC1 phosphorylation has been associated with both an increase and a decrease in enzymatic activity along with disruption of HDAC1 protein complexes. Although not well delineated, it is generally regarded that phosphorylation of HDAC1 promotes its activity, while de-phosphorylation decreases its activity. Thus, our observation that I/R and hypoxia caused de-phosphorylation of HDAC1 and a concomitant decrease in HDAC activity supports the idea that de-phosphorylation of HDAC1 decreases its enzymatic activity. In contrast, HDAC4 is a class II HDAC enzyme and is regulated by cytoplasmic shuttling (25). In response to upstream signaling pathways, nuclear HDAC4 is phosphorylated and binds 14-3-3 proteins which facilitate its export from the nucleus (26). As a result, phosphorylated HDAC4 is exported from the nucleus and inhibited from acting on its nuclear substrates. Thus the regulation of HDAC activities are dynamic and depend on a multitude of factors including specific cell type and the initiating stimulus. While our findings point to HDAC1 and HDAC4 as the isoforms associated with HMGB1 acetylation and release, other HDACs may participate in the process, and our laboratory will continue to investigate this in future studies.
Whereas this report examines the role of HDACs in regulating the acetylation status of HMGB1, it is also known that histone acetyltransferases (HATs) also participate in HMGB1 acetylation. Increased acetylation and decreased deacetylation are not mutually exclusive, and HAT and HDAC enzymes can dynamically regulate HMGB1 acetylation. Indeed, a number of studies have studied the role of HAT on HMGB1 acetylation in vitro (43, 44).
Whereas this study did not examine the role of HAT enzymes in hepatic I/R, recent findings from our laboratory suggest that HAT activation during I/R also contributes to HMGB1 acetylation (45). Our ongoing investigations are focused on understanding the balance between both HAT and HDAC activities in the mobilization and release of HMGB1.
In summary, we have identified the acetylation of HMGB1 as a key mechanism of regulating its active release from hepatocytes undergoing oxidative stress. We also determined the role of histone deacetylates in regulating this phenomenon. Our data further support the idea that HDAC enzymes are participate in cellular responses to ischemic stress by regulating the acetylation status of HMGB1. These novel findings further elucidate signaling pathways governing cellular responses to ischemic stress through HDAC-mediated regulation of HMGB1 release, and may be important for designing therapies to prevent organ damage during ischemic conditions.
Acknowledgments
We thank Nicole Martik and Xinghua Liao for technical assistance in preparing this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM50441 (to T. B.), Howard Hughes Medical Institute Physician-Scientist Award (to A. T.), American College of Surgeons Research Fellowship (to A. T.), Association for Academic Surgery Foundation Research Fellowship (to S. W. C.), and Howard Hughes Medical Institute Research Training for Medical Student Fellowship (to J. E.).
- HMGB1
- high mobility group box 1
- HDAC
- histone deacetylase
- I/R
- ischemia/reperfusion
- TSA
- trichostatin A.
REFERENCES
- 1.Bustin M., Hopkins R. B., Isenberg I. (1978) J. Biol. Chem. 253, 1694–1699 [PubMed] [Google Scholar]
- 2.Wang H., Bloom O., Zhang M., Vishnubhakat J. M., Ombrellino M., Che J., Frazier A., Yang H., Ivanova S., Borovikova L., Manogue K. R., Faist E., Abraham E., Andersson J., Andersson U., Molina P. E., Abumrad N. N., Sama A., Tracey K. J. (1999) Science 285, 248–251 [DOI] [PubMed] [Google Scholar]
- 3.Tsung A., Sahai R., Tanaka H., Nakao A., Fink M. P., Lotze M. T., Yang H., Li J., Tracey K. J., Geller D. A., Billiar T. R. (2005) J. Exp. Med. 201, 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy R. M., Mollen K. P., Prince J. M., Kaczorowski D. J., Vallabhaneni R., Liu S., Tracey K. J., Lotze M. T., Hackam D. J., Fink M. P., Vodovotz Y., Billiar T. R. (2007) Am. J. Physiol. Regul. Integr. Comp. Physiol. 293, R1538–R1544 [DOI] [PubMed] [Google Scholar]
- 5.Hori O., Brett J., Slattery T., Cao R., Zhang J., Chen J. X., Nagashima M., Lundh E. R., Vijay S., Nitecki D. (1995) J. Biol. Chem. 270, 25752–25761 [DOI] [PubMed] [Google Scholar]
- 6.Park J. S., Svetkauskaite D., He Q., Kim J. Y., Strassheim D., Ishizaka A., Abraham E. (2004) J. Biol. Chem. 279, 7370–7377 [DOI] [PubMed] [Google Scholar]
- 7.Sha Y., Zmijewski J., Xu Z., Abraham E. (2008) J. Immunol. 180, 2531–2537 [DOI] [PubMed] [Google Scholar]
- 8.Ivanov S., Dragoi A. M., Wang X., Dallacosta C., Louten J., Musco G., Sitia G., Yap G. S., Wan Y., Biron C. A., Bianchi M. E., Wang H., Chu W. M. (2007) Blood 110, 1970–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian J., Avalos A. M., Mao S. Y., Chen B., Senthil K., Wu H., Parroche P., Drabic S., Golenbock D., Sirois C., Hua J., An L. L., Audoly L., La Rosa G., Bierhaus A., Naworth P., Marshak-Rothstein A., Crow M. K., Fitzgerald K. A., Latz E., Kiener P. A., Coyle A. J. (2007) Nat. Immunol. 8, 487–496 [DOI] [PubMed] [Google Scholar]
- 10.Vardanian A. J., Busuttil R. W., Kupiec-Weglinski J. W. (2008) Mol. Med. 14, 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsung A., Klune J. R., Zhang X., Jeyabalan G., Cao Z., Peng X., Stolz D. B., Geller D. A., Rosengart M. R., Billiar T. R. (2007) J. Exp. Med. 204, 2913–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoppe G., Talcott K. E., Bhattacharya S. K., Crabb J. W., Sears J. E. (2006) Exp. Cell Res. 312, 3526–3538 [DOI] [PubMed] [Google Scholar]
- 13.Youn J. H., Shin J. S. (2006) J. Immunol. 177, 7889–7897 [DOI] [PubMed] [Google Scholar]
- 14.Bonaldi T., Talamo F., Scaffidi P., Ferrera D., Porto A., Bachi A., Rubartelli A., Agresti A., Bianchi M. E. (2003) EMBO J. 22, 5551–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003) Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L., Fischle W., Verdin E., Greene W. C. (2001) Science 293, 1653–1657 [DOI] [PubMed] [Google Scholar]
- 17.Gu W., Roeder R. G. (1997) Cell 90, 595–606 [DOI] [PubMed] [Google Scholar]
- 18.Tsung A., Stang M. T., Ikeda A., Critchlow N. D., Izuishi K., Nakao A., Chan M. H., Jeyabalan G., Yim J. H., Geller D. A. (2006) Am. J. Physiol. Gastrointest. Liver Physiol 290, G1261–G1268 [DOI] [PubMed] [Google Scholar]
- 19.West M. A., Billiar T. R., Curran R. D., Hyland B. J., Simmons R. L. (1989) Gastroenterology 96, 1572–1582 [DOI] [PubMed] [Google Scholar]
- 20.Urbonaviciute V., Fürnrohr B. G., Weber C., Haslbeck M., Wilhelm S., Herrmann M., Voll R. E. (2007) J Leukoc. Biol. 81, 67–74 [DOI] [PubMed] [Google Scholar]
- 21.Henkens T., Papeleu P., Elaut G., Vinken M., Rogiers V., Vanhaecke T. (2007) Toxicol. Appl. Pharmacol. 218, 64–71 [DOI] [PubMed] [Google Scholar]
- 22.Vanhaecke T., Henkens T., Kass G. E., Rogiers V. (2004) Biochem. Pharmacol. 68, 753–760 [DOI] [PubMed] [Google Scholar]
- 23.Pflum M. K., Tong J. K., Lane W. S., Schreiber S. L. (2001) J. Biol. Chem. 276, 47733–47741 [DOI] [PubMed] [Google Scholar]
- 24.Sengupta N., Seto E. (2004) J. Cell. Biochem. 93, 57–67 [DOI] [PubMed] [Google Scholar]
- 25.Wang A. H., Yang X. J. (2001) Mol. Cell. Biol. 21, 5992–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishino T. G., Miyazaki M., Hoshino H., Miwa Y., Horinouchi S., Yoshida M. (2008) Biochem. Biophys. Res. Commun. 377, 852–856 [DOI] [PubMed] [Google Scholar]
- 27.Yang R., Harada T., Mollen K. P., Prince J. M., Levy R. M., Englert J. A., Gallowitsch-Puerta M., Yang L., Yang H., Tracey K. J., Harbrecht B. G., Billiar T. R., Fink M. P. (2006) Mol. Med. 12, 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scaffidi P., Misteli T., Bianchi M. E. (2002) Nature 418, 191–195 [DOI] [PubMed] [Google Scholar]
- 29.Bell C. W., Jiang W., Reich C. F., 3rd, Pisetsky D. S. (2006) Am. J. Physiol. Cell Physiol. 291, C1318–C1325 [DOI] [PubMed] [Google Scholar]
- 30.Semino C., Angelini G., Poggi A., Rubartelli A. (2005) Blood 106, 609–616 [DOI] [PubMed] [Google Scholar]
- 31.Dumitriu I. E., Baruah P., Valentinis B., Voll R. E., Herrmann M., Nawroth P. P., Arnold B., Bianchi M. E., Manfredi A. A., Rovere-Querini P. (2005) J. Immunol. 174, 7506–7515 [DOI] [PubMed] [Google Scholar]
- 32.Xu H., Su Z., Wu J., Yang M., Penninger J. M., Martin C. M., Kvietys P. R., Rui T. (2010) J. Immunol. 184, 1492–1498 [DOI] [PubMed] [Google Scholar]
- 33.Fujii K., Luo Y., Sasahira T., Denda A., Ohmori H., Kuniyasu H. (2009) Cell Prolif. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X., Wheeler D., Tang Y., Guo L., Shapiro R. A., Ribar T. J., Means A. R., Billiar T. R., Angus D. C., Rosengart M. R. (2008) J. Immunol. 181, 5015–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sterner R., Vidali G., Allfrey V. G. (1979) J. Biol. Chem. 254, 11577–11583 [PubMed] [Google Scholar]
- 36.Oh Y. J., Youn J. H., Ji Y., Lee S. E., Lim K. J., Choi J. E., Shin J. S. (2009) J. Immunol. 182, 5800–5809 [DOI] [PubMed] [Google Scholar]
- 37.Urbonavicuite V., Fürnrohr B. G., Meister S., Munoz L., Heyder P., De Marchis F., Bianchi M. E., Kirschning C., Wagner H., Manfredi A. A., Kalden A. A., Schett G., Rovere-Querini P., Herrmann M., Voll R. E. (2008) J. Exp. Med. 205, 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armeanu S., Pathil A., Venturelli S., Mascagni P., Weiss T. S., Göttlicher M., Gregor M., Lauer U. M., Bitzer M. (2005) J Hepatol. 42, 210–217 [DOI] [PubMed] [Google Scholar]
- 39.Papeleu P., Loyer P., Vanhaecke T., Elaut G., Geerts A., Guguen-Guillouzo C., Rogiers V. (2003) J. Hepatol. 39, 374–382 [DOI] [PubMed] [Google Scholar]
- 40.Herold C., Ganslmayer M., Ocker M., Hermann M., Geerts A., Hahn E. G., Schuppan D. (2002) J. Hepatol. 36, 233–240 [DOI] [PubMed] [Google Scholar]
- 41.Carlisi D., Vassallo B., Lauricella M., Emanuele S., D'Anneo A., Di, Leonardo E., Di, Fazio P., Vento R., Tesoriere G. (2008) Int. J. Oncol. 32, 177–184 [DOI] [PubMed] [Google Scholar]
- 42.Brandl A., Heinzel T., Krämer O. H. (2009) Biol. Cell 101, 193–205 [DOI] [PubMed] [Google Scholar]
- 43.Pasheva E., Sarov M., Bidjekov K., Ugrinova I., Sarg B., Lindner H., Pashev I. G. (2004) Biochemistry 43, 2935–2940 [DOI] [PubMed] [Google Scholar]
- 44.Topalova D., Ugrinova I., Pashev I. G., Pasheva E. A. (2008) Int. J. Biochem. Cell Biol. 40, 1536–1542 [DOI] [PubMed] [Google Scholar]
- 45.Dhupar R., Klune J. R., Evankovich J., Cardinal J., Zhang M., Ross M., Murase N., Geller D. A., Billiar T. R., Tsung A. (2010) SHOCK, doi: 10.1097/SHK.0b013e3181f6aab0 [DOI] [PubMed] [Google Scholar]