Abstract
High mobility group A1 (HMGA1) proteins belong to a group of architectural transcription factors that are overexpressed in a range of human malignancies, including pancreatic adenocarcinoma. They promote anchorage-independent growth and epithelial-mesenchymal transition and are therefore suggested as potential therapeutic targets. Employing in vitro selection techniques against a chosen fragment of HMGA1, we have generated biostable l-RNA oligonucleotides, so-called Spiegelmers, that specifically bind HMGA1b with low nanomolar affinity. We demonstrate that the best binding Spiegelmers, NOX-A50 and NOX-f33, compete HMGA1b from binding to its natural binding partner, AT-rich double-stranded DNA. We describe a formulation method based on polyplex formation with branched polyethylenimine for efficient delivery of polyethylene glycol-modified Spiegelmers and show improved tissue distribution and persistence in mice. In a xenograft mouse study using the pancreatic cancer cell line PSN-1, subcutaneous administration of 2 mg/kg per day NOX-A50 formulated in polyplexes showed an enhanced delivery of NOX-A50 to the tumor and a significant reduction of tumor volume. Our results demonstrate that intracellular targets can be successfully addressed with a Spiegelmer using polyethylenimine-based delivery and underline the importance of HMGA1 as a therapeutic target in pancreatic cancer.
Keywords: Anticancer Drug, Chromatin Remodeling, Drug Action, Transcription Factors, Tumor Therapy, Spiegelmer, Aptamer, High Mobility Group Protein, Intracellular Delivery, Pancreatic Carcinoma
Introduction
The mammalian nonhistone chromatin high mobility group A1 proteins HMGA1a,3 HMGA1b, and HMGA1c belong to one protein family encoded by the HMGA1 gene. They act as architectural transcription factors by binding via AT hook motifs to the minor groove of AT-rich DNA, leading to modulation of chromatin and nucleosome remodeling. Their ability to regulate the activity of several genes through the recognition and alteration of the DNA double helix and chromatin substrates has been linked to the development of human cancers (1–3). The overexpression of HMGA proteins is associated with different types of tumors, e.g. pancreas, breast, prostate, cervical, gastric, colorectal cancer as well as lymphomas and neuroblastic malignancies (4–6). The level of expression of HMGA1a/b is low or almost undetectable in most differentiated or nonproliferating normal cells (7–9) but is rapidly up-regulated in response to growth-stimulatory factors (8, 10). Thus, HMGA1 is considered a promising target to treat cancer development, progression, and metastasis.
The aim of our study was to evaluate whether the intracellular HMGA1 protein family can be successfully targeted with a Spiegelmer, a large structured molecule that is comparable with a monoclonal antibody in terms of specific target binding. Spiegelmers (German: Spiegel = mirror) are structured biostable l-oligonucleotides that differ from their aptamer counterparts in their sugar moiety, which consists of mirror-image l-(deoxy)ribose rather than d-(deoxy)ribose and makes Spiegelmers highly resistant to nucleases (11, 12). Spiegelmers have already been described to act potently as inhibitors in vivo (13–15) and have proven to be exceptionally safe in two Phase I clinical studies.4
Here, we report the generation of a Spiegelmer, binding to HMGA1 with high affinity. Employing the SELEX process (16), we first isolated RNA aptamers that bind to the mirror-image configuration of a 21-amino acid-long HMGA1a/b fragment. The chiral counterpart to the aptamer, the corresponding Spiegelmer (l-RNA), binds to recombinant full-length HMGA1b and also competes HMGA1b from its naturally occurring binding partner (AT-rich double-stranded DNA).
To target intracellular HMGA1 we established a delivery system based on polyethylenimine (PEI). Branched PEI has been described to facilitate cellular uptake by condensing nucleic acids and the cationic carrier into complexes suitable for endocytosis (17–19). PEGylation of the nucleic acids sterically prevents aggregation of polyplexes and balances binding to negatively charged proteoglycans on cell surfaces, thus enhancing transfection efficiency and reducing unspecific effects (20).
Using the PEI-based delivery system, we formulated HMGA1-binding Spiegelmers as polyplexes. The polyplexes were characterized in vitro and also used in a xenograft tumor growth model using the pancreatic adenocarcinoma cell line PSN-1 (21). Distribution of the formulated Spiegelmers to the tumor was strongly enhanced, and the tumor volume of the treated animals was significantly attenuated compared with control groups.
EXPERIMENTAL PROCEDURES
Peptides and Oligonucleotides
The biotinylated all-d-21-mer HMGA1b fragment (Ac-HMGA1/b36–56(Lys-AEEAc-AEEAc-biotin)OH) was custom-synthesized (BACHEM, Bubendorf, Switzerland). Oligonucleotides were synthesized using standard phosphoramidite chemistry: DNA library, 5′-GGAGCTCAGACTGGCACGCTG-N40-CAGCACAAGTTGTCGGTTCCAC-3′; forward primer, 5′-TCTAATACGACTCACTGAGCTCGACTGGCACGC-3′; reverse primer, 5′-GTGGAACCGACAACTTGTGC-3′; NOX-A50, 5′-GGCUGAUACGUGGGUGGAUAUGGGGCAGUCAGUGGGUGUUUCAGCC-3′; NOX-f33, 5′-GGAUCGCAGGGGCGUGGCUGGGGUGGGCGAUCC-3′. As control substances the respective reverse sequences (5′- and -3′-end exchanged) called revNOX-A50 and revNOX-f33 as well as an l-RNA with an arbitrary sequence 5′-UAAGGAAACUCGGCUGAUGCGGUAGCGCUGUGCAGAGCU-3′ (control SPM) were used. NOX-A50 and the control Spiegelmers were modified with a 2-kDa polyethylene glycol (PEG) moiety via a 3′-aminohexyl linker as described (15) when used for polyplex formation. The biodistribution analysis was done employing a plate-based sandwich hybridization assay. Therefore, a capture-probe (5′-NH2-TTTTTTTTTAGCTCTGCACAGCGCT-3′) and a detect probe (5′-CCGCATCAGACCGAGTTTCCTTATTTTT-TTT-Biotin-3′) complementary to the 5′- and 3′-ends of the control SPM, respectively, were prepared. For NOX-A50 5′-CCCATATCCACCCACGTATCAGCCTTTTTTTT-NH2-3′) (capture probe) and 5′-Biotin-TTTTTTTTGGCTGAAACCACCCACATGG-3′ (detect probe) were used.
Preparation of Recombinant HMGA1b
HMGA1b from the BD-FreedomTM ORF clone GH00552L1.0 (BioCat, Heidelberg, Germany) was cloned into the pHO2d vector (a kind gift from Dr. Henning Otto, Freie Universität Berlin, Germany) to generate a C-terminally His6-tagged form under control of a T7-promoter (22). It was expressed in Escherichia coli strain BL21 and purified using HIS-Select-columns (Sigma-Aldrich). The HMGA1b-His6 fusion protein differs from the native HMGA1b in position 2 (Ser to Gly exchange), and Leu-Gly-Ser-Leu-Asn-Ser-His6 was added to the C terminus.
In Vitro Selection and Pulldown Assays
Automated and manual in vitro selection experiments were performed against the biotinylated 21-amino acid mirror-image peptide, representing a central sequence of HMGA1a/b in selection buffer resembling intracellular ion conditions and pH (25 mm Tris-HCl, pH 7.0, 140 mm KCl, 12 mm NaCl, 0.8 mm MgCl2, and 0.1% Tween 20) at 37 °C (13, 23, 24). Pulldown assays were carried out as described (13, 23, 24).
Surface Plasmon Resonance Measurements
The Biacore 2000 instrument (General Electric, Fairfield, CT) was set to 37 °C 500–1000 response units (RU) of recombinant HMGA1b-His6 were immobilized on a CM5 sensor chip by NHS/EDC coupling (Amine Coupling kit, General Electric). Kinetic parameters and dissociation constants were determined after a series of Spiegelmer injections at concentrations of 1000, 500, 250, 125, 62.5, 31.25, 15.63, 7.8, 3.9, 1.95, and 0 nm in selection buffer or in “extracellular buffer” (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1 mm CaCl2, and 0.1% Tween 20). The assays were double referenced. Using the BIA-evaluation 3.0 software applying Langmuir 1:1 stoichiometric fitting algorithm the association (ka) and dissociation rate constants (kd) were determined by local fitting and the dissociation constants (KD) were calculated accordingly.
Isothermal Titration Calorimetry (ITC) Measurement
Recombinant HMGA1b (24 μm) was titrated to NOX-A50 or revNOX-A50 (2.4 μm) in a VP-ITC machine (General Electric) in selection buffer at 37 °C. The number of injections was set to 40 with a volume of 3 μl, a duration of 6 s, a spacing of 300 s, and a filter period of 16 s with a stirring speed of 300 rpm. Data analysis was performed with VP-ITC 2000 Evaluation software.
Competitive Assay for the Inhibition of the HMGA1b-dsDNA Interaction
Spiegelmer from 1 nm to 10 μm was incubated at 37 °C with 0.36 μg/ml recombinant HMGA1b in binding buffer (25 mm Tris-HCl, pH 7.0, 140 mm KCl, 12 mm NaCl, 0.8 mm MgCl2, 0.25 mg/ml BSA, 1 mm DTT, 18–20 μg/ml poly(dG·dC) (Sigma-Aldrich) (9), and 0.05% Tween 20) for 30 min. Biotinylated double-stranded DNA (dsDNA, equimolar mixture of 5′-biotin-TCGAAAAAAGCAAAAAAAAAAAAAAAAAACTGGC-3′ and 5′-GCCAGTTTTTTTTTTTTTTTTTTGCTTTTTT-3′ (25) in 150 mm NaCl were added to a final concentration of 150 nm and incubated at 37 °C for 4 h. The mixture was transferred to a streptavidin-coated 96-well plate (Reacti-Bind; Pierce), incubated under shaking at 37 °C for 30 min, and finally washed three times with 200 μl of TBSTCM (20 mm Tris-HCl, pH 7.6, 137 mm NaCl, 1 mm MgCl2, 1 mm CaCl2, and 0.05% Tween 20). Bound recombinant HMGA1b was detected after adding 50 μl of a 1:1000 dilution of Ni2+-HRP (ExpressDetector Nickel-HRP; KPL, Gaithersburg, MD) in TBSTCM with 10 mg/ml BSA at room temperature for 1 h, washing, and finally adding 100 μl of fluorogenic HRP substrate (QuantaBlue, Pierce) for 15 min. Fluorescence (λex/em = 340/405 nm) was measured on a FluoStar Optima plate reader (BMG, Offenburg, Germany); results were plotted as percent recombinant HMGA1b bound versus Spiegelmer concentration.
Spiegelmer-PEI Polyplexes
PEGylated Spiegelmer at a final concentration of 20–100 μg/ml was rapidly mixed in HBG buffer (5% (w/w) glucose, 20 mm HEPES, pH 7.1) with 25-kDa branched PEI (Sigma-Aldrich) at various nitrogen/phosphate (N/P) ratios (N/P = molar ratio of PEI nitrogen to RNA phosphate). Polyplex formation was measured by RiboGreen (R-11491; Molecular Probes) exclusion assay as described (26).
Proliferation Assay
0.5–1 × 104 PSN-1 cells (ECACC, Salisbury, UK) (21) were seeded in 96-well plates (Sigma-Aldrich) 16–24 h prior to Spiegelmer polyplex treatment. The cells were cultivated for 0, 1, 2, or 4 days at 37 °C and 5% CO2. 10 μl of 0.44 mm Alamar-Blue (Resazurin) (Sigma-Aldrich) in PBS was added and incubated at 37 °C for additional 2 h, and fluorescence was measured using a plate reader at λex/em = 544/590 nm.
Tissue Distribution in Tumor-bearing Mice
Studies were performed at EPO (Berlin, Germany). 107 PSN-1 cells were injected subcutaneously in the flank of nude mice (NMRInu/nu, age 6–8 weeks, n = 4/group). The 3′-PEGylated control SPM and polyplexes thereof were injected subcutaneously daily near the tumor site from days 5 to 25 at a dose of 10 mg/kg (Spiegelmer part only). Animals were killed 24 and 96 h after last treatment (n = 4/time point). EDTA plasma was prepared from terminal blood, and tumor and selected organs were dissected. The tissues were homogenized in hybridization buffer (0.05 m trisodium citrate, pH 7.0, 0.5% (w/v) SDS) and centrifuged. The supernatant was used for determination of Spiegelmer levels. Therefore, DNA-BIND plates (Corning Co-star, Corning, NY) were preincubated with capture probe (0.75 nm) in coupling buffer (500 mm Na2HPO4, pH 8.5, 0.5 mm EDTA) at 4 °C overnight, washed with coupling buffer, blocked with 0.5% BSA at 37 °C for 1 h, and finally washed with hybridization buffer. Plasma or homogenized tissue supernatant was incubated with 2 μm detection probe, denatured at 95 °C for 10 min, transferred to the prepared DNA-BIND plate, and incubated at 40 °C for 45 min. After washing the plate with hybridization buffer and Tris-buffered saline containing 0.1% Tween 20 (TBST), the bound Spiegelmer was detected by adding a 1:5000 dilution of streptavidin-HRP (Promega, Mannheim, Germany), incubating at 20 °C for 1 h, and washing two times with TBST and assay buffer (20 mm Tris-HCl, pH 9.8, 1 mm MgCl2). Next, 100 μl of READY-to-use substrate (Applied Biosystems) was added and incubated at 20 °C for 30 min, and chemoluminescence was measured using a plate reader.
Xenograft Tumor Growth Model
Studies were performed at EPO. 107 PSN-1 cells were injected subcutaneously in the flank of nude mice (NMRInu/nu, age 6–8 weeks, n = 8/group). NOX-A50–3′-PEG or revNOX-A50–3′-PEG Spiegelmers (100 μl, 2 mg/kg) in PEI-polyplexes (N/P 2.5) or vehicle (PBS) were injected subcutaneously near the tumor cell inoculation site daily from days 6 to 22. Tumor volume and body weight were measured three times a week. Animals were killed 24 h after the last injection, and EDTA plasma was prepared from terminal blood. Furthermore, kidneys, liver, and tumor were harvested to determine Spiegelmer concentrations.
Statistical Analyses
Nonlinear regression analysis and statistical significance evaluation (Student's t test) were performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA). IC50 values and rate and equilibrium constants are reported as mean ± S.E. of multiple independent experiments. The tumor sizes in the xenograft treatment study are presented as box-and-whisker plots (27).
RESULTS
In Vitro Selection of Aptamers Binding to the Mirror-image Peptide Representing the Central DNA Binding Domain of HMGA1a/b
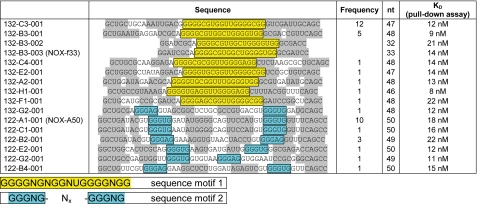
After 12 rounds of in vitro selection performed with the d-peptide fragment of HMGA1a/b (Glu36–Lys56 of HMGA1b) library affinities of 10–20 nm were measured (data not shown). Forty individual sequences were determined by cloning and sequencing. Sequence alignment revealed 22 sequences sharing the common motif 1 Nx-GGGGNGNGGNUGGGGNGG-Nx and 18 sequences motif 2 Nx-GGGNG-Nx-GGGNG-Nx (Fig. 1). Secondary structure predictions (28) suggest a stabilizing stem structure formed by the 5′ and 3′ termini flanking these motifs. Based on this prediction we truncated the primer binding sites. The affinities of these truncated aptamers to the selection target range from 8–22 nm and are comparable with the full-length aptamers. Without substantial loss in affinity sequence 132-B3-001 could be further reduced to the smallest aptamer (132-B3-003), again confirming the predicted stem structure. Further truncations to stems shorter than seven nucleotides, however, resulted in considerably reduced affinity.
FIGURE 1.
Analysis of sequences resulting from in vitro selection against a the mirror-image peptide representing the central DNA binding domain of HMGA1a/b. Frequency refers to the number of occurrences within the 40 sequences obtained. nt = nucleotides. KD refers to the dissociation constants of the truncated aptamers. Shaded regions indicate predicted stem structures.
Determination of Spiegelmer Affinity to Recombinant HMGA1b
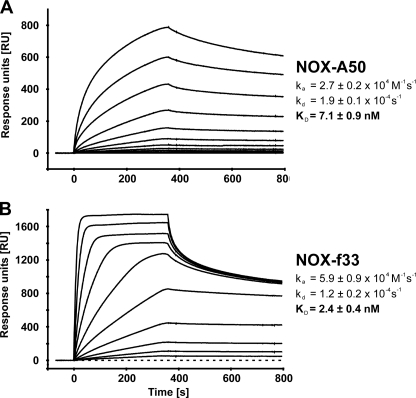
The most frequent motif 2 aptamer (NOX-A1-001) and the shortest motif 1 aptamer (NOX-B3-003) were synthesized as Spiegelmers (NOX-A50 and NOX-f33, respectively). The affinities of NOX-A50 and NOX-f33 to recombinant HMGA1b are comparable with those of the corresponding aptamers to the biotinylated d-HMGA1b(Glu36–Lys56) fragment as determined by surface plasmon resonance measurements at 37 °C under intracellular buffer conditions: NOX-A50 binds to immobilized recombinant HMGA1b with an association rate of 2.7 ± 0.2 × 104 m−1s−1 and a dissociation rate of 1.9 ± 0.1 × 10−4 s−1 whereas NOX-f33 displays a faster association rate with 5.9 ± 0.9 × 104 m−1s−1 and a slower dissociation rate of 1.2 ± 0.2 × 10−4 s−1. The resulting dissociation constants are 7.1 ± 0.9 nm for NOX-A50 and 2.4 ± 0.4 nm for NOX-f33 (Fig. 2). Binding evaluation of the Spiegelmers under extracellular buffer conditions did not show any difference in binding kinetics or affinity. Both Spiegelmers showed a 1:1 Langmuir binding kinetic with little unspecific binding contribution. Control Spiegelmers revNOX-A50 and revNOX-f33 showed only weak unspecific binding with an approximate dissociation constants of 3–5 μm (data not shown). The specific binding of NOX-A50 to recombinant HMGA1b was also confirmed by ITC measurement at 37 °C. In this assay, which does not require immobilization of one binding partner, NOX-A50 binds HMGA1b with a KD of 18 ± 9 nm, whereas revNOX-A50 shows no binding (supplemental Fig. S1).
FIGURE 2.
Biacore measurements of NOX-A50 and NOX-f33 binding to HMGA1b. Spiegelmers were injected at various concentrations at 37 °C. NOX-A50 (A) and NOX-f33 (B) bind to immobilized recombinant HMGA1b with equilibrium dissociation constants of 7.1 ± 0.9 nm and 2.4 ± 0.4 nm, respectively.
Inhibition of Recombinant HMGA1b Binding to AT-rich dsDNA
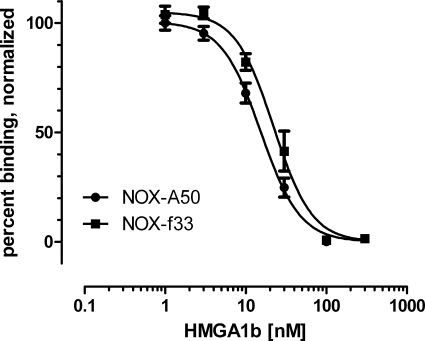
Employing a competitive plate-based assay, NOX-A50 and NOX-f33 were characterized for their ability to inhibit binding of recombinant HMGA1b to AT-rich dsDNA, the natural binding partner of HMGA1b. Binding of 20 nm HMGA1b to biotinylated AT-rich dsDNA was inhibited dose-dependently by NOX-A50 and NOX-f33 with IC50 of 15.35 ± 1.20 nm and 22.21 ± 2.20 nm, respectively (Fig. 3). revNOX-A50 and rev-NOX-f33 showed no significant inhibition up to a concentration of 300 nm (supplemental Fig. S2). Because of its slightly better inhibition, NOX-A50 was selected for further studies.
FIGURE 3.
Inhibition of HMGA1b binding to AT-rich dsDNA in a competitive plate-based assay. The binding of an AT-rich dsDNA oligonucleotide to recombinant HMGA1b was inhibited dose-dependently with NOX-A50 showing an IC50 of 15.4 ± 1.2 nm (n = 25) and NOX-f33 an IC50 of 22.2 ± 2.2 nm (n = 8). Data are means ± S.E.
Spiegelmer-PEI Polyplex Formulation and in Vivo Delivery
To target intracellularly localized HMGA1 proteins with Spiegelmers, a protocol for intracellular delivery based on the cationic polymer PEI was established. For proof of concept purposes a nonfunctional Spiegelmer with a covalently attached 2-kDa PEG moiety was used. This Spiegelmer was complexed with branched 25-kDa PEI at various N/P ratios The complexation rate was determined by a RiboGreen exclusion assay, which indicated that >95% of the Spiegelmer can be trapped in Spiegelmer polyplexes at an N/P ratio of >2 (supplemental Fig. S3). For subsequent studies, an N/P ratio of 2.5 was adjusted to achieve maximal complexation at minimal use of PEI.
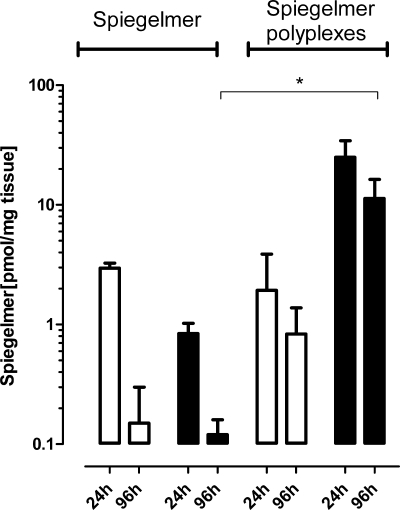
To compare tissue distribution and persistence of pure Spiegelmer and Spiegelmer polyplexes regardless of any target interaction, a multiple dose pilot study with control SPM was conducted in PSN-1 cell tumor-bearing mice. Animals showed no injection site reactions, no significant loss of body weight, or influence on tumor growth over the treatment period (data not shown). 24 h after the last injection Spiegelmer and Spiegelmer polyplexes show comparable plasma levels. However, regarding tumor distribution Spiegelmer polyplexes lead to approximately 30-fold higher levels of Spiegelmer than administration of Spiegelmer alone. At 96 h, a prolonged elimination time from plasma and tumor for Spiegelmer polyplexes compared with Spiegelmer alone was observed (Fig. 4). No significant amounts of Spiegelmer were detected in other tissues with the exception of slightly elevated levels in kidneys (Spiegelmer and Spiegelmer polyplexes) and liver (Spiegelmer alone) (supplemental Table S1).
FIGURE 4.
Multiple dose tissue distribution study with nonfunctional Spiegelmer in tumor-bearing mice. 10 mg/kg control SPM and control SPM in polyplexes were injected s.c. near the tumor site on days 5–25. Open bars show the amount of Spiegelmer in plasma and filled bars the amount in tumor 24 and 96 h after the last injection. The values are log-transformed and accordingly analyzed with an unpaired t test (*, p < 0.05).
Inhibition of Cell Proliferation by Spiegelmer Polyplexes in Vitro
NOX-A50 and revNOX-A50 polyplexes were prepared to characterize their biological activity in vitro. The 3′-terminal 2-kDa PEG modification desired for the polyplex tolerability (29) does not influence the binding of NOX-A50 to recombinant HMGA1b as shown by the competitive assay for the inhibition of the HMGA1b-dsDNA interaction (supplemental Fig. S4). Although Western blots confirm high levels of HMGA1a/b in the nucleus or perinuclear region of PSN1 cells, NOX-A50 polyplexes showed no significant reduction of PSN-1 cell proliferation compared with revNOX-A50 polyplexes. The observed (dose-dependent) effects were considered formulation-dependent and thus unspecific (data not shown).
Tumor Growth Study in Mice with HMGA1b-binding Spiegelmer NOX-A50
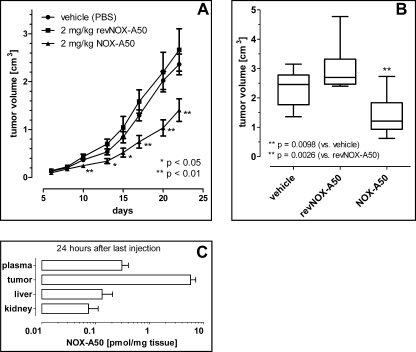
The effects of NOX-A50 polyplexes on tumor growth were evaluated in a xenograft mouse model with PSN-1 cell-derived tumors. In all animals an aggressive, but homogeneous tumor growth was observed. After 22 days the mean tumor volume in the vehicle-treated group was 2.4 ± 0.2 cm3 (n = 8). Daily subcutaneous injections of 2 mg/kg NOX-A50 in polyplexes on days 5–22 showed a significant effect on the course of tumor growth, whereas nonfunctional revNOX-A50 polyplexes showed no significant influence (Fig. 5A). After 23 days the animals were killed for ethical reasons (some tumors in the control groups exceeded 3 cm3). Statistical end point analysis on day 22 revealed a significant (p = 0.0098) reduction of tumor growth by NOX-A50 polyplex treatment (tumor volume 1.4 ± 0.2 cm3) compared with treatment with vehicle (tumor volume 2.4 ± 0.2 cm3). This effect was sequence-specific for NOX-A50 because revNOX-A50 polyplexes showed no influence on tumor growth (tumor volume 3.0 ± 0.4 cm3, p = 0.0026 versus NOX-A50 polyplex group) (Fig. 5B). The analysis of NOX-A50 tissue distribution 24 h after the last injection showed results comparable with the delivery study with the control SPM polyplexes: high amounts of NOX-A50 were observed in the tumor, whereas only little amounts were detected in liver and kidney tissue (Fig. 5C and supplemental Table S1).
FIGURE 5.
PSN-1 xenograft study in mice treated with NOX-A50 and revNOX-A50 polyplexes. A, the course of tumor volumes in mice (n = 8) showed a significant effect of NOX-A50 polyplexes that were administered on days 5–22. *, ** significance versus vehicle. B, statistical end point analysis of tumor volume on day 22 by box-and-whisker plots. C, tissue distribution of NOX-A50 on the 23rd day 24 h after the last injection. The numbers are compared with the control Spiegelmer tissue distribution study in supplemental Table S1.
DISCUSSION
Employing an in vitro selection approach using a central fragment of the architectural transcription factors HMGA1a/b as a selection target, we have identified Spiegelmers that bind to recombinant HMGA1b with high affinity in the low nanomolar range and effectively prevent the binding of HMGA1b to its nuclear binding partner, i.e. AT-rich dsDNA. For targeting intracellularly localized HMGA1, an efficient delivery system based on complexation of Spiegelmers with branched PEI was established. Compared with unformulated Spiegelmers, such Spiegelmer polyplexes show enhanced delivery to the tumor tissue in vivo. The persistence of Spiegelmer polyplexes in tumor and plasma compared with unformulated Spiegelmer was efficiently prolonged. Most importantly, NOX-A50 polyplexes were capable of significantly reducing tumor volume of PSN-1 pancreatic adenocarcinoma cells in a mouse xenograft study.
This finding describes the first approach of targeting the intracellular protein HMGA1b with a large molecule using a formulation technique for an efficient intracellular delivery. HMGA1 proteins have already been suggested as therapeutic targets (5), and several studies demonstrated the importance of HMGA1 in tumor progression and metastasis (3, 6). HMGA1a/b overexpression is a common feature of human malignant neoplasias, plays a crucial role in cell transformation, and possesses oncogenic properties. Some controversy remains, however, because the appearance of a neoplastic phenotype in Hmga1−/− and Hmga1+/− mice revealed that HMGA proteins can also have a tumor suppressor function (30).
Approaches with an adenovirus carrying the HMGA1 gene in antisense orientation resulted in reduced proliferation and apoptosis of cancer cells (31, 32); the adenoviral expression of antisense RNA particularly reduced the proliferation of human pancreatic carcinoma cell lines in vitro and dramatically inhibited the growth of cognate tumors after transplantation of the transfected cells into nude mice (33). Down-regulation of HMGA1 by RNA interference resulted in accelerated repair of UV-induced DNA lesions in intact cells. This finding suggests that HMGA1 overexpression might play an important role in the accumulation of mutations and genomic instabilities associated with many human cancers (34). However, these studies failed to show an approach that is transferable to pharmaceutical development so far.
Spiegelmer antagonists to a number of extracellular targets have been described (11, 14, 24, 35). Two Spiegelmers have proven to be safe and well tolerated in Phase I clinical studies4 providing evidence that the Spiegelmer technology is suitable to generate human medicines. However, the mirror-image approach that is needed to identify Spiegelmers includes the synthesis of a d-peptide as a target for the in vitro selection process. Due to the current limitations in solid phase peptide synthesis technology, larger d-peptide or d-protein targets are not always easily accessible as full-length molecules. The use of epitopes or fragments for in vitro selection can provide a solution. We have now shown that Spiegelmers selected against a central 21-amino acid-long fragment of HMGA1a/b also bind to recombinant full-length HMGA1b with the same high affinity.
HMGA1 binding to AT-rich dsDNA is driven by three cationic “AT hooks.” The central AT hook is always involved (5, 36). Because of the choice of a fragment that includes this pivotal region, the affinity of the Spiegelmers to the HMGA1 fragment could directly translate into inhibition of HMGA1b-dsDNA binding. By further reasons of analogy, it is most likely that besides HMGA1b, the alternative splice product HMGA1a which also comprises the 21-amino acid fragment chosen for Spiegelmer identification, is recognized. Only few such fragment approaches for aptamers have been published to date (24, 37, 38).
Like antibodies, aptamers if intended for therapeutic purposes, generally target extracellular or membrane-bound proteins. Their use as intracellular inhibitors is so far limited, apart from the use of transient expression of RNA aptamers (so called intramers) inside mammalian cells involving gene therapy methods (39). There are a number of hurdles to efficient intracellular action of oligonucleotides: Distribution, crossing of the cell membrane, endosomal acidic pH, endosomal escape while maintaining integrity against ubiquitous endo- and exonucleases. l-RNA-based Spiegelmers have the advantage of being nuclease-resistant and not as susceptible to acidic pH as DNA-based oligonucleotides so that distribution, crossing the cell membrane and endosomal escape were the remaining challenges.
PEI was identified as an effective transfection reagent for oligonucleotides, also allowing an efficient escape of the payload from the endosome that is most likely driven by the “proton sponge” effect (40). The combination of PEI formulation with PEGylation, either through a PEGylated oligonucleotide or the construction of a PEI-PEG copolymer, was shown to reduce PEI-inherent toxicity and distribution issues (41–45). On the basis of data published for modified antisense oligonucleotides (29, 42), we established a protocol for intracellular delivery of 3′-PEGylated Spiegelmers by complexation with 25-kDa branched PEI.
First, we report here the results of a multiple dose tissue distribution study with a non-target-binding control Spiegelmer (control SPM) in tumor-bearing nude mice. The PEI-PEG-formulated Spiegelmer was administered subcutaneously for 21 days. Bioanalytical analysis revealed successful delivery to the tumor and prolonged circulation in plasma compared with uncomplexed Spiegelmer. Only low amounts of the Spiegelmers delivered by polyplexes were detected in kidney, liver, and spleen tissue, reflecting partly the route of clearance. These results are in good agreement with observations published for other oligonucleotides (29, 41, 46). No accumulation of Spiegelmer was observed in lung or heart tissue, further supporting the finding that the functionality of PEI as a nucleotide carrier was improved by incorporating the non-ionic PEG into PEG-PEI copolymers, thus avoiding unspecific accumulation of polyplexes, e.g. in pulmonary capillaries (47, 48). Furthermore, we have not observed any local or evident systemic toxic effects in the mice after the 21 daily injections.
In the treatment study, PEI-PEG-formulated NOX-A50 was not only well tolerated but also slowed down growth of already established PSN-1-derived tumors in nude mice, thus suggesting efficient delivery of the Spiegelmer payload into the tumor cells. As expected, no effect on tumor growth was observed in animals that received revNOX-A50. Trapasso et al. reported that down-regulation through an antisense oligodeoxynucleotide led to decreased HMGA1 protein expression and to a reduced proliferation rate of pancreatic carcinoma cells in vitro. These PSN-1 cells pretreated with HMGA1 antisense oligodeoxynucleotides failed to form xenograft tumors in nude mice (33). Liau et al. showed that down-regulation of HMGA1 in pancreatic cancer cells by stably transfected shRNA vectors reduced proliferation in vitro and tumor growth in vivo (49). They hypothesized that tumor HMGA1 status represents an independent prognostic marker and HMGA1 proteins are therapeutic targets in pancreatic adenocarcinoma.
Despite in vivo efficacy of NOX-A50 polyplexes, dose-dependent inhibition of PSN-1 cell proliferation in vitro could not be delineated to be HMGA1-specific because similar effects were observed with control Spiegelmer polyplexes. This may be due to unspecific cytotoxic effects of PEI potentially masking specific effects of NOX-A50 or NOX-f33. An association between HMGA1 expression and cell cycling in vitro had been shown by others using antisense constructs, i.e. phosphorothioate DNA without transfection reagents or adenoviral transcripts (3, 33). Nevertheless, it is still under discussion whether HMGA1 has a direct influence on cell proliferation because others could find such strong association only with HMGA2 expression but not with HMGA1 (50).
Spiegelmer NOX-A50 has been identified as a potent HMGA1 inhibitor. Using a PEI-based formulation approach the compound was successful in reducing tumor growth in a pancreatic adenocarcinoma xenograft mouse model, suggesting efficient delivery of the s.c. administered compound to the tumor and into the tumor cells. Thus, Spiegelmer-based HMGA1 blockade could potentially add to the so-far limited therapeutic armamentarium to fight pancreatic carcinoma in man. Further work will elucidate whether Spiegelmer polyplexes would also distribute to distant tumor metastases and inhibit the growth of further, preferentially orthotopic, tumors in xenograft studies.
Acknowledgments
We thank Christiane Bohnes, Stefanie Hoffmann, and Kathrin Schindele for excellent technical assistance.
This work was supported by the State of Berlin/Investitionsbank Berlin Grant 10018314. All authors are employees of NOXXON Pharma AG, a company that has a commercial interest in Spiegelmers.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
D. Eulberg, F. Fliegert, M. Humphrey, S. Klussmann, F. Morich, W. G. Purschke, C. Schmidt, A. Vater, S. Vonhoff, D. Vossmeyer, and S. Zöllner, unpublished results.
- HMGA1
- high mobility group protein A1
- ITC
- isothermal titration calorimetry
- N/P
- nitrogen/phosphate
- PEI
- polyethyleneimine
- SPM
- spiegelmer.
REFERENCES
- 1.Martinez Hoyos J., Fedele M., Battista S., Pentimalli F., Kruhoffer M., Arra C., Orntoft T. F., Croce C. M., Fusco A. (2004) Cancer Res. 64, 5728–5735 [DOI] [PubMed] [Google Scholar]
- 2.Battista S., Pentimalli F., Baldassarre G., Fedele M., Fidanza V., Croce C. M., Fusco A. (2003) FASEB J. 17, 1496–1498 [DOI] [PubMed] [Google Scholar]
- 3.Reeves R., Edberg D. D., Li Y. (2001) Mol. Cell. Biol. 21, 575–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fedele M., Fusco A. (2010) Biochim. Biophys. Acta 1799, 48–54 [DOI] [PubMed] [Google Scholar]
- 5.Reeves R., Beckerbauer L. M. (2003) Prog. Cell Cycle Res. 5, 279–286 [PubMed] [Google Scholar]
- 6.Evans A., Lennard T. W., Davies B. R. (2004) J. Surg. Oncol. 88, 86–99 [DOI] [PubMed] [Google Scholar]
- 7.Johnson K. R., Lehn D. A., Elton T. S., Barr P. J., Reeves R. (1988) J. Biol. Chem. 263, 18338–18342 [PubMed] [Google Scholar]
- 8.Johnson K. R., Lehn D. A., Reeves R. (1989) Mol. Cell. Biol. 9, 2114–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giancotti V., Pani B., D'Andrea P., Berlingieri M. T., Di Fiore P. P., Fusco A., Vecchio G., Philp R., Crane-Robinson C., Nicolas R. H., et al. (1987) EMBO J. 6, 1981–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson K. R., Disney J. E., Wyatt C. R., Reeves R. (1990) Exp. Cell Res. 187, 69–76 [DOI] [PubMed] [Google Scholar]
- 11.Eulberg D., Jarosch F., Vonhoff S., Klussmann S. (2006) in The Aptamer Handbook (Klussmann S. ed), pp. 417–442, John Wiley and Sons, New York [Google Scholar]
- 12.Klussmann S., Nolte A., Bald R., Erdmann V. A., Fürste J. P. (1996) Nat. Biotechnol. 14, 1112–1115 [DOI] [PubMed] [Google Scholar]
- 13.Helmling S., Maasch C., Eulberg D., Buchner K., Schröder W., Lange C., Vonhoff S., Wlotzka B., Tschöp M. H., Rosewicz S., Klussmann S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13174–13179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayyed S. G., Hägele H., Kulkarni O. P., Endlich K., Segerer S., Eulberg D., Klussmann S., Anders H. J. (2009) Diabetologia 52, 2445–2454 [DOI] [PubMed] [Google Scholar]
- 15.Wlotzka B., Leva S., Eschgfäller B., Burmeister J., Kleinjung F., Kaduk C., Muhn P., Hess-Stumpp H., Klussmann S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8898–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuerk C., Gold L. (1990) Science 249, 505–510 [DOI] [PubMed] [Google Scholar]
- 17.Lungwitz U., Breunig M., Blunk T., Göpferich A. (2005) Eur. J. Pharm. Biopharm. 60, 247–266 [DOI] [PubMed] [Google Scholar]
- 18.Neu M., Fischer D., Kissel T. (2005) J. Gene Med. 7, 992–1009 [DOI] [PubMed] [Google Scholar]
- 19.Ogris M., Wagner E. (2002) Drug Discov. Today 7, 479–485 [DOI] [PubMed] [Google Scholar]
- 20.Sonawane N. D., Szoka F. C., Jr., Verkman A. S. (2003) J. Biol. Chem. 278, 44826–44831 [DOI] [PubMed] [Google Scholar]
- 21.Yamada H., Yoshida T., Sakamoto H., Terada M., Sugimura T. (1986) Biochem. Biophys. Res. Commun. 140, 167–173 [DOI] [PubMed] [Google Scholar]
- 22.Fasshauer D., Otto H., Eliason W. K., Jahn R., Brünger A. T. (1997) J. Biol. Chem. 272, 28036–28041 [DOI] [PubMed] [Google Scholar]
- 23.Eulberg D., Buchner K., Maasch C., Klussmann S. (2005) Nucleic Acids Res. 33, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purschke W. G., Eulberg D., Buchner K., Vonhoff S., Klussmann S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5173–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fashena S. J., Reeves R., Ruddle N. H. (1992) Mol. Cell. Biol. 12, 894–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen H., Kunath K., Martin A. L., Stolnik S., Roberts C. J., Davies M. C., Kissel T. (2002) Biomacromolecules 3, 926–936 [DOI] [PubMed] [Google Scholar]
- 27.Tukey J. W. (1977) Science 198, 679–684 [DOI] [PubMed] [Google Scholar]
- 28.Zuker M. (2003) Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogris M., Walker G., Blessing T., Kircheis R., Wolschek M., Wagner E. (2003) J. Controlled Release 91, 173–181 [DOI] [PubMed] [Google Scholar]
- 30.Fedele M., Fidanza V., Battista S., Pentimalli F., Klein-Szanto A. J., Visone R., De Martino I., Curcio A., Morisco C., Del Vecchio L., Baldassarre G., Arra C., Viglietto G., Indolfi C., Croce C. M., Fusco A. (2006) Cancer Res. 66, 2536–2543 [DOI] [PubMed] [Google Scholar]
- 31.Masciullo V., Baldassarre G., Pentimalli F., Berlingieri M. T., Boccia A., Chiappetta G., Palazzo J., Manfioletti G., Giancotti V., Viglietto G., Scambia G., Fusco A. (2003) Carcinogenesis 24, 1191–1198 [DOI] [PubMed] [Google Scholar]
- 32.Scala S., Portella G., Fedele M., Chiappetta G., Fusco A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4256–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapasso F., Sarti M., Cesari R., Yendamuri S., Dumon K. R., Aqeilan R. I., Pentimalli F., Infante L., Alder H., Abe N., Watanabe T., Viglietto G., Croce C. M., Fusco A. (2004) Cancer Gene Ther. 11, 633–641 [DOI] [PubMed] [Google Scholar]
- 34.Reeves R., Adair J. E. (2005) DNA Repair 4, 926–938 [DOI] [PubMed] [Google Scholar]
- 35.Kulkarni O., Pawar R. D., Purschke W., Eulberg D., Selve N., Buchner K., Ninichuk V., Segerer S., Vielhauer V., Klussmann S., Anders H. J. (2007) J. Am. Soc. Nephrol. 18, 2350–2358 [DOI] [PubMed] [Google Scholar]
- 36.Reeves R., Nissen M. S. (1990) J. Biol. Chem. 265, 8573–8582 [PubMed] [Google Scholar]
- 37.Proske D., Gilch S., Wopfner F., Schätzl H. M., Winnacker E. L., Famulok M. (2002) ChemBioChem 3, 717–725 [DOI] [PubMed] [Google Scholar]
- 38.Xu W., Ellington A. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7475–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Famulok M., Blind M., Mayer G. (2001) Chem. Biol. 8, 931–939 [DOI] [PubMed] [Google Scholar]
- 40.Pack D. W., Hoffman A. S., Pun S., Stayton P. S. (2005) Nat. Rev. Drug Discov. 4, 581–593 [DOI] [PubMed] [Google Scholar]
- 41.Fischer D., Osburg B., Petersen H., Kissel T., Bickel U. (2004) Drug. Metab. Dispos. 32, 983–992 [PubMed] [Google Scholar]
- 42.Jeong J. H., Kim S. W., Park T. G. (2003) J. Controlled Release 93, 183–191 [DOI] [PubMed] [Google Scholar]
- 43.Ogris M., Brunner S., Schüller S., Kircheis R., Wagner E. (1999) Gene Ther. 6, 595–605 [DOI] [PubMed] [Google Scholar]
- 44.Ogris M., Steinlein P., Carotta S., Brunner S., Wagner E. (2001) AAPS Pharm. Sci. 3, E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sirsi S. R., Williams J. H., Lutz G. J. (2005) Hum. Gene Ther. 16, 1307–1317 [DOI] [PubMed] [Google Scholar]
- 46.Kunath K., von Harpe A., Petersen H., Fischer D., Voigt K., Kissel T., Bickel U. (2002) Pharm. Res. 19, 810–817 [DOI] [PubMed] [Google Scholar]
- 47.Zou S. M., Erbacher P., Remy J. S., Behr J. P. (2000) J. Gene Med. 2, 128–134 [DOI] [PubMed] [Google Scholar]
- 48.Goula D., Becker N., Lemkine G. F., Normandie P., Rodrigues J., Mantero S., Levi G., Demeneix B. A. (2000) Gene Ther. 7, 499–504 [DOI] [PubMed] [Google Scholar]
- 49.Liau S. S., Rocha F., Matros E., Redston M., Whang E. (2008) Cancer 113, 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarhadi V. K., Wikman H., Salmenkivi K., Kuosma E., Sioris T., Salo J., Karjalainen A., Knuutila S., Anttila S. (2006) J. Pathol. 209, 206–212 [DOI] [PubMed] [Google Scholar]