Abstract
A single amino acid change, F580Y (Legs at odd angles (Loa), Dync1h1Loa), in the highly conserved and overlapping homodimerization, intermediate chain, and light intermediate chain binding domain of the cytoplasmic dynein heavy chain can cause severe motor and sensory neuron loss in mice. The mechanism by which the Loa mutation impairs the neuron-specific functions of dynein is not understood. To elucidate the underlying molecular mechanisms of neurodegeneration arising from this mutation, we applied a cohort of biochemical methods combined with in vivo assays to systemically study the effects of the mutation on the assembly of dynein and its interaction with dynactin. We found that the Loa mutation in the heavy chain leads to increased affinity of this subunit of cytoplasmic dynein to light intermediate and a population of intermediate chains and a suppressed association of dynactin to dynein. These data suggest that the Loa mutation drives the assembly of cytoplasmic dynein toward a complex with lower affinity to dynactin and thus impairing transport of cargos that tether to the complex via dynactin. In addition, we detected up-regulation of kinesin light chain 1 (KLC1) and its increased association with dynein but reduced microtubule-associated KLC1 in the Loa samples. We provide a model describing how up-regulation of KLC1 and its interaction with cytoplasmic dynein in Loa could play a regulatory role in restoring the retrograde and anterograde transport in the Loa neurons.
Keywords: Dynein, Kinesin, Molecular Motors, Neurodegeneration, Neurological Diseases, Disease, Loa, Motor Neuron, Mouse, Sensory Neuron
Introduction
Cytoplasmic motor dynein is a multisubunit motor protein involved in the retrograde (minus end) transport of membranous organelles, signaling endosomes, kinetochores, and other cargos along microtubules (MTs)2 (1, 2). It consists of two ∼530-kDa homodimerized heavy chains (DHC, encoded by a single gene Dync1h1), and several intermediate chains (DYNC1I1, here referred to as DIC, encoded by two genes Dync1i1 and Dync1i2), light intermediate chains (DYNC1L1, here referred to as DLIC, encoded by two genes Dync1li1 and Dync1li2), and light chains (here referred to as DLC, encoded by at least six genes Dynlt1, Dynlt3, Dynlrb1, Dynlrb2, Dynll1, and Dynll2), which assemble into different dynein subcomplexes (3–8). DHC is composed of a carboxyl-terminal head domain with ATPase activity for motility along MTs and an amino-terminal tail domain, which mediates DHC-DHC homodimerization and binding of DIC and DLIC subunits to the complex (4). DLCs bind to the complex through their association with DICs (9–11). DICs also mediate the association of dynein with dynactin, another multisubunit complex composed of p150Glued, p50, and several other polypeptides (12, 13). Dynactin mediates the binding of some cargos to the dynein complex, and it also regulates dynein activity (14, 15).
The compositional complexity of dynein plays a key role in dynein function, and it is essential in targeting dynein to its cargos. The subunit diversity is thought to be responsible for the wide range of interacting binding partners and cargos to the dynein complex, but it has made the elucidation of dynein subunit composition and organization, and their roles in dynein functions, more challenging. In addition, splice variations and phosphorylation of DICs and DLICs increase the diversity of cytoplasmic dynein subcomplexes (16–20). In vitro assays have shown that the dynein complex could be separated into two distinct subpopulations. One is a stable subcomplex composed of DHC and DLICs, and the second subcomplex contains DICs and DLCs (10, 21).
We have previously shown that autosomal dominant point mutations causing F580Y and Y1055C substitutions in DHC in the Legs at odd angles (Loa) and Cramping 1 (Cra1) mice, respectively, give rise to a progressive motor deficit in heterozygous Dync1h1+/Loa (referred to hereafter as +/Loa) and Dync1h1+/Cra1 mice (22). In addition, we showed that in cultured motor neurons, isolated from E13.5 homozygous Dync1h1Loa/Loa (referred to as Loa/Loa from this point onward) embryos, this mutation impairs retrograde axonal transport leading to motor neuron degeneration and death of the homozygous pups within a day after birth (22). Subsequent studies by Chen et al. (23) and Ilieva et al. (24) showed significant loss of spinal cord γ motor neurons, as well as proprioceptive sensory neurons in +/Loa mice. Moreover, point mutations in the p150Glued subunit of dynactin have been linked to motor neuron disease in humans (25, 26).
In this study, we have used a cohort of biochemical methods combined with the in vivo mammalian-protein-protein interaction trap (MAPPIT) system (27) to examine the effects of the Loa mutation on the interactions between dynein/dynactin components and on the assembly of dynein. We found that a lighter subcomplex of dynein is enriched in the Loa, at the expense of a heavier subcomplex, and that the association of mutant dynein to dynactin is reduced. In contrast, the binding affinities of dynein to DICs, DLIC1, and Tctex-1 (through DIC) are significantly increased in the Loa mutant protein compared with those in the wild type. These data suggest that the Loa mutation changes the conformation of DHC, modulating its interactions with DICs (and Tctex-1), DLIC1, and dynactin. In addition, we show that the kinesin light chain 1 (KLC1) expression and its interaction with the dynein complex is modified, possibly as a compensatory mechanism in response to impaired retrograde transport in Loa.
EXPERIMENTAL PROCEDURES
The experiments were performed under license from the UK Home Office (Animals Scientific Procedures Act 1986), following local ethical review.
Mice and Tissue Preparation
+/Loa heterozygote female and male mice were intercrossed to produce wild type and Loa/Loa mice, which were identified by genotyping for mutations in the Dync1h1 gene from tail DNA (22). Brains isolated from two or more E13 embryos or newborn pups were homogenized in homogenization buffer, PBS without calcium or magnesium (PBS−) (Invitrogen), supplemented with 1× protease inhibitors (Roche Applied Science), and 10 μl ml−1 phosphatase inhibitors 1 and 2 (Sigma). A volume 9× weight of tissue was used. Following homogenization, samples were centrifuged for 10 min at 16,000 × g at 4 °C for immunoblotting and 20 min for immunoprecipitation. The supernatants were collected and protein concentrations determined using a BCA protein assay kit (Pierce).
Sucrose Density Gradient Centrifugation
Mouse brains were homogenized on ice in a Dounce homogenizer in 4 volumes of PBS− buffer containing protease inhibitor mixture (Roche Applied Science) and phosphatase inhibitor mixture (Sigma), followed by centrifugation at 800 × g. Supernatants were isolated, and their protein contents were quantified using the BCA protein assay kit (Pierce). Density purification experiments used 2 mg of total protein in soluble extracts of mouse embryo brains to sediment through 5 ml of linear density gradient consisting of 5–20% sucrose prepared in PBS− buffer as described in Collins and Vallee (28). The gradients were centrifuged at 237,000 × g for 4 h in an SW55Ti rotor (Beckman Instruments) at 4 °C. Gradient fractions (each 0.45 ml) were collected and stored at −80 °C for subsequent analysis.
Immunoprecipitation
Immunoprecipitation from brain extracts was carried out as described previously (29, 30). Briefly, primary antibody was first allowed to bind protein A-Sepharose beads (Zymed Laboratories Inc.) and then incubated with equal amounts of brain extract of Loa/Loa and wild type overnight at 4 °C. For immunoprecipitation of DHC, anti-DHC antibody was cross-linked to cyanogen bromide-activated Sepharose 4B beads (Sigma) and then incubated with brain extract overnight at 4 °C as above. After washing three times with PBS buffer, proteins were eluted into SDS-PAGE sample loading buffer by boiling for 5 min, and the equal volumes were loaded onto a gel for immunoblot analysis.
For λ-protein phosphatase treatment, the immunoprecipitated dynein was precipitated with an acidified acetone/methanol method to remove SDS. The pellet was treated with λ-protein phosphatase (New England Biolabs) for 1 h at 30 °C or mock-treated in the supplied buffer as a control, in accordance with the manufacturer's instructions.
Preparation of MT-associated Proteins
Cytoplasmic dynein was prepared from brains of newborn mice by a modified taxol-based procedure (28). The tissue was minced in 4 volumes of PHEM buffer (30 mm PIPES, 50 mm HEPES, pH 7.1, 1 mm EGTA, and 2 mm MgCl2) containing 250 mm sucrose, protease inhibitor mixture (Roche Applied Science), phosphatase inhibitor mixture (Sigma), and 0.5 mm dithiothreitol. The homogenate was centrifuged at 16,000 × g for 30 min at 4 °C. The supernatant was recovered and centrifuged at 192,000 × g for 1 h in an SW55Ti rotor (Beckman Instruments) at 4 °C. The supernatant (cytosolic extract) was recovered, and taxol and AMP-PNP were added to 20 μm and 1 mm, respectively. The sample was warmed to 37 °C for 5 min. MT-associated proteins were sedimented at 30,000 × g at 4 °C for 30 min through a cushion of 7.5% sucrose in PHEM buffer. The MTs were washed twice with 500 μl of PHEM sample extract buffer as above but containing 5 μm Taxol and recentrifuged to sediment MTs at 30,000 × g at 4 °C for 30 min. The pellet was resuspended in 200 μl of SDS-PAGE sample loading buffer for subsequent analysis.
RNA Isolation and Illumina Bead Arrays
Two wild type and homozygous Loa E13 mouse brains were collected, and RNA was isolated using the TRIzol reagent (Invitrogen) following the manufacturer's protocol with some modifications. Prior to homogenization with TRIzol, the brains were weighed, snap-frozen in liquid nitrogen, and crushed using a porcelain pestle and mortar until they turned into a fine powder. 1 ml of TRIzol was added per 100 mg of brain weight, and the samples were homogenized using a tight glass Teflon homogenizer. After a 5-min incubation at 25 °C, chloroform was added at a ratio of 1 to 5 to the amount of TRIzol previously used, and the samples were shaken by hand for 15 s and incubated at 25 °C for 3 min. The RNA from each sample was collected after centrifugation at 12,000 × g for 15 min at 6 °C and was transferred in fresh tubes where it was mixed with isopropyl alcohol at a ratio of 1 to 2 to the amount of TRIzol used for the initial homogenization. After 10 min of incubation at 25 °C and centrifugation at 12,000 × g for 10 min at 6 °C, the RNA pellet was washed by vortexing once with 75% ethanol at equal amounts to the TRIzol used in the initial homogenization and centrifuged at 7,500 × g for 5 min at 6 °C. The resulting RNA pellets were air-dried for 10 min, resuspended in 20 μl of RNase-free water, and finally incubated at 57 °C for 10 min before being snap-frozen in liquid nitrogen and stored at −80 °C. 2 μl of each sample were run on a 1% agarose gel alongside a 100-bp ladder to test the integrity of the RNA.
Illumina mouse oligonucleotide arrays (Mouse-6_V1) were used according to the manufacturer's instructions as described previously (31). Briefly, 500 ng of total RNA was processed for each sample, and standard Quality Control was performed prior to analysis. Differential expression values were derived using the Illumina Beadstudio software suite.
Western Blotting
12.5% SDS-PAGE or precast NuPAGE® 4–12% gels were used in the immunoblotting experiment. Electrophoresis was carried out on an XCell SurelockTM mini-cell device (Invitrogen) at room temperature. Proteins were then transferred to polyvinylidene difluoride membrane (GE Healthcare). Blots were probed with anti-p150Glued/135 (1:300; Santa Cruz Biotechnology), anti-DHC polyclonal (1:200; Santa Cruz Biotechnology), anti-p150Glued (1:400; BD Transduction Laboratories), anti-DIC (1:1000; gift from Dr. Kevin Pfister, University of Virginia), anti-DIC1 (1:2000), anti-DLIC1 monoclonal ab72 (1:2000), anti-DLIC1/2 monoclonal ab77 (1:2000), anti-DIC2 (1:200; Santa Cruz Biotechnology), anti-Tctex-1 monoclonal (1:200; gifts from Dr. Kevin Pfister), and anti-KLC1 polyclonal antibody (1:200; Santa Cruz Biotechnology). Proteins were detected using alkaline phosphatase-linked secondary antibodies with the CDP-STAR chemiluminescence system (Sigma) or horseradish peroxidase-linked secondary antibodies with the SuperSignal West Dura extended duration substrate (Pierce).
MAPPIT
MAPPIT was used to quantify the interactions of DIC isoforms with wild type or Loa DHCs, according to the method described by Eyckerman et al. (27, 32). In MAPPIT, a heterologous “bait” polypeptide is fused to a receptor chimera consisting of the extracellular domain of the erythropoietin receptor fused to the transmembrane and cytosolic domains of a leptin receptor variant F3 (LR-F3) that carries tyrosine to phenylalanine mutations that eliminate the functional STAT3 recruitment. A heterologous “prey” polypeptide was fused to a fragment of the gp130 cytokine receptor component, and it contains four functional STAT3 recruitment sites. If bait and prey interact then, upon EPO-induced receptor activation, the gp130 receptor fragment was phosphorylated, and a STAT3-dependent signal was obtained that can be measured using the Firefly luciferase reporter gene. Furthermore, the Renilla luciferase gene, whose expression is independent of prey-bait protein interactions, is also used to account for variances in transfection efficiency.
In this study, DIC isoforms were subcloned into the pMG1-SVT vector (prey). The DHC fragments encompassing the coding sequences for residues 268–992 of wild type and mutant DHCwt and DHCLoa, respectively, were subcloned into the pSel1-FKBP12 vector (bait). All constructs were sequence verified.
HEK293T cells were cultured in 35-mm dishes at a density of 4 × 105 cells/dish and standard DMEM in a 37 °C, 5% CO2 incubator. The MAPPIT assay was performed using the Dual-Luciferase reporter assay system (Promega), following the manufacturer's protocol. The constructs were cotransfected with the following constructs: pSell-DHCwt or pSell-DHCLoa and individual pMG1-DIC isoforms 1A, 1C, 1D, 1E, 2A, 2B, or 2C; Firefly luciferase reporter gene and Renilla luciferase (as a transfection-efficiency control), using Lipofectamine 2000 (Invitrogen). Transfections using the pSel1-p53, pMG1-SVT, Firefly, and Renilla luciferases were used as positive controls for protein-protein interaction. 24 h after transfection, the cells were trypsinized with 0.25% trypsin and were split in 6 wells of equal cell numbers in 96-well plates where half of the samples were stimulated with erythropoietin (EPO) (1 unit per well), and the rest were left untreated to serve as negative controls. Cell lysis was achieved by incubation of the cells with 20 μl per well passive lysis buffer for 15 min on a rocking plate at room temperature. The Firefly and Renilla luciferase activities were measured using LARII and Stop and Glo (Promega), respectively, as priming agents on a Lucy Luminometer (Labtec Instruments). The results were analyzed using the Stingray software.
Immunocytochemistry
HEK293T cells were cultured on glass coverslips in 35-mm dishes at a density of 2 × 105 cells per dish containing standard DMEM in a 37 °C, 5% CO2 incubator. The cells were fixed and permeabilized with 0.5% glutaraldehyde and 0.1% Triton X-100 for 1 min in warmed up CB buffer (137 mm NaCl, 5 mm KCl, 1.1 mm Na2HPO4, 0.4 mm KH2PO4, 2 mm MgCl2, 2 mm EGTA, 5 mm PIPES, 5.5 mm glucose, pH 6.1) to preserve the cytoskeleton, rinsed twice in CB buffer, and incubated for 15 min at room temperature with 1% glutaraldehyde in warmed CB buffer. The cells were rinsed twice in warmed CB buffer followed by a 5-min incubation in 0.5 mg ml−1 sodium borohydride at room temperature with period agitation. The recombinant FLAG-DHCwt or FLAG-DHCLoa were visualized using the anti-FLAG mouse primary antibody (1:400; Sigma) followed by the Alexa Fluor 456 goat anti-mouse IgG secondary antibody (1:200; Molecular Probes). The cells were visualized using a Delta Vision microscope.
Quantitative PCR (qPCR)
For qPCR, RNA was extracted using the NucleoSpin II RNA extraction kit, and cDNAs were generated using the Promega reverse transcription system. Gene sequences were found in the National Center for Biotechnology Information (NCBI) data base and were cross-referenced using a Basic Local Alignment Search Tool (BLAST) to those found on the Protein Knowledgebase (NniProtKB) data base. Primers were designed from this information using the primer design tools (Invitrogen). qPCRs were carried out by adding 2× QuantiTect SYBR Green PCR master mix (Qiagen) to 1 mm each of forward and reverse primers as follows: KLC1 tcttcccaaatgacgaggac and ctgtacaccagggccaagat, and 18S rRNA gccgctagaggtgaaattctt and cattcttggcaaatgctttcg, respectively. 1.5 μg of cDNA was required per reaction, and the overall reaction volume was made up to 30 μl with RNase-free water. A Stratagene Mx4000 qPCR machine was set to denature at 95 °C for 15 min before 40 cycles of 94 °C denaturation for 30 s, annealing at 56 °C for 30 s, and elongation at 72 °C for 30 s. A dissociation curve followed each real time cycle. All data sets were collected from at least three brains for each genotype, and each qPCR was carried out in triplicate.
For analysis, the mean 18S CT for each genotype was subtracted from the CT triplicates of the gene of interest to give a normalized ΔCT. The mean ΔCT of the wild type was then subtracted from the triplicate ΔCT values of the gene of interest for +/Loa and Loa/Loa to give a calibrated ΔΔCT. To calculate a fold change, the ΔCT of the wild type gene of interest triplicates was divided by the mean ΔCT of the wild type resulting in a mean value of 1. A fold change for the other genotypes was calculated by 2−ΔΔCT for each triplicate. Fold changes were analyzed across the experiments using GraphPad and a Mann-Whitney U test of significance.
Quantifications and Statistical Analysis
Film exposures used for quantification were below saturation, as established by multiple exposures. Images were scanned for quantification using an image scanner with software ImageMaster Labscan version 3.01 (Amersham Biosciences). Signal density was summed in the area of the bands, and the intensity of film background was subtracted using ImageQuant TL 2005 (Amersham Biosciences). Statistical analysis was performed using GraphPad Prism. Data were analyzed by Mann-Whitney U test or two-way ANOVA followed by Bonferroni post-tests. Significance was set at p < 0.05.
RESULTS
Loa Mutation Alters the Composition of Cytoplasmic Dynein Complex
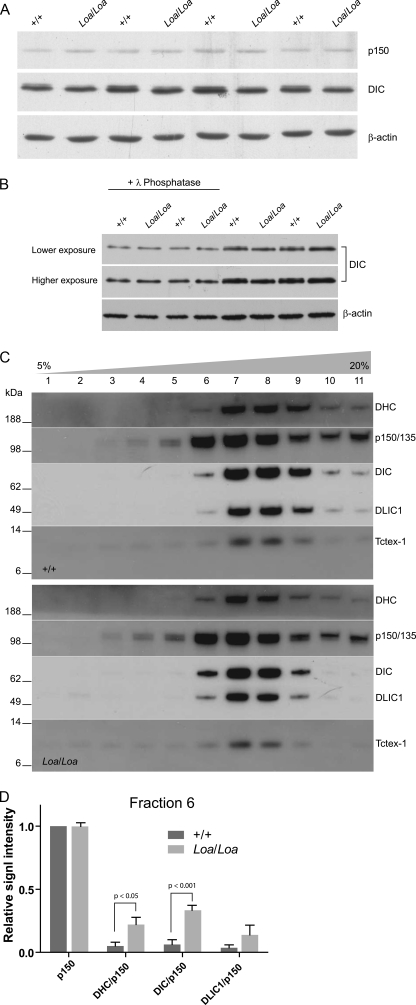
In our Western blot analysis of the brain homogenates for the levels of DHC, DIC, and DLIC, we did not find any difference in protein levels between the wild type and Loa (data not shown). However, we detected a subtle reduction in the intensity of the higher molecular weight variant of DIC in homozygous Loa when compared with the wild type (Fig. 1A). As DIC undergoes phosphorylation, we treated these samples with λ-protein phosphatase to examine whether the observed difference is a result of altered phosphorylation modification of DIC in the Loa. As shown in Fig. 1B, following the phosphatase treatment the higher molecular weight DIC variant almost disappears in both genotypes, indicating that the post-translational phosphorylation of DIC is reduced in Loa.
FIGURE 1.
Loa mutation alters the composition of cytoplasmic dynein complex. A, newborn mouse brains were homogenized in homogenization buffer and then centrifuged at 16,000 × g for 10 min at 4 °C. Equal quantities were loaded onto 12.5% gel for immunoblotting. The blot was probed with antibodies against p150Glued, DIC, and β-actin. There is a subtle reduction in the intensity of the higher molecular weight variant of DIC in homozygous Loa. B, newborn mouse brains were homogenized in homogenization buffer without phosphatase inhibitor and then centrifuged at 16,000 × g for 10 min at 4 °C. The supernatants were treated with (first 4 lanes) or without (last 4 lanes) λ-phosphatase, and the equal quantities were loaded on to 12.5% gel for immunoblotting. The blot was probed with antibodies against DIC and β-actin. This result revealed that the subtle reduction of the higher molecular weight DIC variant in Loa is due to post-translational phosphorylation. C and D show that the Loa mutation alters the composition of cytoplasmic dynein complex. C, 5–20% sucrose gradients were loaded with 2 mg of protein of wild type (+/+, top) and Loa/Loa (bottom) cytosolic extracts of newborn mouse brain. Equal volumes of fractions were examined in 4–12% gradient SDS-PAGE followed by immunoblotting. The blot was probed with antibodies against DHC, p150Glued/135, DIC, DLIC1, and Tctex-1. Denser fractions are to the right. The results reveal that the dynein complex slightly shifts to lighter fractions in Loa. D, bar graph shows the densitometry quantification and analysis of the bands in fraction 6 in C normalized against p150Glued signals.
Phosphorylation of DIC plays an important role in the regulation of cytoplasmic dynein function. Thus, we asked whether the Loa mutation affects the organization of dynein and the integrity of the dynein-dynactin complex. Therefore, we performed sucrose density gradient (5–20%) sedimentations on equal amounts of brain homogenates, isolated from 1-day-old (P1) Loa/Loa and wild type mice. Western blot analysis of the gradient fractions revealed that as expected most of the dynein complex sedimented at 20 S (Fig. 1C). However, we consistently observed a slight shift of DHC, DIC, and DLIC components of the dynein complex toward the lighter fraction 6 in the Loa/Loa homogenates (Fig. 1C, lane 6). In contrast, fractions 9 and 10 showed clear reductions of the signal intensities in these proteins in the Loa/Loa samples compared with the wild type (Fig. 1C, lanes 9 and 10). Distribution and levels of p150Glued, however, remained similar between the genotypes in all fractions.
As p150Glued levels appeared to be almost identical giving a value of 1.06 for the p150Loa/Loa/p150+/+ ratio in fraction 6, we normalized the signals against p150Glued for each genotype (Fig. 1D). This analysis revealed that the relative signal intensities for DHC were 22.0 ± 5.7% (mean ± S.E., n = 4) in Loa/Loa but 5.1 ± 3.0% (mean ± S.E., n = 4) in the wild type (Fig. 1D). Relative DIC signal intensities were 33.3 ± 4.0% (mean ± S.E., n = 4) in Loa/Loa but only 6.2 ± 3.8% (mean ± S.E., n = 4) in the wild type (Fig. 1D). Moreover, DLIC1/p150Glued ratios showed the same trend, 13.8 ± 7.7% (mean ± S.E., n = 4) in Loa/Loa and 3.6 ± 2.4% (mean ± S.E., n = 4) in the wild type (Fig. 1D). Two-way analysis of variance (ANOVA) followed by Bonferroni post-tests revealed that there is a significant difference between the genotypes (p = 0.0001, residual degrees of freedom = 24) and that the differences in DHC and DIC were statistically significant (p < 0.05 and p < 0.001, respectively). We also analyzed the DIC/DHC and DLIC1/DHC ratios between the genotypes, but they did not show any significant changes in Loa/Loa versus wild type samples in this fraction (data not shown), suggesting that the stoichiometry of these subunits in fraction 6 of the Loa samples is similar to that in the wild type. Because dynein light chain Tctex-1 was present in low amounts in fraction 6, reliable quantitative data could not be obtained from this subunit in these assays.
Impaired Interactions of DHC/DIC and Dynein/Dynactin in the Loa
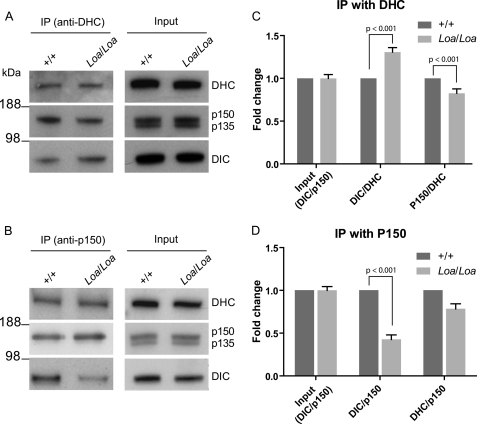
The above data prompted us to examine the DHC-DIC associations and the interaction of the p150Glued subunit of dynactin with the dynein complex in more detail. We used anti-DHC and anti-p150Glued antibodies to pull down these polypeptides and their associated proteins in a series of immunoprecipitation assays (Fig. 2). Western blot analysis revealed that when we immunoprecipitated the dynein-dynactin complex with anti-DHC antibody there was consistently more DIC but less p150Glued present in the Loa/Loa samples compared with the wild type (Fig. 2A). On the other hand, when we used anti-p150Glued antibody to pull down the complex, we reproducibly detected less DIC and DHC in Loa/Loa samples compared with the wild type (Fig. 2B).
FIGURE 2.
Altered interactions of DHC/DIC and dynein/dynactin in Loa. A, antibodies against DHC were cross-linked to cyanogen bromide-activated Sepharose 4B beads to coimmunoprecipitate both the dynein DIC subunits and dynactin from cytosolic extracts of newborn mouse brains. There was no difference between +/+ and Loa/Loa in the precleaned brain homogenates, as control (right panel, Input), but differences between +/+ and Loa/Loa in IP DIC and p150Glued are present (left panel). B, antibody against dynactin p150Glued coimmunoprecipitated both DHC and DIC subunits from cytosolic extracts of newborn mouse brains. There was no difference between +/+ and Loa/Loa in the precleaned brain homogenates (right panel), but immunoprecipitated DIC and DHC were different between +/+ and Loa/Loa (left panel). C, ratios of DIC/DHC and p150Glued/DHC in Loa/Loa were compared with those of +/+ by densitometry quantification of DIC and p150Glued bands in A. Two-way analysis of variances (ANOVA) and Bonferroni post-tests indicated that there is a significant difference between the genotypes (p < 0.0001, residual degrees of freedom = 19) and that the fold changes in Loa/Loa DIC/DHC and p150Glued/DHC ratios are significantly different relative to +/+ (p < 0.001 for both ratios). D, DIC/p150Glued and DHC/p150Glued ratios in Loa/Loa were compared with those in +/+ by densitometry quantification of DIC, DHC, and p150Glued bands in B. ANOVA and Bonferroni post-tests showed that the genotype significantly affects the ratios (p < 0.0001, residual degrees of freedom = 22) and that the fold change in Loa/Loa DIC/p150Glued ratio is significantly different relative to +/+ (p < 0.001). DHC/p150Glued ratios were not significantly different (p > 0.05), but they showed the same trend as DIC/p150Glued ratios.
Quantitative analysis of the input samples was carried out by taking the DIC/p150Glued ratios for each genotype and measuring fold changes against the wild type. As shown in Fig. 2, C and D, for input control, this ratio for the Loa is 0.998 ± 0.05 (mean ± S.E.) of that in the wild type, indicating that same amount of proteins were used in the IPs. For quantification analysis of the signals in the IP with anti-DHC antibody, we took the DIC/DHC and p150Glued/DHC ratios for each genotype and measured the fold changes against those in the wild type. This analysis showed that the DIC/DHC ratio was 1.3 ± 0.05 (mean ± S.E., n = 3)-fold higher in the Loa/Loa than that in the wild type (Fig. 2C) and that this difference was statistically significant (p < 0.001). Conversely, the p150Glued/DHC ratio was 1.2 ± 0.05 (mean ± S.E.; n = 3, p < 0.001)-fold lower in the Loa/Loa relative to that in the wild type (Fig. 2C).
Analysis of the signals in the IP with p150Glued antibody, using the same approach but applying DIC/p150Glued and DHC/p150Glued ratios, revealed that these ratios were 2.3 ± 0.05 (mean ± S.E.; n = 8, p < 0.001)- and 1.3 ± 0.06 (mean ± S.E.; n = 2, p > 0.05)-fold lower in Loa/Loa compared with that in the wild type, respectively (Fig. 2D). These differences were statistically significant except for DHC/p150Glued ratios, which nonetheless showed the same trend as the DIC/p150Glued ratios.
Collectively, these data suggest that the Loa mutation in DHC enhances the affinity of this subunit to DIC, and consequently, the DIC-p150Glued interaction is compromised in the Loa.
Mutated DHC Enhanced the Interaction of DIC and DLIC1 with DHC
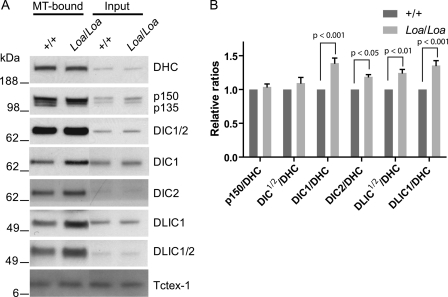
To further investigate the interactions between DHC and its protein partners, we cosedimented the cytoplasmic dynein-dynactin complex with MTs from the cytosolic fraction of newborn mouse brain tissues. We used taxol to stabilize the MTs and AMP-PNP to promote binding of dynein to the MTs. Equal total protein quantities were examined in 4–12% gradient SDS-PAGE followed by comparative Western blot analysis of dynein components in Loa/Loa versus wild type. The blots were probed with antibodies against DHC, p150Glued/p135, DIC1/2, DIC1, DIC2, DLIC1, and DLIC1/2. As shown in Fig. 3A, cytoplasmic dynein and p150Glued/p130 subunits of dynactin were successfully cosedimented with the MTs.
FIGURE 3.
Mutated DHC enhanced the interaction of DIC and DLIC1 with DHC. A, microtubule-associated proteins were prepared from newborn mouse brains. Dynein complex was enriched in the microtubule pellet (MT proteins). Equal total protein was loaded in each lane (MT proteins in first two lanes; input in last two lanes). The blot was probed with antibodies against DHC, p150Glued/135, DIC1/2, DIC1, DIC2, DLIC1, DLIC1/2, and Tctex-1. B, quantification of dynein-complex composition in MT-associated proteins and comparison of +/+ with Loa/Loa in A. Two-way ANOVA followed by Bonferroni post-tests revealed that the fold change in the p150Glued/DHC ratio in MT-bound dynein-dynactin complex is not statistically different between +/+ and Loa/Loa; but those of DIC1/2/DHC, DIC1/DHC, DIC2/DHC, DLIC1/2/DHC, and DLIC1/DHC ratios are significantly higher in Loa/Loa compared with those in +/+.
Signals from the “input” indicated that equal amounts of protein were used in the pulldown. We analyzed the amounts of the MT-associated dynein and dynactin subunits in Loa/Loa or wild type by taking their ratios over DHC levels for each sample, and we quantified them relative to those in the wild type (Fig. 3B). Two-way ANOVA analysis showed that the genotype has a significant effect (p = 0.0001, residual degrees of freedom = 48) on the overall fold changes in protein ratios. There were no significant differences (p > 0.05; n = 5) in the p150/DHC ratios between the genotypes (Fig. 3, A and B). Using the DIC74.1 pan-antibody that recognizes all DIC polypeptides (DIC1/2), we observed a trend in enhanced affinity of mutant DHC to DICs manifested as a 1.1 ± 0.08 (mean ± S.E.)-fold increase in Loa/Loa DIC/DHC ratio relative to the wild type, but Bonferroni post-tests did not show this difference as statistically significant (p > 0.05; n = 5) (Fig. 3, A and B).
However, when we used anti-DIC1- and anti-DIC2-specific antibodies, the ratios were 1.4 ± 0.07 (mean ± S.E.)- and 1.2 ± 0.03 (mean ± S.E.)-fold, respectively, higher in Loa/Loa compared with the wild type. These differences were statistically significant (p < 0.001 for DIC1 and p < 0.05 for DIC2; n = 5). In addition, DLIC1/2/DHC and DLIC1/DHC ratios were 1.25 ± 0.05 (mean ± S.E.)- and 1.36 ± 0.07 (mean ± S.E.)-fold higher in the Loa/Loa compared with those in the wild type, respectively (Fig. 3B). Statistical analysis of these data also revealed significant differences between the genotypes (p < 0.01 for DLIC1/2 and p < 0.001 for DLIC1; n = 5). These data are consistent with the results from the immunoprecipitations with anti-DHC antibody (Fig. 2), supporting the notion that the DIC-DHC and DLIC-DHC interactions are enhanced in Loa/Loa.
Dynein Light Chain Tctex-1 Is More Tightly Associated with DIC in Loa/Loa
The assembly of dynein subunits is an interdependent process, and there is evidence implicating DIC and DLCs in stabilizing the dynein complex (10). The above data suggested that the Loa mutation in the overlapping DIC and DLIC binding domains of DHC compromises not only the DIC-DHC and DLIC-DHC interactions but also those of dynein-dynactin. Moreover, some studies have suggested that the stability of DIC dimers and their binding efficiencies to dynactin and DHC are enhanced by conformational changes to the DIC polypeptides, which are brought about by DLCs (11, 13, 33–36).
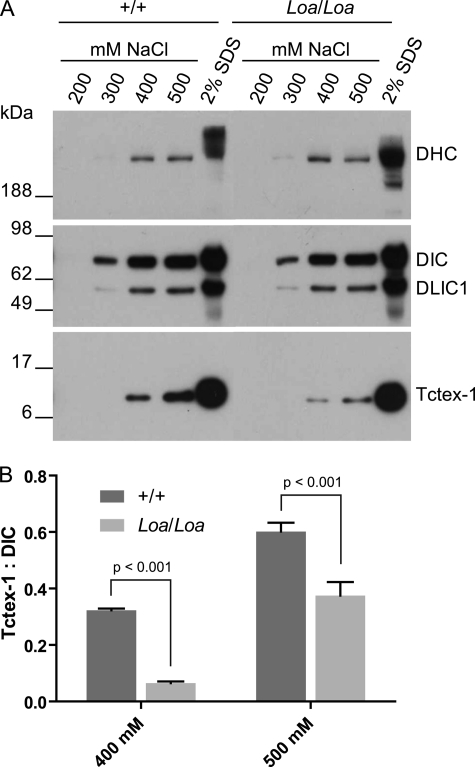
Thus we asked whether the altered DIC-DHC binding properties have any effects on the DLC-DIC interactions. To this end, we carried out salt extraction assays on dynein complex immunoprecipitated with the anti-DIC74.1 antibody followed by Western blot analysis detecting for Tctex-1, DHC, DIC, and DLIC. We used 200–500 mm NaCl for stepwise extraction of immunoprecipitated dynein subunits. The results of these assays revealed that the dynein complex pulled down by the anti-DIC antibody was stable in 200 mm NaCl and that even in 300 mm NaCl only a small part of dynein polypeptides were released from the beads (Fig. 4A). Interestingly, we observed differences between Loa/Loa samples and the wild type at higher salt concentrations (Fig. 4A).
FIGURE 4.
Dynein light chain Tctex-1 is more tightly associated with DICs in Loa/Loa. A, dynein complex was immunoprecipitated from cytosolic extracts isolated from newborn mouse brains using anti-DIC antibody. Proteins were sequentially eluted from the Sepharose 4B beads by stepwise increases in NaCl concentration from 200 to 500 mm, followed by a final extraction with 2% SDS-PAGE loading buffer. Equal volumes of the eluates were loaded on to 4–12% gradient SDS-polyacrylamide gels followed by electrophoresis and immunoblotting to detect DHC, DIC, DLIC1, and Tctex-1. The results revealed that the dynein complex is stable in 200 mm NaCl, and only a small part of dynein polypeptides was released from the beads in 300 mm NaCl. However, much more Tctex-1 was released from the beads at 400 and 500 mm NaCl in +/+ than in Loa/Loa. B, Tctex-1/DIC ratio of Loa/Loa was compared with that of +/+ by densitometry quantification of Tctex-1 and DIC bands. The Tctex-1/DIC ratios are significantly different between Loa/Loa and +/+ at 400 mm NaCl (p < 0.001) and 500 mm NaCl (p < 0.001), n = 4.
Quantification analysis of these data showed that the amounts of extracted DHC and DLIC relative to DIC were not significantly different between the genotypes. But the Tctex-1/DIC ratios were significantly different between the Loa/Loa and wild type at 400 (p < 0.001) and 500 mm (p < 0.001) salt concentrations. They were 0.32 ± 0.01 (mean ± S.E.) in wild type compared with 0.07 ± 0.01 (mean ± S.E.) in Loa/Loa at 400 mm and 0.60 ± 0.03 (mean ± S.E.) in wild type compared with 0.38 ± 0.05 (mean ± S.E.) in Loa/Loa at 500 mm salt concentrations (Fig. 4B). These data therefore suggest that the Tctex-1 polypeptides are more stably associated with DIC in Loa/Loa compared with that in the wild type and that the Loa mutation indeed affects the organization of dynein complex.
Loa Mutation Enhances the DHC-DIC Interactions in Vivo
To confirm the above biochemical data, we utilized the in vivo MAPPIT system and the luciferase activity as a reporter for protein-protein interactions in this system, as described under “Experimental Procedures.” We examined the interactions of a fragment encompassing residues 268–992 of mutant or wild type DHC with a set of DIC isoforms (37) that we had cloned into the MAPPIT vectors. This approach allowed us to quantify the level of interactions of DIC isoforms with the wild type and mutant fragments of DHC (DHCwt and DHCLoa, respectively), under physiological conditions. We used these DHC fragments as they span the mutation site at residue 580 and because the full-length DHC is 4644 amino acids long, making the cloning and transfection of mutant and wild type recombinant constructs extremely complicated.
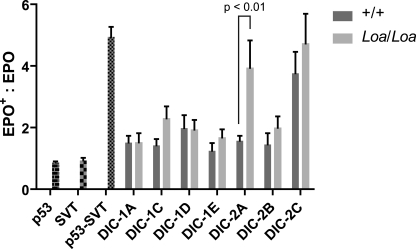
We used the Renilla luciferase construct, which constitutively expresses Renilla luciferase, as a control for transfection efficiencies and against which we could normalize the expression of EPO-induced Firefly luciferase in the transfected cells. Analysis of the levels of the Firefly luciferase activation in HEK293T cells cotransfected with the bait-DHCwt or bait-DHCLoa fragments, and the prey-DIC1 isoforms A and C–E or prey-DIC2 isoforms A–C showed reporter gene activation after EPO stimulation (supplemental Fig. 1). For quantification analysis of the protein-protein interactions in these assays, we measured the ratios of Firefly luciferase activity in the cells stimulated with EPO (EPO+) over those in unstimulated (EPO−) cells. As shown in Fig. 5, there is a trend in higher Firefly luciferase activity in the cells cotransfected with individual DICs and the mutant DHC, compared with those cotransfected with the wild type DHC (Fig. 5).
FIGURE 5.
Mutant DHC interactions with DIC-2A isoform is significantly enhanced in vivo. Firefly luciferase reporter construct was cotransfected with p53 bait alone, SV40 large T antigen (SVT) prey alone, or both constructs together in HEK293T cells as controls. Cotransfection of p53 bait and SV40 large T antigen prey resulted in marked increase in the reporter gene activity. Cotransfection of the constructs harboring wild type or mutant DHC bait with individual DIC isoform preys produced varying levels of the reporter gene activation, but mutant DHC interaction with DIC-2A isoform gave rise to a significant increase (p < 0.01) in the reporter gene activation when compared with the interaction between wild type DHC and this isoform.
ANOVA and Bonferroni post-test analyses on these data confirmed this trend and revealed a significant overall effect exerted by the genotype (p = 0.0073, residual degrees of freedom = 87). Moreover, cells that harbor the mutant DHC fragment and DIC-2A isoform showed the most striking (∼2.5-fold) and statistically significant (p < 0.01; n = 9) increase in the Firefly luciferase activity, compared with those harboring the wild type DHC (Fig. 5). These data indicate that the affinity of mutant DHC to DIC isoforms, in general, and that of mutant DHC to DIC2A, in particular, is enhanced. In addition, the overall trend in increased affinity between DIC and the mutant DHC versus the wild type supports the above biochemical data (Fig. 2A).
Interaction of Cytoplasmic Dynein with Kinesin Is Altered in the Loa
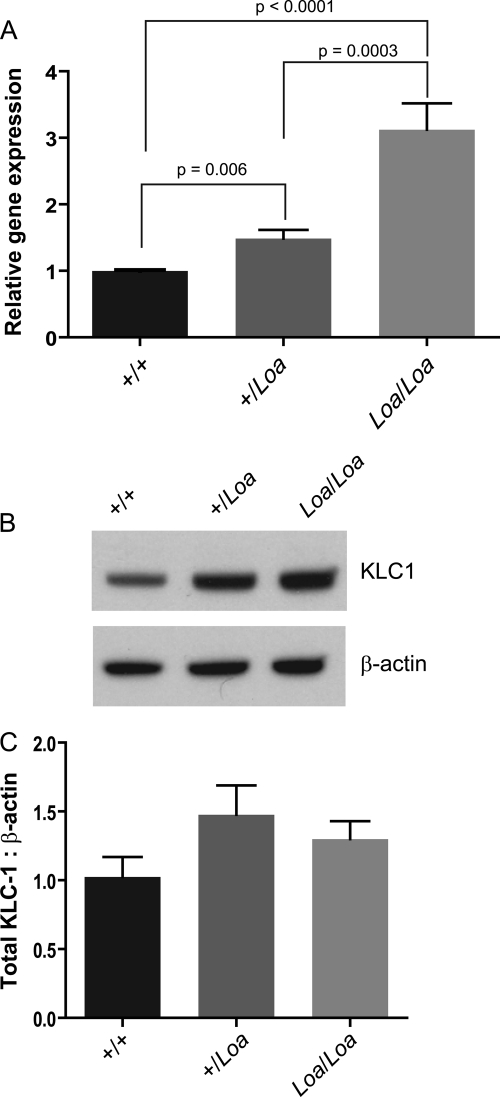
Interaction between cytoplasmic dynein and the plus end-directed motor kinesin I has been shown to be mediated by direct interactions between DIC and kinesin light chains (KLCs) (38, 39). Our microarray analysis of brain tissues isolated from E13.5 homozygous Loa versus that of the age-matched wild type embryos highlighted the modified expression of KLC1 in the Loa. We confirmed the data by performing qPCR and showed that there is a Loa mutation dose-dependent increase in KLC1 transcripts in Loa/Loa and +/Loa versus that of the wild type (Fig. 6A). Protein expression analysis of KLC1 in E13 brains also showed a trend for up-regulation in both +/Loa and Loa/Loa (Fig. 6, B and C).
FIGURE 6.
KLC1 is up-regulated in Loa. A, RNA was extracted using a NucleoSpin II RNA extraction kit from at least three E13 brains from each genotype. cDNA was generated with the Promega reverse transcription system, SYBR Green master mix, and a Stratagene Mx4000 qPCR system was used for analysis. qPCR analysis revealed up-regulation of KLC1 in +/Loa (p = 0.006) and Loa/Loa (p < 0.0001) compared with wild type and between +/Loa and Loa/Loa (p = 0.0003). B and C, protein analysis also revealed a trend for up-regulation in +/Loa and Loa/Loa; n = 4. E13 brains were homogenized in homogenization buffer and then centrifuged at 16,000 × g for 10 min at 4 °C. Equal quantities were loaded onto NuPAGE 4–12% gradient gel for immunoblotting.
As our biochemical and in vivo data had shown altered interactions between mutant DHC and other dynein-dynactin components, we asked whether the Loa mutation affected the interaction between DIC and KLC1 as well. To this end, we carried out MT binding and immunoprecipitation assays and detected for DIC and KLC1 polypeptides.
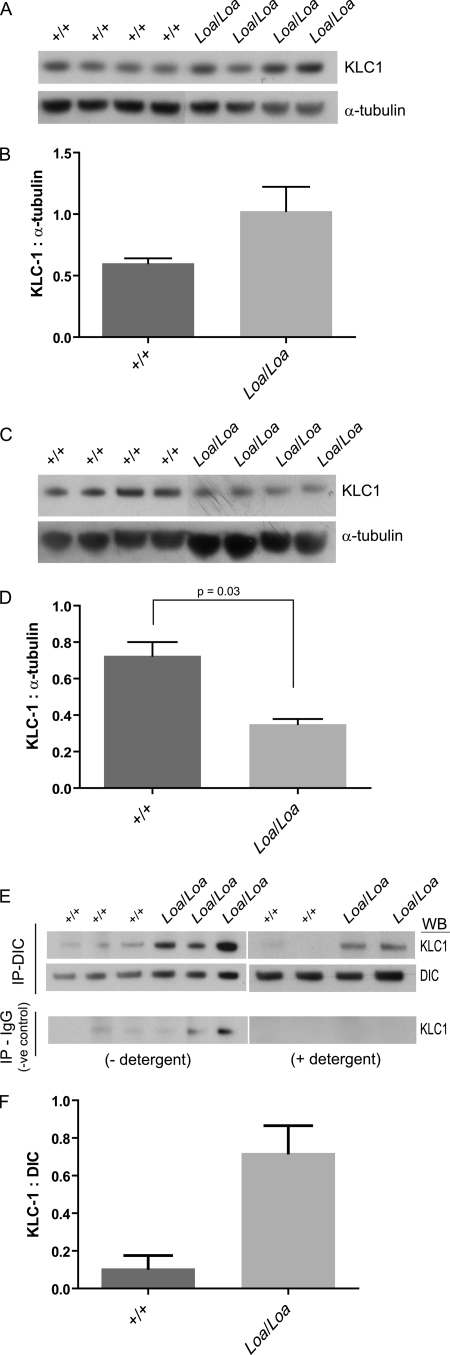
KLC1 present in the supernatant (unbound) of P1 brain homogenates following a MT binding assay was greater in Loa/Loa than in wild type, almost to significance (p = 0.0530) (Fig. 7, A and B). Complementary to this, the KLC1 pulled down with the MTs was significantly less (p = 0.03) in the Loa/Loa compared with wild type (Fig. 7, C and D). Surprisingly, immunoprecipitation of KLC1 with DIC showed a higher amount of KLC1 cosedimenting with DIC in Loa/Loa when compared with wild type (Fig. 7, E and F), indicating that there are perhaps both local and global mechanisms at work to restore the balance of inadequately distributed organelles, neurotrophic factors, and motor proteins in Loa/Loa.
FIGURE 7.
Association of KLC1 with microtubules is reduced in Loa/Loa. Tissue was prepared in PHEM buffer (see “Experimental Procedures”). The homogenates were then centrifuged at 16,000 × g for 30 min at 4 °C. The supernatant was recovered and centrifuged at 192,000 × g for 1 h at 4 °C. Taxol and subsequently AMP-PNP were added. Microtubule-associated proteins were sedimented at 30,000 × g at 4 °C for 30 min through a cushion of 7.5% sucrose in PHEM buffer. Microtubules were resuspended in PHEM sample extract buffer containing 5 μm Taxol and recentrifuged to sediment the microtubules. The pellet was resuspended in SDS-PAGE sample loading buffer, and equal volumes were loaded for Western blot (WB) analysis. A and B, there was more KLC1 in the supernatant of Loa/Loa following microtubule-associated protein purification when compared with wild type. C and D, KLC1 pulled down with microtubules was greater in wild type compared with Loa/Loa (p = 0.03). E and F, immunoblotting of KLC1 pulled down with DIC and with mouse IgG, as negative control. Homogenized E13 brains were centrifuged at 16,000 × g for 10 min at 4 °C before supernatants were incubated with Sepharose protein A beads with agitation for 1 h to pre-clear. Anti-DIC or mouse IgG (as negative control) antibodies were bound to Sepharose protein A beads at a concentration of 1.2 μg per 1.3 mg of protein. Beads were then washed in PBS and incubated overnight with the pre-cleared homogenates. Beads were washed with either PBS or PBS− plus 0.01% detergent (Tween 20) to eradicate nonspecific binding. Detergent washes did, however, reduce the overall KLC1 detected. There was no DIC or KLC1 detected bound to the IgG negative controls with detergent washes. More KLC1 was pulled down with DIC in the Loa/Loa compared with wild type (F).
Population of DHC and DLIC1 Polypeptides Form Phosphorylation-dependent Supercomplexes
In our investigations into the dynein complex stability, we noticed that when the complex is immunoprecipitated with anti-DIC 74.1 antibody, the anti-DLIC1 antibody detects large protein species that are similar in size to DHC in transfer blots from SDS-PAGE assays (supplemental Fig. 2A). Conversely, probing the blots with the anti-DHC antibody revealed a band in the DLIC region (supplemental Fig. 2B). We suspected that a fraction of the DLIC1 polypeptides are strongly associated with DHC in the form of supercomplexes, which migrate differently to denatured/linear DHC. We therefore analyzed the protein contents of gel slices along a lane of the SDS-PAGE from these IPs using mass spectrometry Q-TOF. We detected DIC and DLCs (including Tctex-1 and LC8) only in the gel slices that corresponded to the molecular weights of these polypeptides. Interestingly, however, there were peptide hits for DLIC1/2 and DHC in the high molecular weight region of the gel as well as in the 50–60-kDa region corresponding to DHC and DLIC polypeptides, respectively (data not shown). Moreover, the number of DHC peptide hits in the 50–60-kDa region was a small fraction of those in the high molecular weight region, but they span the full length of the DHC, suggesting that a small population of the DHC polypeptides forms supercomplexes with DLIC polypeptides. To ascertain whether this interaction is phosphorylation-dependent we treated the pulldowns with λ-protein phosphatase under native conditions. Interestingly, almost all the DLIC signals in the high molecular weight region disappeared and that of the DLIC1 increased (supplemental Fig. 3A). Similarly, the DHC signal in the lower band shifted upwards (asterisk in supplemental Fig. 3B), but the DHC signal in the 50–60-kDa region remained unchanged. However, there was no difference between Loa and wild type in these assays.
DISCUSSION
The neurodegenerative consequence of the F580Y substitution in the Loa mouse is intriguing as it is within the highly conserved tail domain of DHC, the largest subunit of the cytoplasmic dynein complex and where DHC homodimerization as well as DIC and DLIC binding occur (2, 22, 40). DICs function as adaptors for dynactin, hence mediating membranous-cargo transport, and they also act as intermediary subunits for DLC and other small protein binding.
The location of the Loa mutation suggests that this mutation could impair the homodimerization of the dynein complex. Our data, however, do not support this possibility, because in density gradient assays we did not observe sedimentation of any dissociated heavy chains at ∼12 S (41) in the mutants, but the dynein complex sedimented at the 20 S region in both wild type and mutant samples as expected (Fig. 1) (28, 42).
Biochemical studies on dynein components have shown that the DIC-DLC subcomplex could be isolated as two pools as follows: one almost exclusively composed of IC2 gene products and few DLCs, and the other containing equal amounts of DIC1 and DIC2 gene products plus four DLCs (10). These multiple DIC phosphorylations, alternative splice sites, and multiple possible DLC compositions result in a heterogeneous population of dynein molecules for specific interactions with a wide range of cargos. Impairment of components of these pools could explain the unique phenotype observed in Loa mice (22).
The shift in the sedimentation of dynein components from denser sucrose gradient fractions to lighter fraction 6 in the Loa samples suggests that a fraction of a subpopulation of dynein complexes with larger components is depleted in the Loa. Moreover, the stepwise dissociation of the dynein subunits with NaCl indicated that the dynein light chain Tctex-1 is more tightly associated with DIC in Loa than in wild type and that consequently and/or concurrently these compromise the p150Glued binding, and thus association of dynactin, to dynein in Loa.
Previously, we used the GST-pulldown method to investigate the interaction of mutant versus wild type DHC with DIC isoforms, using 342-amino acid fragments of the DHC encompassing the Loa mutation site (43). These nonquantitative assays, however, did not show any significant difference between the mutant and wild type DHC interactions with the DIC isoforms (43). Our current in vivo analysis using MAPPIT, however, supports increased affinity of the DICs with DHC in Loa relative to the wild type.
MAPPIT requires subcloning of interacting polypeptides into bait and prey vectors. As subcloning and expression of the large DHC transcripts, wild type and Loa, are exceptionally problematic, we subcloned an ∼2-kb fragment of the wild type and Loa cDNA, encoding 725-amino acid polypeptides and spanning the Loa mutation site, into the MAPPIT vectors. Interaction between p53 and SV40-large-T antigen, used as controls in these assays, produces a significant activation of the reporter gene Firefly luciferase. In the samples, however, the activation of the reporter gene was not as much as the controls. This could be caused by the obligatory use of a nonfunctional fragments of the DHC in these assays.
Interestingly, when we switched the DHCs to the prey vector, for cytosolic expression under the strong SRα promoter, and the DICs to the bait vector, for SV40-promoter-driven expression and translocation to the plasma membrane, we observed no activation of the reporter gene. An explanation for the lack of the reporter gene activation came to light after immunocytochemical analysis revealed aggregation of the DHC fragments in the transfected cells, which predictably would not be interacting with the bait-DICs on the plasma membrane (supplemental Fig. S3). The MAPPIT data, however, indicate a general trend in increased affinity of all, except IC-1A and IC-1D, to mutant DHC. Moreover, the interaction between mutant DHC and DIC-2A was markedly significant (Fig. 5).
King et al. (10) have demonstrated that there are strong affinities between dynein DHCs and DLICs to form a stable and exclusive dynein subcomplex. Our data support this finding, and we show that a phosphatase-sensitive DHC-DLICs subcomplex remains intact even after boiling in Laemmli sample buffer (supplemental Fig. 3). Interestingly, it appears that the Loa mutation enhances the affinity of DLIC1 and possibly DLIC2 to the dynein complex (Fig. 3).
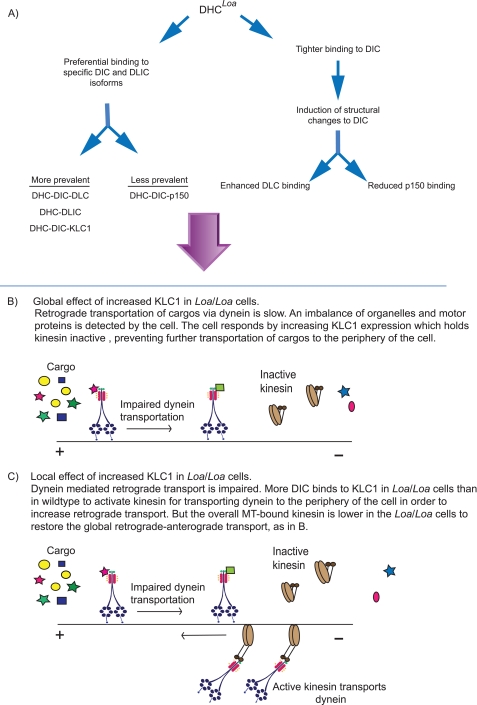
Our immunoprecipitation, MT binding, and in vivo assays demonstrate increased affinity of mutant DHC to DICs-Tctex1 and DLICs but reduced interactions with the p150Glued subunit of dynactin. Collectively, these data support the notion that the Loa mutation confers structural changes to DHC to induce two possible consequences, with regard to the assembly of dynein complex, as depicted in Fig. 8A. 1) The mutant DHC preferentially binds to specific DIC and DLIC isoforms, and the DHCLoa-bound DIC isoforms are mainly associated with DLCs and KLC1 than with p150Glued, leading to an imbalance in abundance of dynein subcomplexes and subsequently the inadequate presence of dynactin-associated dynein in the Loa. 2) Upon tighter binding of the DIC to mutant DHC, the Loa mutation confers structural changes to the DIC and promotes enhanced affinity to Tctex-1 and reduced binding to p150Glued. This is a plausible possibility as the binding domains for Tctex-1 and p150Glued on DIC are in close proximity (2).
FIGURE 8.
Model. Figure shows a schematic representation of the effects of the Loa mutation on the dynein complex and its likely consequences on the regulation of the anterograde transport.
Indeed, disruption of dynactin in postnatal motor neurons impairs retrograde transport and the long term survival of motor neurons in vivo (44). In addition, dynactin is proved to improve dynein motor activity by increasing its processivity in vitro (45), and isolated DHC-DLIC subcomplex reduced the microtubule gliding activity (46). Hence, the results of impaired dynein-dynactin and higher affinity of DHC-DLIC in this study may explain the results found by Hafezparast et al. (22), which showed the frequency of the high speed carriers was reduced and the stationary pauses were increased in retrograde transport of a fluorescently labeled fragment of the tetanus toxin in Loa/Loa motor neurons.
qPCR analysis showed a significant up-regulation of KLC1 expression in both +/Loa and Loa/Loa when compared with wild type. Protein expression has also shown a trend in this direction. The majority of kinesin is found in the soluble cytosolic pool where, at a physiological ionic state, it is held in an inactive, folded conformation, where the tail domain binds the heavy chain head, preventing unnecessary energy expenditure by inhibiting ADP release and interactions with microtubules (47, 48). KLCs have been shown to have a role in stabilizing and regulating this state by pushing apart the two dimerized heavy chain motor domains (47). KLC1 binds kinesin heavy chain stalk and tail through its amino-terminal heptad repeats. Cargo bound to KLC through the tetratricopeptide repeats of KLCs at the carboxyl terminus may induce the inactive conformation of kinesin to unfold into an active state allowing microtubule binding and hydrolysis of ATP (47, 49, 50).
In the Loa/Loa mouse, retrograde transport kinetics are impaired (22), likely leading to an imbalance of neurotrophic factors and an increase of dynein at the microtubule plus end. An elevated expression of KLC1 to hold kinesin heavy chain inactive may compensate for this imbalance by inhibiting further plus end transportation (Fig. 8B). Our data from purified microtubule-bound proteins support this theory as more KLC1 is present in the supernatants of Loa/Loa compared with wild type, and there is more KLC1 bound to microtubules for transportation in the wild type (Fig. 7, A–D).
KLC isoforms have been shown to determine specificity for cargos, and dynein has been shown to interact with KLC through DIC (38, 51). Immunoprecipitation revealed a greater association of KLC1 with DIC in Loa/Loa compared with wild type (Fig. 7, E and F). Thus, at a local level in the Loa/Loa mouse, detection of reduced retrograde transport/trophic signaling may signal for the cell to send more dynein to the cell periphery in preparation of retrograde transport. The interaction between KLC1 and DIC would enable kinesin complex activation for this to occur (Fig. 8C). A wider regulatory mechanism may prevent the anterograde transport of further cargo to the cell periphery to try and restore the global balance as discussed above across the cell as a whole (Fig. 8B).
Future structural analysis of the mutant DHC and its interactions with DIC and DLIC, plus identification of specific cargos whose transport are compromised in the Loa, will provide clues into the molecular basis of the motor and sensory neurodegeneration phenotype in the Loa mice. In addition, further examination of kinesin dynamics and the anterograde transport in the Loa mice could highlight the global regulatory mechanisms that control axonal transport.
Supplementary Material
Acknowledgments
We are grateful to Professor Jan Tavernier and in particular Dr. Sam Lievens (Cytokine Receptor Lab, Albert Baertsoenkaai, 9000 Ghent, Belgium) for the MAPPIT constructs and technical advice regarding the MAPPIT technique. We also thank Dr. Kevin Pfister (Department of Cell Biology, School of Medicine, University of Virginia) for the anti-DIC and anti-Tctex-1 antibodies and Dr. Mark Willett (School of Life Sciences, University of Sussex) for advice on qPCR analysis.
This work was supported by Biotechnology and Biological Sciences Research Council (to M. H., W. D., and C. G.), the Wellcome Trust (to G. B.), the ENDOCYTE Research and Training Network funded by the European Union (to E. M. C. F.), and the University of Sussex (to V. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- MT
- microtubule
- ANOVA
- analysis of variance
- DHC
- cytoplasmic dynein heavy chain
- DIC
- cytoplasmic dynein intermediate chain
- DLC
- cytoplasmic dynein light chain
- DLIC
- cytoplasmic dynein light intermediate chain
- EPO
- erythropoietin
- KLC1
- kinesin light chain 1
- MAPPIT
- mammalian-protein-protein interaction trap
- SVT
- SV40 T large antigen
- qPCR
- quantitative PCR
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- IP
- immunoprecipitation.
REFERENCES
- 1.Pfister K. K. (1999) Mol. Neurobiol. 20, 81–91 [DOI] [PubMed] [Google Scholar]
- 2.Vallee R. B., Williams J. C., Varma D., Barnhart L. E. (2004) J. Neurobiol. 58, 189–200 [DOI] [PubMed] [Google Scholar]
- 3.Höök P., Vallee R. B. (2006) J. Cell Sci. 119, 4369–4371 [DOI] [PubMed] [Google Scholar]
- 4.King S. M. (2000) J. Cell Sci. 113, 2521–2526 [DOI] [PubMed] [Google Scholar]
- 5.Levy J. R., Holzbaur E. L. (2006) Int. J. Dev. Neurosci. 24, 103–111 [DOI] [PubMed] [Google Scholar]
- 6.Pfister K. K., Fisher E. M., Gibbons I. R., Hays T. S., Holzbaur E. L., McIntosh J. R., Porter M. E., Schroer T. A., Vaughan K. T., Witman G. B., King S. M., Vallee R. B. (2005) J. Cell Biol. 171, 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfister K. K., Shah P. R., Hummerich H., Russ A., Cotton J., Annuar A. A., King S. M., Fisher E. M. (2006) PLoS Genet. 2, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King S. M., Barbarese E., Dillman J. F., 3rd, Benashski S. E., Do K. T., Patel-King R. S., Pfister K. K. (1998) Biochemistry 37, 15033–15041 [DOI] [PubMed] [Google Scholar]
- 9.Susalka S. J., Nikulina K., Salata M. W., Vaughan P. S., King S. M., Vaughan K. T., Pfister K. K. (2002) J. Biol. Chem. 277, 32939–32946 [DOI] [PubMed] [Google Scholar]
- 10.King S. J., Bonilla M., Rodgers M. E., Schroer T. A. (2002) Protein Sci. 11, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makokha M., Hare M., Li M., Hays T., Barbar E. (2002) Biochemistry 41, 4302–4311 [DOI] [PubMed] [Google Scholar]
- 12.Vaughan K. T., Vallee R. B. (1995) J. Cell Biol. 131, 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King S. J., Brown C. L., Maier K. C., Quintyne N. J., Schroer T. A. (2003) Mol. Biol. Cell 14, 5089–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroer T. A. (2004) Annu. Rev. Cell Dev. Biol. 20, 759–779 [DOI] [PubMed] [Google Scholar]
- 15.Burkhardt J. K., Echeverri C. J., Nilsson T., Vallee R. B. (1997) J. Cell Biol. 139, 469–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King S. M. (2000) Biochim. Biophys. Acta 1496, 60–75 [DOI] [PubMed] [Google Scholar]
- 17.Susalka S. J., Pfister K. K. (2000) J. Neurocytol. 29, 819–829 [DOI] [PubMed] [Google Scholar]
- 18.Susalka S. J., Hancock W. O., Pfister K. K. (2000) Biochim. Biophys. Acta 1496, 76–88 [DOI] [PubMed] [Google Scholar]
- 19.Pfister K. K., Salata M. W., Dillman J. F., 3rd, Vaughan K. T., Vallee R. B., Torre E., Lye R. J. (1996) J. Biol. Chem. 271, 1687–1694 [DOI] [PubMed] [Google Scholar]
- 20.Dell K. R., Turck C. W., Vale R. D. (2000) Traffic 1, 38–44 [DOI] [PubMed] [Google Scholar]
- 21.Tynan S. H., Purohit A., Doxsey S. J., Vallee R. B. (2000) J. Biol. Chem. 275, 32763–32768 [DOI] [PubMed] [Google Scholar]
- 22.Hafezparast M., Klocke R., Ruhrberg C., Marquardt A., Ahmad-Annuar A., Bowen S., Lalli G., Witherden A. S., Hummerich H., Nicholson S., Morgan P. J., Oozageer R., Priestley J. V., Averill S., King V. R., Ball S., Peters J., Toda T., Yamamoto A., Hiraoka Y., Augustin M., Korthaus D., Wattler S., Wabnitz P., Dickneite C., Lampel S., Boehme F., Peraus G., Popp A., Rudelius M., Schlegel J., Fuchs H., Hrabe de Angelis M., Schiavo G., Shima D. T., Russ A. P., Stumm G., Martin J. E., Fisher E. M. (2003) Science 300, 808–812 [DOI] [PubMed] [Google Scholar]
- 23.Chen X. J., Levedakou E. N., Millen K. J., Wollmann R. L., Soliven B., Popko B. (2007) J. Neurosci. 27, 14515–14524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ilieva H. S., Yamanaka K., Malkmus S., Kakinohana O., Yaksh T., Marsala M., Cleveland D. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12599–12604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Münch C., Sedlmeier R., Meyer T., Homberg V., Sperfeld A. D., Kurt A., Prudlo J., Peraus G., Hanemann C. O., Stumm G., Ludolph A. C. (2004) Neurology 63, 724–726 [DOI] [PubMed] [Google Scholar]
- 26.Puls I., Jonnakuty C., LaMonte B. H., Holzbaur E. L., Tokito M., Mann E., Floeter M. K., Bidus K., Drayna D., Oh S. J., Brown R. H., Jr., Ludlow C. L., Fischbeck K. H. (2003) Nat. Genet. 33, 455–456 [DOI] [PubMed] [Google Scholar]
- 27.Eyckerman S., Verhee A., der Heyden J. V., Lemmens I., Ostade X. V., Vandekerckhove J., Tavernier J. (2001) Nat. Cell Biol. 3, 1114–1119 [DOI] [PubMed] [Google Scholar]
- 28.Collins C. A., Vallee R. B. (1989) Cell Motil. Cytoskeleton 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 29.Dillman J. F., 3rd, Pfister K. K. (1994) J. Cell Biol. 127, 1671–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Kadi A. M., Bros-Facer V., Deng W., Philpott A., Stoddart E., Banks G., Jackson G. S., Fisher E. M., Duchen M. R., Greensmith L., Moore A. L., Hafezparast M. (2010) J. Biol. Chem. 285, 18627–18639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Brug M. P., Blackinton J., Chandran J., Hao L. Y., Lal A., Mazan-Mamczarz K., Martindale J., Xie C., Ahmad R., Thomas K. J., Beilina A., Gibbs J. R., Ding J., Myers A. J., Zhan M., Cai H., Bonini N. M., Gorospe M., Cookson M. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10244–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eyckerman S., Lemmens I., Lievens S., Van der Heyden J., Verhee A., Vandekerckhove J., Tavernier J. (2002) Sci. STKE 2002, pl18. [DOI] [PubMed] [Google Scholar]
- 33.Barbar E. (2008) Biochemistry 47, 503–508 [DOI] [PubMed] [Google Scholar]
- 34.Nyarko A., Hare M., Hays T. S., Barbar E. (2004) Biochemistry 43, 15595–15603 [DOI] [PubMed] [Google Scholar]
- 35.Hall J., Karplus P. A., Barbar E. (2009) J. Biol. Chem. 284, 33115–33121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma S., Triviños-Lagos L., Gräf R., Chisholm R. L. (1999) J. Cell Biol. 147, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuta A., Deng W., Morsi El-Kadi A., Banks G. T., Hafezparast M., Pfister K. K., Fisher E. M. (2010) PLoS One 5, e11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ligon L. A., Tokito M., Finklestein J. M., Grossman F. E., Holzbaur E. L. (2004) J. Biol. Chem. 279, 19201–19208 [DOI] [PubMed] [Google Scholar]
- 39.Yamada M., Toba S., Takitoh T., Yoshida Y., Mori D., Nakamura T., Iwane A. H., Yanagida T., Imai H., Yu-Lee L. Y., Schroer T., Wynshaw-Boris A., Hirotsune S. (2010) EMBO J. 29, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tynan S. H., Gee M. A., Vallee R. B. (2000) J. Biol. Chem. 275, 32769–32774 [DOI] [PubMed] [Google Scholar]
- 41.Neely M. D., Erickson H. P., Boekelheide K. (1990) J. Biol. Chem. 265, 8691–8698 [PubMed] [Google Scholar]
- 42.Schroer T. A., Sheetz M. P. (1991) J. Cell Biol. 115, 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers K. R., Lo K. W., Lye R. J., Kogoy J. M., Soura V., Hafezparast M., Pfister K. K. (2007) J. Neurosci. Res. 85, 2640–2647 [DOI] [PubMed] [Google Scholar]
- 44.LaMonte B. H., Wallace K. E., Holloway B. A., Shelly S. S., Ascaño J., Tokito M., Van Winkle T., Howland D. S., Holzbaur E. L. (2002) Neuron 34, 715–727 [DOI] [PubMed] [Google Scholar]
- 45.King S. J., Schroer T. A. (2000) Nat. Cell Biol. 2, 20–24 [DOI] [PubMed] [Google Scholar]
- 46.Gill S. R., Cleveland D. W., Schroer T. A. (1994) Mol. Biol. Cell 5, 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai D., Hoppe A. D., Swanson J. A., Verhey K. J. (2007) J. Cell Biol. 176, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hackney D. D., Stock M. F. (2008) Biochemistry 47, 7770–7778 [DOI] [PubMed] [Google Scholar]
- 49.Dietrich K. A., Sindelar C. V., Brewer P. D., Downing K. H., Cremo C. R., Rice S. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8938–8943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhey K. J., Lizotte D. L., Abramson T., Barenboim L., Schnapp B. J., Rapoport T. A. (1998) J. Cell Biol. 143, 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.WoŸniak M. J., Allan V. J. (2006) EMBO J. 25, 5457–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.