Abstract
Par-3 is a component of Par complex, which is critical for the integrity of tight junction. We previously reported that TGF-β down-regulated Par-3 expression in rat proximal tubular epithelial cells, but the underlying mechanism remains unknown. In the present study, we demonstrated by a luciferase reporter assay that miR-491-5p down-regulated the luciferase activity through a binding site in the 3′ UTR of Par-3. Overexpression of miR-491-5p dramatically decreased the expression of endogenous Par-3, disrupted tight junction, and resulted in decreased transepithelial resistance. Moreover, miR-491-5p expression was induced by TGF-β1 through the MEK/p38 MAPK pathway. Importantly, miR-491-5p levels were increased significantly in a rat model of obstructive nephropathy, in parallel with decreased Par-3 levels. Taken together, we conclude that up-regulation of miR-491-5p contributes to TGF-β-regulated Par-3 expression. Our study uncovered a novel mechanism by which TGF-β disrupts cell junction.
Keywords: Epithelial Cell, Kidney, Micro-RNA, Tight Junction, Transforming Growth Factor β (TGFβ)
Introduction
In epithelial cells, apical-basal polarity is maintained through the formation of several intercellular adhesion systems consisting of tight junctions, adherens junctions, and desmosomes. The tight junction regulates paracellular diffusion and functionally segregates the plasma membrane into two compartments, which is a requirement for full polarization of epithelial cells (1). In mammalian epithelia, the Par3-Par6-aPKC complex (Par complex) is essential for the formation and function of the tight junction (2, 3). It localizes to the tight junction and regulates its formation and positioning with respect to basolateral and apical membrane domains (4, 5). Par-3 protein is the key component of the Par complex that recruits the complex to the primordial tight junction through interacting with Tiam1 (6). In endothelial cells, it has been reported that Par-3 associated directly with adhesion junction protein-vascular endothelial cadherin (VE-cadherin) (7). Suppression of Par-3 expression by RNA interference caused a dramatic disruption of tight junction assembly (8, 9).
Disruption of cell junction is the initial step of renal tubular epithelial cell injury (10). Various growth factors, cytokines, hormones, and extracellular cues could cause the renal tubular epithelial cell injury, and TGF-β1 is the most potent (11–13). However, the underlying mechanism by which TGF-β disrupts cell-cell junctions still remains unclear.
Micro-RNAs are a class of 22-nucleotide noncoding RNAs that are evolutionarily conserved and function as negative regulators of gene expression. Micro-RNA precursors (pre-miRNAs) are ∼70 nucleotides in length processed into a stem-loop structure by the Drosha-DGCR8 microprocessor complex and cleaved into ∼22-nucleotide duplexes by Dicer after export to the cytoplasm. One strand of the duplex is incorporated into an RNA-induced silencing complex and guides the complex to regions of complementarity in the 3′ untranslated region of target mRNAs, and triggers either their degradation or the inhibition of translation (14–16). Accumulating evidence demonstrates that micro-RNAs play important roles in a wide range of biological functions, including cellular differentiation, embryonic development, and apoptosis (17, 18). Recently, several studies have implicated micro-RNAs in TGF-β signaling. MiR-192 is found to be induced by TGF-β in mouse mesangial cells and plays an important role in diabetic nephropathy (19). MiR-155 is a direct transcriptional target of the TGF-β/Smad4 pathway and mediates TGF-β-induced epithelial to mesenchymal transition through targeting RhoA (20).
We previously reported that TGF-β1 down-regulated Par-3 expression in rat proximal tubular epithelial cells in a time- and dose-dependent manner (21). In the present work, we identified Par-3 as a target of miR-491-5p. Overexpression of miR-491-5p decreased Par-3 expression, disrupted tight junction, and resulted in decreased transepithelial resistance (TER).2 Moreover, we demonstrated that TGF-β1 induced miR-491-5p expression through the MEK/p38 kinase pathway. This study revealed a novel mechanism by which TGF-β disrupts epithelial cell junction.
EXPERIMENTAL PROCEDURES
Animal Experiment
Adult male Sprague-Dawley rats weighing 150 to 180 g were obtained from the Experiment Animal Center at the Northern Campus of Sun Yat-sen University. All animal experiments were approved by the Committee on Animal Experimentation of Sun Yat-sen University and performed in compliance with the Guidelines for the Care and Use of Laboratory Animals of the university. Fifteen male Sprague-Dawley rats were randomly allocated into three groups (five rats in each group): rats in the sham group served as control. Unilateral ureteral obstruction (UUO) was performed using an established procedure (22). Rats were killed at the indicated time points after surgery, and kidneys were removed. Par-3 and miR-491-5p expression was examined by real time PCR of whole kidney lysates.
Reagents
pEm-GFP-miR-491-5p expression vector and scrambled negative control vector were purchased from Genepharm (Shanghai, China). The information regarding the expression vector could be obtained from the Genepharm website. The miR-491-5p expression sequence was inserted into the C-terminal of GFP. The miR-491-5p inhibitor (2′-O-methyl-modified RNA oligonucleotides) designed to inhibit the mature miR-491-5p and the negative control inhibitors were obtained from Thermo (CA). Small interfering RNA for silencing Smad4 was purchased from Qiagen (Hilden, Germany). The target sequence was 5′-AGCAATTGAGATCTGGTAAA-3′. Specific inhibitors for PI3K (wortmannin), MEK (PD 98059), p38 MAPK (SC 68376), PKA (PKAI), and PKC (Ro-31-8220) were purchased from Calbiochem. Polyclonal rabbit anti-Par-3 antibody was purchased from Upstate (New York). Rabbit anti-ZO-1 antibody was from Zymed Laboratories Inc. Rabbit anti-Smad4 antibody and mouse anti-GAPDH were from Cell Signaling. HRP-conjugated goat anti-rabbit or HRP-conjugated goat anti-mouse secondary antibodies were purchased from Cell Signaling (MA). Alexa-546 goat anti-rabbit antibody was from Invitrogen. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and trypsin/EDTA solution for cell culture were all purchased from Invitrogen.
Plasmid Construction
3′ UTR reporter plasmids were constructed via insertion of the Par-3 3′ UTR into the XbaI site 3′ to the firefly luciferase coding region in the pGL3-control vector. For the 3′ UTR of Par-3, an 1161-bp fragment of the Par-3 3′ UTR (gi|3868777) was amplified from genomic DNA of the NRK52E cell by PCR. Mutations to the 3′ UTR of Par-3 were made by replacing two or four nucleotides in the miR-491-5p binding site using the site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene). Primers used for PCR were as follows: pGL3 S1-5, 5′-CAATGGAGGATGGCAGCAT-3′ and 5′-GATTCTCAGGCACTTTAGCA-3′; pGL3 S1mt, 5′-GGACCCCATCCCCATCCGATACCCCAACCT-3′ and 5′-AGGTTGGGGTATCGGATGGGGATGGGGTCC-3′; pGL3 S1′mt, 5′-CACGCGTATATCTCGTTTTTGGACCCCATCCCC-3′ and 5′-GGGGATGGGGTCCAAAAACGAGATATACGCGTG-3′; pGL3 S2mt, 5′-CAAGGCATCTACTCCGTGATTCTCAGCAG-3′ and 5′-CTGCTGAGAATCACGGAGTAGATGCCTTG-3′; pGL3 S3mt, 5′-AGGGGACCCCCCCTCCCCAGTACACGTGTT-3′ and 5′-AACACGTGTACTGGGGAGGGGGGGTCCCCT-3′; pGL3 S4mt, 5′-GCCGGTCTATCCAGTCCTATAGTTACTTCG-3′ and 5′-CGAAGTAACTATAGGACTGGATAGACCGGC-3′; pGL3 S5mt, 5′-TGAGCACCGGGAGTCCGTGACTCCGTGCA-3′ and 5′-TGCACGGAGTCACGGACTCCCGGTGCTCA-3′.
Par-3 3′ UTR Luciferase Reporter Assay
NRK52E cells were co-transfected with the reporter plasmid pRL-TK (Promega). 48 h after transfection, cells were collected by PLB buffer supplied in dual luciferase reporter assays kit (Promega). The luciferase activity was measured by Luminometer (Berthold, Pforzheim, Germany). The data are given as mean ± S.D. of three independent experiments and shown as the ratio of firefly to Renilla luciferase activity.
Cell Cultures and Transfection
HK-2 cells and NRK52E cells were cultured in DMEM supplemented with 10% fetal bovine serum and maintained at 37 °C in a humidified 5% CO2 incubator. Unless otherwise specified, cells were seeded in 6-well plates for 24 h, all plasmids were transfected at a final amount of 1 μg and inhibitors at 50 nm using the Lipofectamine LTX kit according to the manufacturer's instructions (Invitrogen).
Calcium Switch Assay
NRK52E cells were transfected with either pEm-GFP-miR-491-5p or scrambled negative control vector. 48 h after transfection, cells were incubated in DMEM supplemented with 1 mm EDTA for 30 min and switched back to regular medium for the indicated time periods.
RNA Extraction and Real Time PCR
The MiRNeasy Mini Kit (Qiagen, Hilden, Germany) was used for RNA extraction and miRNA purification. RNA was treated by the Turbo DNA-free kit to remove genomic DNA contamination before reverse transcription. SYBR Green-based real time PCR was performed to detect Par-3 and pri-miR-491. The primer sequences were as follows: Par-3, 5′-ACCACCCATATCACAGCGATT-3′ and 5′-CTCAGCTCCTATCTCCTTCCT-3′; pri-miR-491, 5′-CCGCTGTGGAAATTGACTTAG-3′ and 5′-GAGCAGGATCTGACTTCAACC-3′; GAPDH, 5′-CCCGCAGCCTCGTCTCATAGA-3′ and 5′-CTTCGGCCACCCTATCCAC-3′. Reaction was performed on a ABI 7000 real time PCR system (Applied Biosystems). Program parameters were 50 °C for 2 min, 95 °C for 10 min, and then 40 °C cycles at 95 °C for 15 s, 52 °C for 15 s, extended to 72 °C for 31 s.
For mature miR-491-5p detection, real time PCR was performed as described in the TaqMan Micro-RNA Assays protocol using 4.5 S RNA as inner control(Applied Biosystems). Briefly, PCR mixtures that included the TaqMan hsa-miR-491-5p or 4.5 S RNA probe were diluted in TaqMan Universal PCR Master Mix (Applied Biosystems) at a total volume of 20 μl. Cycling parameters were 50 °C for 2 min, 95 °C for 10 min, and then 40 °C cycles of 95 °C for 15 s, extended to 60 °C for 1 min. Relative quantification of gene expression was performed using the 2−ΔΔCt method based on Ct values for both target and reference genes (23). Data are given as mean ± S.D. of three independent experiments.
Western Blot
NRK52E cells were washed with PBS and scraped into lysis buffer (50 mm Hepes, pH 7.5, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 1.5 mm MgCl2, 1 mm EGTA, 10 mm NaF, 10 mm Na4P2O7, 1 mm Na3VO5, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml of leupeptin, and 20 μg/ml of aprotinin). Protein was quantified by the Bradford assay (Bio-Rad) and samples were heated at 100 °C for 10 min before loading. An equal amount of protein was separated on SDS-polyacrylamide gels and transferred onto nitrocellulose membranes (Amersham Biosciences). After blocking in 5% skim milk for 1 h at room temperature, the membranes were incubated with the indicated primary antibodies at 4 °C overnight followed by horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature and detected by chemiluminescence (Amersham Biosciences). Quantification of the Western blot data were performed by measuring the intensity of the hybridization signals using the Fluorchem Image analysis program (24).
Immunofluorescence Staining
NRK52E cells were cultured on 10-mm coverslips in a 24-well plate (EM Science, NJ). Cells were fixed in PBS containing 4% paraformaldehyde and 0.1% Triton X-100 for 10 min at room temperature and 10 min at 4 °C, then in −20 °C methanol for 1 min (25). After washing with PBS, cells were blocked in solution with PBS containing 5% BSA and 10% goat serum for 30 min at 37 °C. Cells were incubated overnight at 4 °C with the indicated primary antibodies diluted in blocking solution, and then incubated with Alexa-546 goat anti-rabbit antibody (Invitrogen) for 1 h at 37 °C. DAPI was subsequently used for nuclei staining for 5 min and samples were mounted in mounting medium (R & D Systems) last. Images were analyzed and collected with 160 Zeiss LSM 510 Confocal Imaging System (Zeiss, Jena, Germany).
In Situ Hybridization
8-μm paraffin sections of normal rat kidney were fixed in 4% paraformaldehyde in PBS for 20 min, and then treated with acetylation solution for 10 min. After a gentle wash in PBS, tissue sections were treated with 5 μg/ml of Proteinase K solution for 10 min, and then prehybridized in hybridization buffer (65% formamide, 5× SSC, 500 mg/ml of yeast tRNA, 50 μg/ml of heparin, 0.1% Tween 20, sodium citrate, pH 6.0) at 42 °C for 4 h. MiR-491-5p expression was detected by the 1 pm digoxigenin-labeled miR-491-5p miRCURYTM LNA detection probe (Exiqon, Vedbaek, Denmark). After overnight hybridization at 55 °C, tissue sections were washed in 5× SSC at 60 °C and then 0.2× SSC at 60 °C for 1 h. After stringency wash, tissue sections were incubated in blocking buffer (0.1 m Tris, pH 7.5, 0.15 m NaCl, and 10% sheep serum) for 30 min and then incubated with anti-digoxigenin-AP Fab fragments (1:1000) overnight at 4 °C. Color reaction was detected by nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution for 30 min (Roche Applied Science). The images were analyzed on Zeiss Axio Plan 2 microscope (Zeiss).
Transepithelial Electrical Resistance Measurements
1 × 105 cells were plated on a 6.5-mm Transwell filter with 0.4-μm pore size (Corning, Corning, NY). A millicell-ERS (Millipore) was used to determine TER quantification. The value was measured at the indicated time point after cell seeding. TER values (Ω × cm2) were calculated by subtraction of the blank value and multiplied by the surface area of the filter. Data were collected from three replicates and represent the mean ± S.D.
Statistical Analysis
Data are shown as the mean ± S.D. from at least three independent experiments. The differences between groups were analyzed using Student's t test, differences were considered statistically significant at p < 0.05.
RESULTS
MiR-491-5p Is Expressed in the Kidney
We previously reported that TGF-β1 down-regulated Par-3 expression in a time- and dose-dependent manner in rat proximal tubular epithelial cells (NRK52E cells), but the underlying mechanism remained unknown (21). Given the important role of micro-RNAs in regulating gene expression, we assessed the 3′ UTR of Par-3 using the online prediction service of the Segal Lab of Computational Biology center and found 5 sites in the 3′ UTR of Par-3 that perfectly match the seed region of miR-491-5p (Fig. 1A) (26). By scanning the genome database of the University of Southern California, we found that pre-miR-491 was located on chromosome 9 in human and chromosome 4 in mouse. Although, pre-miR-491 was not found in the rat assembled genome, we were able to identify it in traces in the NCBI database (gnl ti 26755808), which contained segments from the whole genome short gun sequencing (Fig. 1B).
FIGURE 1.
The 3′ UTR of Par-3 harbors five putative binding sites of miR-491-5p. A, schematic representation of the putative miR-491-5p target sites in the 3′ UTR of Par-3. The bases of the putative miR-491-5p target sites in the Par-3 3′ UTR are indicated in the box. B, alignment of the pre-miR-491 sequence (84 nucleotides length) in human, rhesus, mouse, rat, and dog genome. The sequences of mature miR-491-5p are shown in bold letters.
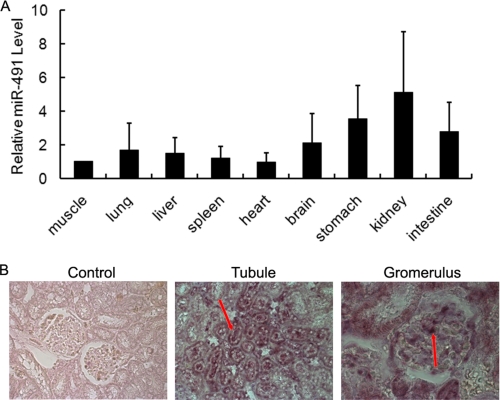
To explore the potential function of miR-491-5p, we first examined the expression of miR-491-5p in rat tissues by real time PCR. As shown in Fig. 2A, miR-491-5p was ubiquitously expressed in various tissues but relatively highly expressed in the kidney and stomach. We next performed in situ hybridization to further dissect localization of miR-491-5p in the kidney. As shown in Fig. 2B, positive staining for miR-491-5p was detected in both glomeruli and tubules.
FIGURE 2.
A, the expression of miR-491-5p is ubiquitous in rat tissues but relatively high in the kidney. Micro-RNAs were reverse transcribed utilizing miR-491-5p and 4.5 S RNA-specific primers, and real time PCR was performed as described under “Experimental Procedures.” The relative expression of the mature miR-491-5p was normalized to 4.5 S RNA. Data are given as mean ± S.D. of four rats. B, the localization of miR-491-5p in the rat kidney was detected by in situ hybridization. Paraffin-fixed sections of normal rat kidney were hybridized with the digoxigenin-labeled miR-491-5p LNA probe. Positive signal (purple blue) for miR-491-5p is indicated by arrows. Original magnification of the images for negative control and tubule was ×200. Original magnification of the image for glomerulus was ×400.
Par-3 Is a Target of miR-491-5p
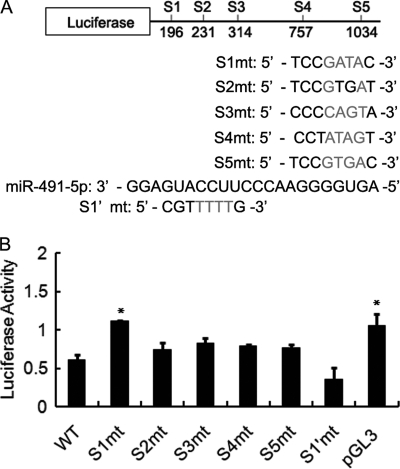
To evaluate the ability of miR-491-5p binding to the 3′ UTR of Par-3, we first synthesized a reporter plasmid bearing a 1.1-kb fragment of the rat Par-3 3′ UTR behind luciferase and co-transfected it with miR-491-5p or negative control micro-RNA into NRK52E cells. Compared with empty vector (pGL3), miR-491-5p significantly inhibited the luciferase activity of the wild type Par-3 3′ UTR reporter by 40% (Fig. 3B). We next mutated the 5 putative target sites of miR-491-5p in the 3′ UTR of Par-3 (Fig. 3A) and then co-transfected them with miR-491-5p, respectively. As shown in Fig. 3B, although all mutations attenuated the effect of miR-491-5p on the activity of the Par-3 3′ UTR reporter, only mutation of the putative target site 1 (S1), which is 196 nucleotides downstream from the stop codon, resulted in a significant increase of luciferase activity compared with that of the wild type Par-3 3′ UTR. Because S1 is perfectly base-paired of both 5′ and 3′ segments of the target sequence, we further tested whether the 5′ segment of S1 is also necessary for miR-491-5p binding. As shown in Fig. 3B, mutation of the 5′ segment of S1 (S1′mt) showed similar luciferase activity as the wild type Par-3 3′ UTR, indicating that the 5′ segment of S1 is not responsible for function of the S1 site.
FIGURE 3.
MiR-491-5p can inhibit reporter activity. A, schematic representation of the luciferase reporter constructs. Six mutant constructs were made by replacing two or four nucleotides in the five potential miR-491-5p binding sites in the Par-3 3′ UTR. Mutated nucleotides are marked in gray. B, the luciferase reporter constructs carrying either wild-type or mutant 3′ UTR of Par-3 was co-transfected with miR-491-5p or negative control into NRK52E cells. Renilla luciferase expressing vector pRL-TK was used as an internal control to correct for the differences in both transfection and harvest efficiencies. Data are shown as the relative luciferase activity normalized to negative control transfected cells. All data are mean ± S.D. of three independent experiments. WT, wild type Par-3 3′ UTR; S1mt to S5mt, mutants of the 3′ segment of the five potential binding sites in the 3′ UTR of Par-3; S1′mt, mutant of the 5′ segment of site 1 in the 3′ UTR of Par-3; pGL3, empty vector. *, p < 0.05 versus the activity of wild type 3′ UTR of Par-3.
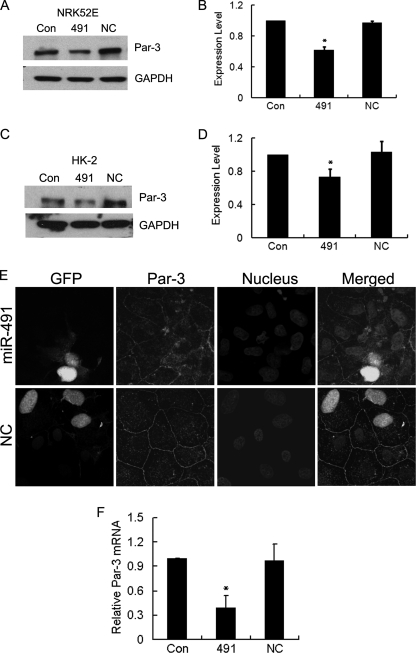
Although the luciferase reporter assay was useful in evaluating the effect of miR-491-5p on regulating Par-3 expression, it is important to demonstrate that miR-491-5p can regulate the expression of endogenous Par-3. To this end, NRK52E cells were transfected with either GFP-tagged miR-491-5p or control micro-RNA. Cell lysates were harvested 48 h after transfection and Western blot was performed to detect the protein level of endogenous Par-3. As shown in Fig. 4, A and B, the protein level of Par-3 was significantly lower in miR-491-5p-transfected cells than in the control cells. To ensure the effect of miR-491-5p on Par-3 expression was not specific to NRK52E cells, we transfected GFP-tagged miR-491-5p in human proximal tubular epithelial cells (HK-2 cells) and demonstrated that miR-491-5p, too, significantly down-regulated Par-3 expression (Fig. 4, C and D). Immunofluorescence microscopy further confirmed the attenuation of Par-3 immunostaining in GFP-tagged miR-491-5p-transfected NRK52E cells (Fig. 4E). Moreover, to investigate whether miR-491-5p could target Par-3 mRNA for degradation, real time PCR was performed. As shown in Fig. 4F, miR-491-5p significantly decreased the mRNA level of Par-3, suggesting that miR-491-5p regulates Par-3 expression through promoting Par-3 mRNA degradation.
FIGURE 4.
MiR-491-5p down-regulated Par-3 expression via promoting Par-3 mRNA degradation. MiR-491-5p was transfected into either NRK52E cells (A) or HK-2 cells (C). Cells were transfected with either miR-491-5p or negative control. 48 h after transfection, cell lysates were harvested and the protein level of Par-3 was analyzed by Western blot. GAPDH was used to verify equivalent loading. Graphic presentation of relative abundance of Par-3 normalized to GAPDH in NRK52E cells (B) and HK-2 cells (D). Data are given as mean ± S.D. of three independent experiments. *, p < 0.05 versus negative control transfected cells. E, immunofluorescence evidence for the decreased expression of Par-3 in miR-491-5p-transfected cells. NRK52E cells were transfected with either miR-491-5p or negative control. 48 h after transfection, cells were fixed and stained with anti-Par-3 antibody (red). Nuclei were stained with DAPI (blue). Images (original magnification ×630) were taken by confocal microscopy. F, real time PCR revealed that miR-491-5p promoted Par-3 mRNA degradation. NRK52E cells were transfected with either miR-491-5p or negative control. 48 h after transfection, mRNA was harvested and real time PCR was performed. Data are the mean ± S.D. of three independent experiments, as normalized to GAPDH expression. *, p < 0.05 versus untransfected cells. Con, untransfected cells. 491, miR-491-5p-transfected cells. NC, negative control transfected cells.
MiR-491-5p Overexpression Disrupts Cell-Cell Junction
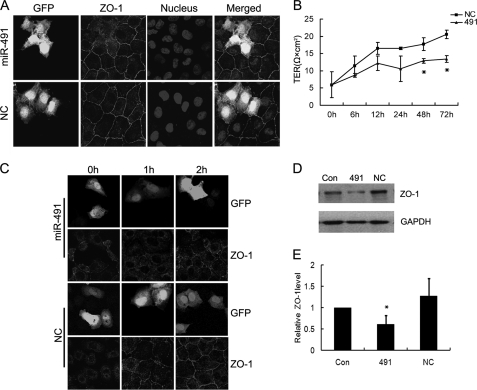
In epithelial cells, the Par complex is important for modulating tight junction homeostasis. We therefore investigated the biological significance of miR-491-5p on integrity of the tight junction. GFP-tagged miR-491-5p was transiently transfected into NRK52E cells. As shown in Fig. 5A, overexpression of miR-491-5p resulted in a dramatic loss of ZO-1, a marker of tight junction, from cell-cell contacts, suggesting a defect in tight junction formation. We next investigated the effect of miR-491-5p on integrity of the tight junction by measuring TER at different time points after transfection. TER was measured up to 72 h post-transfection to ensure full confluence of all monolayers. As shown in Fig. 5B, the decrease of TER in miR-491-5p-transfected cells was detected 6 h after transfection. At 72 h after transfection, miR-491-5p overexpression led to a 34% reduction in TER compared with the control.
FIGURE 5.
Overexpression of miR-491-5p disrupted tight junction of NRK52E cells. A, immunofluorescence staining of ZO-1 (red) in GFP-tagged miR-491-5p-transfected NRK52E cells and negative control transfected cells. Nuclei were stained with DAPI (blue). Images (original magnification ×630) were taken by confocal microscopy. B, TER was measured at the indicated time points after transfection. TER values (Ω × cm2) were calculated by subtraction of blank value and multiplied by the surface area of the filter. Each value is the mean of three independent measurements. C, overexpression of miR-491-5p delayed relocalization of ZO-1 to cell borders during a calcium switch assay. NRK52E cells were transfected with negative control or GFP-tagged miR-491-5p. 48 h after transfection, cells were incubated with a medium with low calcium and stained for ZO-1 (red) after re-addition of calcium at 0, 1, and 2 h. Original magnification ×630. Con, untransfected cells. 491, miR-491-5p-transfected cells. NC, negative control transfected cells. D, miR-491-5p overexpression decreased ZO-1 expression. NRK52E cells were transfected with miR-491-5p or negative control. 48 h after transfection, cell lysates were harvested and ZO-1 expression was analyzed by Western blot. GAPDH was used to verify equivalent loading. E, graphic presentation of relative abundance of ZO-1 normalized to GAPDH. Data are the mean of three independent experiments, *, p < 0.05 versus negative control transfected cells.
Because the depletion of Par-3 resulted in the delay of tight junction assembly, we next investigated the effect of miR-491-5p on formation and maturation of the intercellular contacts by calcium switch assay. NRK52E cells grown in low calcium medium exhibited a rounded morphology and lack tight junctions. Upon switching back to normal culture medium, control cells formed intact tight junctions within 2 h. Whereas, in miR-491-5p-transfected cells, the ZO-1 staining pattern was still fragmented (Fig. 5C). Interestingly, we found a dramatic decrease of ZO-1 protein level in miR-491-5p-transfected cells compared with control cells (Fig. 5, D and E), suggesting that miR-491-5p might regulate ZO-1 expression.
MiR-491-5p Is Up-regulated by TGF-β1 at the Transcriptional Level
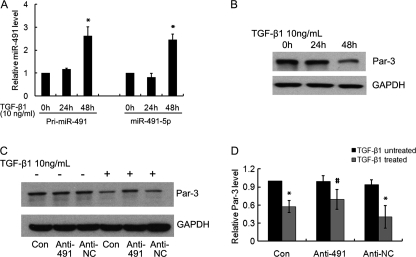
Because we previously reported that TGF-β1 down-regulated Par-3 expression in NRK52E cells, we next examined whether TGF-β1 could modulate miR-491-5p expression. NRK52E cells were treated with 10 ng/ml of TGF-β1. RNA was collected at various time points and real time PCR was performed for pri-miR-491 and mature miR-491-5p. As shown in Fig. 6A, TGF-β1 treatment for 48 h led to a dramatic increase of both pri-miR-491 and mature miR-491-5p. Meanwhile, Par-3 expression was significantly decreased (Fig. 6B). This data indicates that TGF-β1 regulates miR-491-5p at the transcriptional level.
FIGURE 6.
A, TGF-β1 induced miR-491-5p expression. NRK52E cells were harvested 0, 24, and 48 h after TGF-β1 treatment and real time PCR was performed as described under “Experimental Procedures.” Expression of pri-miR-491 and mature miR-491-5p is shown. All PCR were performed in triplicate; data are the mean ± S.D. of three independent experiments. *, p < 0.05 versus TGF-β1 untreated cells. B, NRK52E cells were harvested at 0, 24, and 48 h after 10 ng/ml of TGF-β1 treatment. Western blot analysis was performed to analyze Par-3 expression. GAPDH was used to verify equivalent loading. C, miR-491-5p inhibitor blocked TGF-β1-dependent Par-3 down-regulation. NRK52E cells were transfected with either miR-491-5p inhibitor or negative control inhibitor and then treated with 10 ng/ml of TGF-β1. 48 h after TGF-β1 treatment, cell lysates were harvested and the protein level of Par-3 analyzed by Western blot. GAPDH was used to verify equivalent loading. D, graphic presentation of relative abundance of Par-3 normalized to GAPDH. Data are mean of three independent experiments. *, p < 0.05 versus TGF-β1 untreated cells; #, p < 0.05 versus TGF-β1 treated control cells.
We next examined whether the miR-491-5p inhibitor could block TGF-β1-dependent Par-3 down-regulation. NRK52E cells were transfected with either miR-491-5p inhibitor or control. After 48 h of TGF-β1 treatment, the Par-3 protein level was examined by Western blot. As shown in Fig. 6, C and D, Par-3 expression was significantly reduced by TGF-β1 treatment in control cells. In contrast, in miR-491-5p inhibitor-transfected cells, the Par-3 level did not change obviously.
MiR-491-5p Expression Is Induced in the Fibrotic Kidney
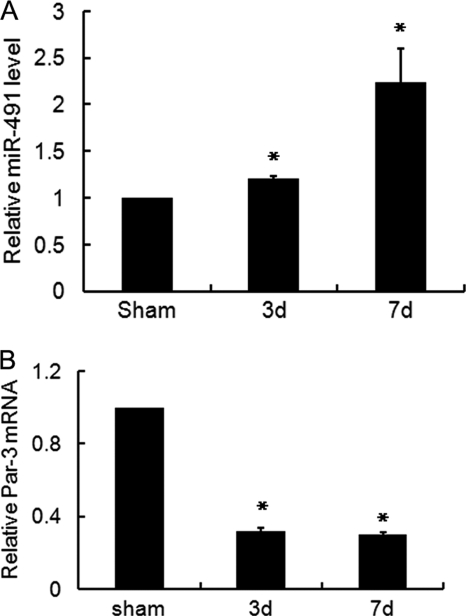
Previous study has demonstrated that TGF-β1 is up-regulated in the process of renal fibrosis. We thus examined the expression of miR-491-5p in the fibrotic kidney induced by UUO. As shown in Fig. 7A, real time PCR revealed that miR-491-5p expression was significantly increased in a time-dependent manner after UUO. Whereas, the mRNA level of Par-3 was dramatically decreased when compared with the sham controls (Fig. 7B). This data indicated that in the fibrotic kidney, miR-491-5p induction was correlated with down-regulation of Par-3 expression.
FIGURE 7.
The expression of miR-491-5p was induced in the fibrotic kidney after UUO. Real time PCR detected a marked induction of the miR-491-5p (A) and a dramatic decrease of Par-3 (B) in the fibrotic kidney induced by UUO in a time-dependent manner. The relative expression of the mature miR-491-5p was normalized to 4.5 S RNA. The expression of Par-3 was normalized to GAPDH. Sham, the sham control; 3 d and 7 d, 3 and 7 days after UUO, respectively. Data are given as mean ± S.D. of five rats. *, p < 0.05 versus the sham group.
TGF-β1-regulated Par-3 and miR-491-5p Expression through p38/MEK Pathway
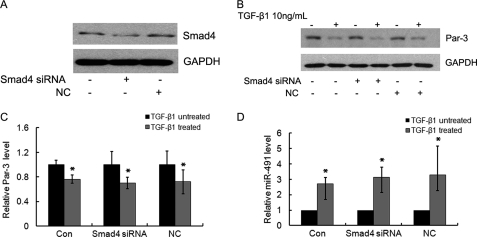
It has been reported that TGF-β1 activated several distinctive signaling pathways such as Smads, p38, MEK, PI3K, PKC, and PKA. Among them, the Smad pathway is the major avenue to transduce TGF-β-specific cellular responses. To unravel the signaling pathway by which TGF-β1 regulates Par-3 and miR-491-5p expression, we first specifically knocked down Smad4 expression by siRNA in NRK52E cells. The reduced Smad4 expression was confirmed by Western blot (Fig. 8A). However, knockdown of Smad4 did not affect TGF-β1-suppressed Par-3 expression (Fig. 8, B and C), nor TGF-β1-induced miR-491-5p expression (Fig. 8D). These results indicated that the Smad pathway is not responsible for TGF-β-regulated Par-3 and miR-491-5p expression.
FIGURE 8.
Smad pathway is not involved in TGF-β1 regulated Par-3 and miR-491-5p expression. A, the protein level of Smad4 was detected by Western blot. GAPDH was used to verify equivalent loading. B, knockdown of Smad4 by siRNA did not affect TGF-β1-suppressed Par-3 expression. NRK52E cells transfected with Smad4 siRNA or negative control siRNA were treated with 10 ng/ml of TGF-β1 for 48 h, and then cell lysates were harvested and Par-3 expression was analyzed by Western blot. GAPDH was used to verify equivalent loading. C, graphic presentation of the relative protein level of Par-3 in Smad4 siRNA or negative control siRNA-transfected NRK52E cells. The expression of Par-3 was normalized to GAPDH. Data are mean ± S.D. of three independent experiments. *, p < 0.05 versus TGF-β1-untreated cells. D, real time PCR analysis of the miR-491-5p level in NRK52E cells transfected with Smad4 siRNA or negative control siRNA. Data are the mean ± S.D. of three independent experiments. *, p < 0.05 versus TGF-β1-untreated cells. Con, untransfected cells; Smad4 siRNA, Smad4 siRNA-transfected cells; NC, scramble siRNA-transfected cells.
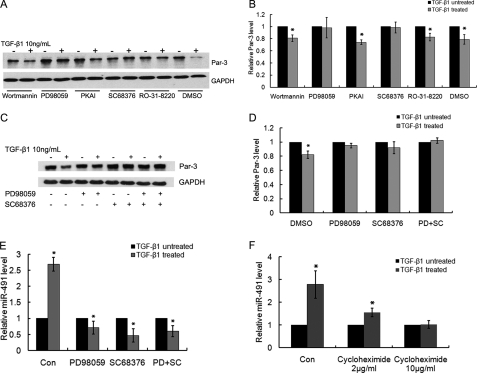
We thus blocked Smad-independent pathways by treating NRK52E cells with various specific chemical inhibitors before TGF-β1 treatment. As shown in Fig. 9, A and B, specific inhibitors for PI3K (wortmannin), PKA (PKAI), and PKC (Ro-31-8220) did not display obvious effect on TGF-β1-induced Par-3 down-regulation. In contrast, inhibition of p38 MAPK (by SC 68376) or MEK (by PD 98059) significantly inhibited TGF-β1-initiated Par-3 down-regulation. However, no further significant effect was detected on the Par-3 protein level when the cells were treated with SC 68376 and PD 98059 together (Fig. 9, C and D).
FIGURE 9.
p38 MAPK and MEK inhibitors blocked TGF-β1-regulated Par-3 and miR-491-5p expression. A, NKR52E cells were treated with 10 μm of various chemical inhibitors or vehicle control (DMSO, dimethyl sulfoxide) for 6 h, followed by 48 h of TGF-β1 (10 ng/ml) treatment. The protein level of Par-3 was detected by Western blot. GAPDH was used to verify equivalent loading. Specific inhibitors for phosphoinositide 3-kinase (wortmannin), MEK (PD 98059), p38 mitogen-activated protein kinase (SC 68376), protein kinase A inhibitor (PKAI), and protein kinase C (Ro-31-8220) were used, respectively. B, graphic presentation of the relative protein level of Par-3 in the phosphoinositide 3-kinase inhibitor (wortmannin), MEK inhibitor (PD 98059), p38 MAPK inhibitor (SC 68376), protein kinase A inhibitor (PKAI), and protein kinase C inhibitor (Ro-31-8220) treated cells normalized to GAPDH. Data are mean ± S.D. of three independent experiments. *, p < 0.05 versus TGF-β1 untreated cells. C, Western blot analysis of the Par-3 level in cells treated with MEK inhibitor (PD 98059), p38 MAPK inhibitor (SC 68376), or PD 98059 combined with SC 68376. After a 6-h treatment, NRK52E cells were treated with 10 ng/ml of TGF-β1 for 48 h. Western blot analysis was performed to examine the expression of Par-3, GAPDH was used to verify equivalent loading. D, graphic presentation of the relative protein level of Par-3 in MEK inhibitor- (PD 98059) and p38 MAPK inhibitor (SC 68376)-treated cells normalized to GAPDH. Data are mean ± S.D. of three independent experiments. *, p < 0.05 versus TGF-β1-untreated cells. E, real time PCR analysis of the miR-491-5p level in cells treated with the MEK inhibitor (PD 98059), p38 MAPK inhibitor (SC 68376), or PD 98059 combined with SC 68376. Data are the mean ± S.D. of three independent experiments. *, p < 0.05 versus TGF-β1-untreated cells. F, cycloheximide inhibited TGF-β1-induced miR-491-5p expression. NRK52E cells were pre-treated with 2 or 10 μg/ml of cycloheximide for 6 h, followed by 10 ng/ml of TGF-β1 for 48 h. Real time PCR was performed to detect the expression of miR-491-5p. Data are the mean ± S.D. of three independent experiments, as measured by TaqMan qRT-PCR and normalized to 4.5 S RNA. *, p < 0.05 versus TGF-β1 untreated cells.
We further examined whether miR-491-5p was regulated by the same pathway. As shown in Fig. 9E, real time PCR revealed that TGF-β1-induced miR-491-5p expression was dramatically blocked by p38 MAPK or MEK inhibitor, and no further effect was observed when cells were treated with SC 68376 and PD 98059 together, which is consistent with that Par-3 regulation. Taken together, these data suggest that TGF-β regulates miR-491-5p and Par-3 expression through the p38/MEK pathway.
It has been demonstrated that MEK and p38 MAPK kinases regulate downstream target genes indirectly through activating AP-1 or ATF2 transcription and translation (27). To test whether TGF-β1 induction of miR-491-5p is direct, we utilized cycloheximide to inhibit protein synthesis. As shown in Fig. 9F, pretreatment with cycloheximide 6 h prior to addition of TGF-β1 effectively blocked miR-491-5p induction in NRK52E cells, suggesting that TGF-β1-induced-miR-491-5p expression requires de novo protein synthesis.
DISCUSSION
In the present study, miR-491-5p was identified to regulate Par-3 expression by binding to the 3′ UTR of Par-3. TGF-β-induced miR-491-5p expression, which in turn down-regulated Par-3 expression by the acceleration of Par-3 mRNA degradation. The miR-491-5p inhibitor dramatically blocked the effect of TGF-β on down-regulation of Par-3 expression. Moreover, we detected a significant increase in the miR-491-5p level in a rat model of obstructive nephropathy, in parallel with the decreased Par-3 level. We dissected the signaling pathway, which mediated the effect of TGF-β on both Par-3 and miR-491-5p regulation and found that blocking the p38 and MEK pathways significantly attenuated TGF-β-regulated miR-491-5p and Par-3 expression. These observations provide a mechanistic explanation for our previous observation that TGF-β down-regulated Par-3 expression.
Using the online prediction service of the Segal Lab of Computational Biology center, we found potential binding sites of miR-491-5p, miR-608, and miR-638 in the 3′ UTR of Par-3. Further analysis revealed that the binding sites of miR-638 are not well conserved across species, and expression of miR-608 was not detected in the kidney (data not shown). Therefore, we focused on the role of miR-491-5p on the regulation of Par-3 expression.
We first assessed the complementarity of miR-491-5p to the 3′ UTR of Par-3 and found 5 potential binding sites. Mutation of either putative site could block the effect of miR-491-5p on the reporter activity, but only S1 showed a statistically significant effect. Further analysis revealed that S1 shows dramatic base pairing of both 5′ and 3′ segments of the micro-RNA, whereas, S2–S5 only shows complementarity to the 3′ segment. However, mutation of the 5′ segment of site 1 did not relieve the inhibitory effect of miR-491-5p on the luciferase activity. These observations are in accordance with a previous report that complementarity of seven or more bases to the 3′ end of micro-RNA is sufficient to confer target regulation (28). The stronger regulation effect of S1 than S2–S5 might due to the accessibility of this site to miR-491-5p. It has been reported that the folding structure of mRNAs may mask some potential micro-RNA target sites and therefore influence micro-RNA target recognition (29). Moreover, our data could not exclude the possibility that miR-491-5p binds to more than one site and combined binding would have a more dramatic effect on the reporter activity.
It has been reported that micro-RNAs either regulated the stability or translational efficiency of target mRNAs (14). We found that overexpression of miR-491-5p led to decreased Par-3 mRNA, indicating that miR-491-5p regulates Par-3 expression through promoting its mRNA degradation. Consistent with our data, Nakano et al. (30) showed that miR-491-5p degraded Bcl-XL mRNA in colorectal cancer cells. Further sequence analysis revealed that the binding sites of miR-491-5p in both the 3′ UTR of Par-3 and Bcl-XL are perfect for base pairing with the seed sequence of miR-491-5p. These data support the idea that micro-RNA induces mRNA degradation when the seed sequence perfectly matches the target 3′ UTR or inhibits translation when the sequences are partially identical (31, 32).
The miR-491-5p target site harbored in the 3′ UTR of Par-3 mRNA is highly conserved through evolution. In addition, miR-491-5p could regulate Par-3 expression in both human and rat proximal epithelial cells, suggesting that miR-491-5p-mediated translational repression of Par-3 is well conserved across species. However, miR-491-5p was not shown to be regulated by TGF-β in human keratinocytes and murine mammary gland epithelial cells using the micro-RNA microarray (33). This discrepancy may be due to the different chromosomal locations of miR-491-5p in different species. Second, the signal transduction pathways that regulate miR-491-5p expression may also vary in different tissues. Therefore, we speculate that TGF-β-regulated miR-491-5p expression may vary with species.
In the present study, miR-491-5p expression in the rat kidney was detected by real time PCR. In situ hybridization further revealed that miR-491-5p was localized in both the glomerulus and the renal tubules. In support of our finding, Tian et al. (34) identified miR-491-5p expression in the cortex and medulla of rat kidneys by miRNA microarray. Moreover, we detected increased miR-491-5p expression in the fibrotic kidney, suggesting a role of miR-491-5p in the pathogenesis of renal fibrosis. We previously reported that knockdown of endogenous Par-3 could promote epithelial to mesenchymal transition of NRK2E cells (21). Nakano et al. (30) reported that miR-491-5p could induce cell apoptosis via targeting Bcl-XL. Taken together, it is plausible to postulate a multifunctional role for miR-491-5p in both renal tubular and glomerular injury.
The expression of micro-RNAs has been shown to be regulated at transcriptional and/or post-transcriptional levels (35, 36). We detected an increase of pri-miR-491 upon TGF-β treatment, suggesting that TGF-β regulates miR-491-5p at the transcriptional level. However, the induction of miR-491-5p by TGF-β was detected 48 h after treatment, implying the effect of TGF-β on miR-491-5p expression might not be direct. Consistent with our speculation, we found that cycloheximide could block TGF-β-induced miR-491-5p expression, suggesting that de novo protein synthesis is required for TGF-β-induced miR-491-5p expression. In addition, we demonstrated that TGF-β induced miR-491-5p expression via p38/MEK kinase activation and it is well known that the p38/MEK kinase pathway regulates downstream target genes indirectly through activating AP-1 or ATF2 transcription and translation (27). Further dissecting the promoter region of miR-491-5p and identification of the transcription factors responsible for miR-491-5p expression will provide additional support to our speculation.
Par-3 is a key factor in the generation and maintenance of cell polarity. In epithelial cells, Par-3 functions as a modulator of tight junction homeostasis and apical-basal polarity. Although Par-3 is critical for maintaining cell polarity, the regulation of Par-3 expression remains largely unknown. Our study, for the first time, provides a mechanism by which Par-3 expression is regulated. Moreover, our findings revealed that miR-491-5p-regulated Par-3 expression is a novel mechanism by which TGF-β disrupts tight junction.
This work was supported by National Science Foundation of China Grant 30871033 and Natural Science Foundation of Guangdong province Grant 8251008901000014.
- TER
- transepithelial resistance
- UUO
- unilateral ureteral obstruction
- miRNA
- micro-RNA.
REFERENCES
- 1.Tsukita S., Furuse M., Itoh M. (2001) Nat. Rev. Mol. Cell Biol. 2, 285–293 [DOI] [PubMed] [Google Scholar]
- 2.Lin D., Edwards A. S., Fawcett J. P., Mbamalu G., Scott J. D., Pawson T. (2000) Nat. Cell Biol. 2, 540–547 [DOI] [PubMed] [Google Scholar]
- 3.Suzuki A., Ohno S. (2006) J. Cell Sci. 119, 979–987 [DOI] [PubMed] [Google Scholar]
- 4.Macara I. G. (2004) Nat. Rev. Mol. Cell Biol. 5, 220–231 [DOI] [PubMed] [Google Scholar]
- 5.Shin K., Fogg V. C., Margolis B. (2006) Annu. Rev. Cell Dev. Biol. 22, 207–235 [DOI] [PubMed] [Google Scholar]
- 6.Chen X., Macara I. G. (2005) Nat. Cell Biol. 7, 262–269 [DOI] [PubMed] [Google Scholar]
- 7.Iden S., Rehder D., August B., Suzuki A., Wolburg-Buchholz K., Wolburg H., Ohno S., Behrens J., Vestweber D., Ebnet K. (2006) EMBO Rep. 7, 1239–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Macara I. G. (2006) J. Cell Biol. 172, 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ooshio T., Fujita N., Yamada A., Sato T., Kitagawa Y., Okamoto R., Nakata S., Miki A., Irie K., Takai Y.(2007) J. Cell Sci. 120, 2352–2365 [DOI] [PubMed] [Google Scholar]
- 10.Liu Y. (2004) J. Am. Soc. Nephrol. 15, 1–12 [DOI] [PubMed] [Google Scholar]
- 11.Fan J. M., Huang X. R., Ng Y. Y., Nikolic-Paterson D. J., Mu W., Atkins R. C., Lan H. Y. (2001) Am. J. Kidney Dis. 37, 820–831 [DOI] [PubMed] [Google Scholar]
- 12.Yang J., Liu Y. (2001) Am. J. Pathol. 159, 1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeisberg M., Hanai J., Sugimoto H., Mammoto T., Charytan D., Strutz F., Kalluri R. (2003) Nat. Med. 9, 964–968 [DOI] [PubMed] [Google Scholar]
- 14.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 15.Calin G. A., Croce C. M. (2006) Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 16.Meister G., Tuschl T. (2004) Nature 431, 343–349 [DOI] [PubMed] [Google Scholar]
- 17.Ambros V. (2004) Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 18.Krützfeldt J., Stoffel M. (2006) Cell Metab. 4, 9–12 [DOI] [PubMed] [Google Scholar]
- 19.Kato M., Zhang J., Wang M., Lanting L., Yuan H., Rossi J. J., Natarajan R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3432–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong W., Yang H., He L., Zhao J. J., Coppola D., Dalton W. S., Cheng J. Q. (2008) Mol. Cell. Biol. 28, 6773–6784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Nie J., Zhou Q., Liu W., Zhu F., Chen W., Mao H., Luo N., Dong X., Yu X. (2008) Biochim. Biophys. Acta 1782, 51–59 [DOI] [PubMed] [Google Scholar]
- 22.Vidyasagar A., Reese S., Acun Z., Hullett D., Djamali A. (2008) Am. J. Physiol. Renal Physiol. 295, F707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 24.Zhu F., Li T., Qiu F., Fan J., Zhou Q., Ding X., Nie J., Yu X. (2010) Am. J. Pathol. 176, 650–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brock R., Hamelers I. H., Jovin T. M. (1999) Cytometry 35, 353–362 [DOI] [PubMed] [Google Scholar]
- 26.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. (2007) Nat. Genet. 39, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 27.Kojima R., Taniguchi H., Tsuzuki A., Nakamura K., Sakakura Y., Ito M. (2010) J. Immunol. 184, 5253–5262 [DOI] [PubMed] [Google Scholar]
- 28.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 29.Doench J. G., Sharp P. A. (2004) Genes Dev. 18, 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano H., Miyazawa T., Kinoshita K., Yamada Y., Yoshida T. (2009) Int. J. Cancer 27, 1072–1080 [DOI] [PubMed] [Google Scholar]
- 31.Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 32.Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R. (2006) Genes Dev. 20, 515–524 [DOI] [PubMed] [Google Scholar]
- 33.Zavadil J., Narasimhan M., Blumenberg M., Schneider R. J. (2007) Cells Tissues Organs 185, 157–161 [DOI] [PubMed] [Google Scholar]
- 34.Tian Z., Greene A. S., Pietrusz J. L., Matus I. R., Liang M. (2008) Genome Res. 18, 404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis B. N., Hilyard A. C., Lagna G., Hata A. (2008) Nature 454, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Q., Zhang Y., Yang G., Chen X., Zhang Y., Cao G., Wang J., Sun Y., Zhang P., Fan M., Shao N., Yang X. (2008) Nucleic Acids Res. 36, 2690–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]