Abstract
The Abl tyrosine kinases, Abl and Arg, play a role in the regulation of the actin cytoskeleton by modulating cell-cell adhesion and cell motility. Deregulation of both the actin cytoskeleton and Abl kinases have been implicated in cancers. Abl kinase activity is elevated in a number of metastatic cancers and these kinases are activated downstream of several oncogenic growth factor receptor signaling pathways. However, the role of Abl kinases in regulation of the actin cytoskeleton during tumor progression and invasion remains elusive. Here we identify the Abl kinases as essential regulators of invadopodia assembly and function. We show that Abl kinases are activated downstream of the chemokine receptor, CXCR4, and are required for cancer cell invasion and matrix degradation induced by SDF1α, serum growth factors, and activated Src kinase. Moreover, Abl kinases are readily detected at invadopodia assembly sites and their inhibition prevents the assembly of actin and cortactin into organized invadopodia structures. We show that active Abl kinases form complexes with membrane type-1 matrix metalloproteinase (MT1-MMP), a critical invadopodia component required for matrix degradation. Further, loss of Abl kinase signaling induces internalization of MT1-MMP from the cell surface, promotes its accumulation in the perinuclear compartment and inhibits MT1-MMP tyrosine phosphorylation. Our findings reveal that Abl kinase signaling plays a critical role in invadopodia formation and function, and have far-reaching implications for the treatment of metastatic carcinomas.
Keywords: Breast Cancer, Chemokines, Nonreceptor Tyrosine Kinase, Signal Transduction, Src, Abl, MT1-MMP, Invadopodia, Invasion, Matrix Degradation
Introduction
Podosomes and invadopodia are specialized protrusive structures consisting of a core assembly of F-actin- and actin-binding proteins that form on the ventral surface of migratory and invading cells. These structures are observed in physiological and pathological processes that involve remodeling of the extracellular environment and are found in endothelial cells during extracellular matrix (ECM)5 degradation (1), transmigrating monocytic cells (2, 3), osteoclasts during bone reabsorption (4), and cancer cells during invasion and metastasis (5). Although podosomes and invadopodia are structurally distinct, they share many common features such as the enrichment of integrins, actin regulatory proteins, matrix metalloproteinases (MMPs), and tyrosine kinases (6–8).
Carcinoma cells utilize invadopodia to degrade the ECM during tumor invasion and metastasis (8). Invadopodia assembly occurs through sequential steps that begin with the assembly of precursor structures containing actin, cortactin, Tsk5, N-WASP, and other actin regulatory proteins, and progress into mature structures with matrix degradation activity (9). Invadopodia were first described in cells transformed with oncogenic v-Src (10), and endogenous Src kinases have been shown to promote podosome/invadopodia formation in response to growth factors and chemokines (1, 11–13). Src phosphorylates several invadopodia components including cortactin, N-WASP, and Tsk5/FISH (14). Cortactin regulates the formation and maturation of invadopodia (9, 15). Tyrosine phosphorylation of cortactin regulates the recruitment of N-WASP, Nck, and Arp2/3-dependent actin polymerization at invadopodia. Moreover, subsequent cortactin dephosphorylation promotes invadopodia maturation and MMP-dependent matrix degradation (16–18). While cortactin was initially identified as a Src substrate, recent reports showed that Abl kinases phosphorylate and interact with cortactin and may mediate Src-induced cortactin phosphorylation in response to growth factor stimulation (19, 20).
The Abl family of non-receptor tyrosine kinases consists of two members, Abl (Abl1) and Abl-related gene (Arg or Abl2). Abl tyrosine kinases were first identified as oncogenes in leukemias harboring aberrant Abl fusion proteins (i.e. Bcr-Abl, Tel-Abl, Tel-Arg) generated by chromosomal translocation events (21). Constitutive activation of Abl kinases promotes proliferation and tumorigenesis (22, 23). Recently, accumulating data suggest that activation of the Abl kinases may play a role in the development of solid tumors as Arg is elevated in advanced and high-grade colorectal and pancreatic cancers (24, 25). Moreover, Abl kinase activity is required for the proliferation, survival and invasion of breast cancer cells (22, 23, 26). Abl kinases have been implicated in the regulation of cytoskeletal dynamics, cell morphology, adhesion, and migration (21, 27). In this regard, in addition to cortactin, Abl kinases phosphorylate and interact with a number of actin-regulatory proteins including N-WASP (20, 28) (29), the Abl-interacting (Abi) proteins (30, 31), and WAVE2 (32). We previously showed that Abl kinases are activated downstream of Src kinases in response to growth factor stimulation and are required for cytoskeletal remodeling downstream of cell surface receptors (12, 33, 34). Here we examine whether the effects of Src and extracellular factors on invadopodia formation and maturation are mediated, in part, by Abl kinases. We show that the Abl kinases are required for Src-induced invadopodia formation and MMP-dependent matrix degradation and function downstream of chemokines in these processes.
EXPERIMENTAL PROCEDURES
Antibodies
Anti-Abl (clone 8E9), anti-phosphotyrosine (clone 4G10), and anti-CrkL were purchased from BD Biosciences. Anti-Arg (clone 9H5) and anti-GFP antibodies were purchased from Santa Cruz Biotechnology. Anti-cortactin (4F11) was purchased from Upstate. Anti-phospho-CrkL (Tyr207), anti-phospho-Akt (Ser-473), anti-Akt, and anti-NA, K-ATPase were purchased from Cell Signaling Technology. Anti-MT1-MMP (clone 3G4.2) used for Western blot analysis and anti-MT1-MMP (clone 113-5B7) used for immunoprecipitation were purchased from Millipore/Chemicon. Recombinant human SDF-1α was from R&D Systems.
Cell Culture
The human breast adenocarcinoma cell line, MDA-MB-231 was purchased from American Type Culture Collection (ATCC, Manassas, VA). MDA-1833 (bone metastasis) and MDA-4175 cells (lung metastasis) are organ-specific metastases derived from in vivo selection from parental MDA-MB-231 cells and were a generous gift from Dr. Joan Massague (Memorial Sloan-Kettering Cancer Center). MDA-MB-231 and their derivative cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 4 mm l-glutamine (Invitrogen). The respective non-metastatic and metastatic mouse melanoma cell lines, B16F-0 and B16F-10 (ATCC), were cultured in DMEM containing 10% FBS and 4 mm l-glutamine. The human embryonic kidney cell line, HEK-293T (ATCC), was cultured in Eagle's Minimum Essential Medium (MEM, Invitrogen) supplemented with 10% FBS. The embryonic mouse fibroblast cell line, NIH3T3 (ATCC), was cultured in DMEM supplemented with 10% Bovine Calf Serum (Hyclone). The human non-tumorigenic mammary epithelial cell line, MCF10A (ATCC) was cultured in DMEM supplemented with 5% heat inactivated horse serum (Sigma), 4 mm l-glutamine, 10 μg/liter insulin (Invitrogen), 20 ng/ml epithelial growth factor (Sigma), 500 ng/ml hydrocortisone (Sigma), and 100 ng/ml cholera toxin (Sigma). All cultures were maintained at 37 °C in humidified air containing 5% CO2.
Drug Treatment
STI571/Gleevec was a generous gift from Novartis. In brief, cells were cultured in serum-free medium overnight and incubated with either the vehicle dimethysulfoxime (DMSO, Sigma) or 10 μm STI571 for a period of 1–16 h as indicated.
Transfections and Retroviral Transduction
Various pcDNA3.1 constructs (MT1-MMP, Abl, Arg) and PX1-Arg-YFP fusion constructs were expressed in HEK293T cells using CaCl2 phosphate-mediated transfection. Cells were used for experimental procedures 48 h following transfection. Retroviral transduction of PX1-Arg-YFP fusion and MIGR1 constructs expressing constitutively active SrcY527F, Abl, and/or Arg (WT, wild type; PP, constitutively active; and KR, dominant inactive) were performed by transfecting HEK293T with along CMV-VSVG and gag/pol packaging constructs. Medium containing virus was collected, filtered, incubated on cells with polybrene (8 μg/ml) for a period of 6–24 h. Cells were then sorted for GFP+ expression and cultured and used accordingly.
RNA Interference
B16F10 and NIH3T3SrcY527F cells were transfected with small interfering RNA (siRNA) (Dharmacon). Either a non-targeted control or mouse specific siRNAs targeting Abl (GAAGGAAAUCAGUGACAUAUU) and Arg (GAAAUGGAGCGAACAGAUAUU) were transfected into cell using Oligofectamine as previously described (35).
Transduction of Lentiviral MicroRNAs
Lentiviral microRNAs (miRNAs) were transduced into various cell lines as previously described (35). Constructs encoding human-specific Abl miRNA (GGTGTATGAGCTGCTAGAGAA) and Arg miRNA (CCTTATCTCACCCACTCTGAA) were cloned in tandem into the pFc-EmGFPW vector and transfected into HEK293T cells along with pCMV-VSVG, pRSV-Rev and pMDL plasmids as described (36). Lentiviral supernatants were collected, filtered, and placed on cells (MDA-MB-231 and derivative cell lines) with 8 μg/ml polybrene. Cells were then sorted by flow cytometry to collect GFP+ populations and either cultured or immediately used under “Experimental Procedures.” Similarly, the mouse cell line, NIH3T3SrcY527F was transduced with mouse-specific miRNAs to individually and simultaneously target Abl (CAGTTTGACATCCACCTTT) and/or Arg (CCGTTTGGGTGATGCTGAA).
Western Blotting
Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in Nonidet P-40 lysis buffer (50 mm Tris hydrochloride, pH 8.0, 150 mm sodium chloride, and 1% Nonidet-P40) containing protease and phosphatase inhibitors (2 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μg/ml pepstatin A (Thermo Scientific), 10 mm sodium fluoride, 1 mm phenylmethanesulfonyl fluoride, and 10 mm sodium orthovanadate). Cell debris was removed by microcentifugation and protein concentrations were quantified using the Bradford Protein Assay (Bio-Rad) according to the manufacturer's instructions. Cell lysates were then diluted in sample buffer (100 mm Tris, pH 6.8, 200 mm dithiothreitol, 4% sodium dodecyl sulfate, 0.2% bromphenol blue, and 20% glycerol) and boiled for 5 min. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked in Tris-buffered saline Tween-20 (TBST) containing either 5% (w/v) nonfat milk powder or 5% (w/v) bovine serum albumin (BSA) for 1 h and then incubated with in blocking solution containing primary antibody overnight at 4 °C. Blots were washed several times with TBST and incubated with secondary Horseradish peroxidase (HRP)-conjugated antibody (Santa Cruz Biotechnology) for 1 h followed by several washes with TBST. Proteins were visualized using ECL Western blotting Detection Reagent (GE Healthcare).
Immunoprecipitations
Cells were washed in cold PBS and lysed in Nonidet P-40 Lysis Buffer. Protein concentration was quantified and 0.5–1 mg of protein were incubated with 2 μg of primary antibody or control IgG between 4–16 h at 4 °C. Then lysates containing protein-antibody complexes were incubated with 40 μl of protein A/G PLUS-agarose (Santa Cruz Biotechnology) for 4 h at 4 °C. Lysates were centrifuged and agarose-bound immune complexes were washed several times with Nonidet P-40 lysis buffer. Beads were then suspended in sample buffer, boiled, and processed as described for Western blotting.
Biotinylation of Surface Proteins
Cells were pretreated for 1 h with either DMSO or 10 μm STI571. Alternatively, cells were transduced with miRNAs for Abl kinases, sorted by flow cytometry for GFP+ expression, and cultured. Medium was aspirated from cells and cells were washed several times with ice-cold PBS containing MgCl2 and CaCl2 (PBS+/+, Invitrogen). Surface proteins were labeled with 1 mg/ml Sulfo-NHS-SS-Biotin (Pierce) reconstituted in PBS+/+ and incubated for 30 min at 4 °C. Unreacted biotin was quenched with 20 mm Tris, pH 8.0 followed by several washes with ice-cold PBS+/+. Then cells were lysed in RIPA buffer (150 mm sodium chloride, 50 mm Tris, pH 8.0, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) supplemented with protease and phosphatase inhibitors. Then 250 μg of protein from cell lysates were incubated with NeutrAvidin-agarose Resin (ThermoScientific) for 1 h at 4 °C. Bead resin was washed several times with PBS+/+ containing 1% Nonidet P-40 and protease/phosphatase inhibitors. Subsequently, beads were suspended in sample buffer and analyzed by Western blotting for detection of MT1-MMP and Na, K+ ATPase, as a loading control.
Immunofluorescence Microscopy
Cells were cultured on either bare or gelatin-coated glass coverslips. Exponentially growing cells were fixed with 4% (w/v) paraformaldehyde (PFA) prepared in PBS for 15 min and then permeabilized with PBS containing 0.2–0.5% (v/v) Triton X-100 for 10 min. Next, fixed cells were blocked in PBS containing 3% (w/v) BSA for 1 h, incubated with the appropriate primary antibodies for an additional hour, and followed with incubation with either the appropriate Alexa Fluor conjugated secondary antibodies (Santa Cruz Biotechnology). Alternatively, secondary staining procedures included Alexa Fluor-conjugated anti-phalloidin antibodies (Molecular Probes) to stain F-actin. Lastly, cells were stained with Hoechst (Molecular Probes) to identify the nuclei. Fluorescent images were acquired using the Zeiss Axio Imager (Carl Zeiss MicroImaging). Confocal images were gathered using the Leica SP5 confocal scanning microscope. Images were analyzed using MetaMorph software (MDS Analytical Technologies).
Fluorescent in Situ Gelatin Zymography
Glass coverslips were coated with 20 μg/ml OregonGreen-conjugated gelatin (Molecular Probes) suspended in 2% sucrose for 1 h. Gelatin solution was removed, and coverslips were incubated with 0.5% glutaraldehyde in PBS for 15 min. Plates were washed several times and then quenched with 5 mg/ml of sodium borohydride for 3 min followed with several wash in PBS. NIH3T3SrcY527F cells were plated on gelatin for a period of 3 h; MDA-MB-231 and their derivative cell lines were examined after 6 h. Cells were then fixed and stained according to procedures outlined in the section describing immunofluorescence microscopy. Matrix degradation was quantified by counting the total number of cells producing gelatin-degradation patches/nuclei in five individual fields; results are expressed as the mean ± S.E.
Matrigel Invasion
Invasion was evaluated by plating 25,000 cells in the upper chambers of 8.0 μm pore size matrigel chambers (BD Biosciences) in serum-free medium. Cells were allowed to invade for up to 48 h in the presence of either serum-containing medium or serum-free medium containing 150 ng/ml of SDF1-α. Then the remaining cells, medium, and matrigel were removed from the upper chambers and cells on the undersurface of the membrane were fixed, stained with DiffQuik (Dade Behring), and quantified under the 20× objective. Each experiment analyzed cells in at least four different fields performed in triplicates.
Statistical Analysis
Experiments were performed at least three independent times. Unpaired student t-tests were performed to determine statistical significance using the GraphPad software (www.graphpad.com). p < 0.05 was considered statistically significant. Densitometry was performed using the Image J software.
RESULTS
Abl Kinases Regulate Cell Invasion and MMP-dependent ECM Degradation
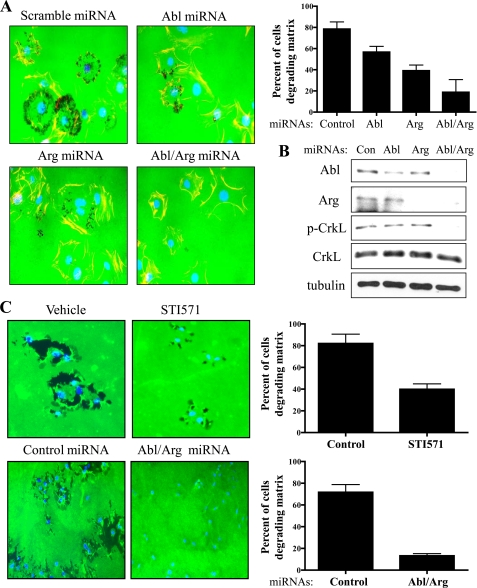
Metastatic tumor cells invade their surrounding microenvironment by employing a variety of mechanisms that include protease-mediated degradation of ECM and tissues. Whereas increased invasiveness correlates with enhanced metastatic activity, the ability of cancer cells to spread from the primary site of tumor growth to specific organs may require the activation of distinct signaling pathways. Elevated Abl kinase activity has been detected in highly invasive breast cancer cell lines (26). However, it is not known whether Abl kinases are required for matrix degradation and invasion of breast cancer cells with organ-specific metastatic capacity. To investigate the requirement for Abl kinases in these processes, we employed MDA-MB-231-derived breast cancer cell lines of varying metastatic potential, which were selected in vivo for their ability to metastasize to either the bone (MDA1833) and lung (MDA4175) (37–40). These breast cancer cells were depleted of Abl and Arg following transduction with lentivirus encoding Abl/Arg-specific microRNAs (miRNAs). Abl and Arg protein levels were decreased by greater than 90%. This reduction in Abl and Arg proteins correlated with a reduction (greater than 75%) in phosphorylation of CrkL at an Abl kinase specific phosphorylation site (tyrosine 207) (Fig. 1C). Silencing of Abl and Arg resulted not only in markedly decreased invasion toward serum, but also dramatically impaired protease-mediated matrix degradation in the parental cell line and the MDA-MB-231-derivative cell lines with ability to metastasize to either bone or lung (Fig. 1, A and B). Furthermore, reconstitution of miRNA-resistant mouse Abl partially restored the invasive phenotype and the ability to degrade the matrix (Fig. 1, D and E). Abl kinases are also required for invasion and matrix degradation of melanoma cells (supplemental Fig. S1). Abl kinase activity was elevated in the metastatic melanoma cell line, B16F10 compared with its non-metastatic counterpart, B16F0 (supplemental Fig. S1A) and silencing of Abl kinases in the B16F10 metastatic melanoma cell line significantly decreased invasion and matrix degradation (supplemental Fig. S1, B and C). Moreover, expression of activated Abl kinases enhanced invasion and matrix degradation in B16F0 melanoma cells with weak metastatic potential (data not shown). Taken together, these findings support a requirement for Abl family kinases in matrix degradation and invasion of metastatic cancer cells.
FIGURE 1.
Abl kinases are required for invasion and MMP-dependent matrix degradation. A, MDA-MB-231, MDA1833, a derivative that metastasizes to the bone, and MDA4175, a derivative that metastasizes to the lung, were transduced with the indicated lentiviral miRNAs, sorted, and plated on the upper wells of matrigel invasion assay chambers in serum-free medium. Invasion toward serum-containing medium was analyzed after 48 h. Cells on the undersurface of the matrigel membrane were stained and quantified below as mean ± S.E. B, cells transduced with lentiviral miRNAs for control or Abl+Arg miRNAs, were sorted and plated on coverslips coated with OregonGreen-conjugated gelatin for 6 h. Cells were fixed and nuclei stained with Hoechst (blue). Areas of gelatin degradation (black area) were quantified as mean ± S.E. Experiments are quantified from five randomly selected fields under a 20× objective. Data are representative of at least three independent experiments. C, Western blot analysis of lysates from control (C) and Abl/Arg (A/A) knockdown cells with antibodies for Abl, Arg, p-CrkL (Y207), and total CrkL. D, MDA-MB-231 cells expressing either control or Abl/Arg lentiviral microRNAs were retrovirally transduced with miRNA-resistant mouse wild type (WT) Abl and analyzed for invasion and gelatinase activity. Gelatin degradation is quantified as described in Fig. 1B. E, Western blot analysis of knockdown and reconstituted cells with indicated antibodies as above, with tubulin staining used as a loading control. Data are representative of three independent experiments.
Src-induced ECM Degradation Is Mediated by Abl Kinases
A role for Src in metastasis has been well established and several Src kinase inhibitors are effective in inhibiting proliferation, invasion, and migration in vitro and in vivo models (41–44). Because Abl kinases function downstream of Src in growth factor receptor signaling (12, 33), we hypothesized that the invasive properties of Src are mediated, in part, by activation of Abl kinases. Silencing of Abl, Arg, or both Abl and Arg together in NIH3T3 cells expressing constitutively active SrcY527F resulted in a dose-dependent decrease in MMP-dependent gelatinase activity (Fig. 2, A and B). Knockdown of both Abl and Arg was required for maximal inhibition of MMP-dependent gelatinase activity (60% compared with controls) while depletion of Abl or Arg alone resulted in partial inhibition of matrix degradation (Fig. 2A). Similarly, inhibition of Abl kinase activity with the pharmacological inhibitor, STI571/Gleevec or knockdown of Abl/Arg in MDA-MB-231 breast cancer cells expressing constitutively active Src markedly decreased gelatinase activity by 43 and 58%, respectively, compared with controls (Fig. 2C). Moreover, expression of activated Arg kinase (Arg-PP) further enhanced matrix degradation in breast cancer cells, and pharmacological inhibition of endogenous Src kinases abrogated matrix degradation in these cells (supplemental Fig. S2). Collectively, these data show that endogenous Src and Abl family kinases are required for efficient ECM degradation.
FIGURE 2.
Abl kinases are required for Src-dependent matrix degradation. A, NIH3T3-SrcY527F cells were transduced with lentiviruses encoding either control scrambled or Abl/Arg miRNAs, sorted by flow cytometry, and plated on cover slips coated with Oregon Green-conjugated gelatin for 3 h. Dark areas of gelatinase activity (black) reveal MMP-mediated degradation. Nuclei are labeled with Hoechst (blue). Matrix degradation was quantified (right) by counting the total number of cells producing gelatin-degradation patches/nuclei in five individual fields; results are expressed as the mean ± S.E. B, Western blot analysis of lysates from cells transduced with scrambled (Scr) control or Abl and Arg miRNAs with the indicated antibodies. C, MDA-MB-231 cells were serum-starved overnight, and then treated with either vehicle (DMSO) or 10 μm STI571 for 1 h to inhibit Abl kinases (top panels) and analyzed for matrix degradation as above. MDA-MB-231 cells were transduced with lentiviruses encoding either control or Abl/Arg miRNAs (lower panels), sorted by flow cytometry, and plated on FITC-gelatin coated coverslips for 3 h and then fixed. Dark areas of gelatinase activity (black) reveal MMP-mediated degradation. Nuclei are labeled with Hoechst (blue). Matrix degradation was quantified (shown right) by counting the number of cells producing gelatin-degradation patches in five individual fields. Results are expressed as the mean ± S.E. Data are representative of at least three independent experiments.
Abl Kinases Are Activated Downstream of SDF1α/CXCR4
SDF1α (CXCL12) is a well-characterized chemokine that plays a critical role in metastasis and invasion in melanoma as well as lung and breast cancer (53–56). SDF1α signals through the CXCR4, a G protein-coupled receptor, to activate signaling pathways required for cell proliferation, survival and migration. Interestingly, Src kinase is required for SDF1α-induced survival of metastatic breast cancer cells (37). Thus, we examined whether Abl kinases were activated in response to SDF1α in MDA-MB-231 breast cancer cells. Stimulating MDA-MB-231 cells with SDF1α induced rapid activation of the endogenous Abl kinases as detected by increased tyrosine phosphorylation of CrkL, which was abolished by treatment with the Abl kinase inhibitor STI571 (Fig. 3A). Notably, inhibition of the Abl kinases did not inhibit activation of Akt by SDF1α, which has been linked to cell survival (Fig. 3A). In contrast, inhibition of endogenous Src with the Src-selective inhibitor SU6656 impaired Akt activation induced by SDF1 α (supplemental Fig. S3). Depletion of Abl and Arg with specific miRNAs impaired SDF1α-induced invasion (Fig. 3B). Moreover, inhibition of Abl kinases with STI571 impaired matrix degradation by 60% compared with control cells (Fig. 3C). Reduced invasion and matrix degradation were not due to differences in CXCR4 expression as treatment with STI571 or silencing of Abl kinases with miRNAs did not alter CXCR4 protein levels or surface expression (data not shown). Further, we found that activation of Abl kinases downstream of SDF1α/CXCR4 is Src-dependent as pharmacological inhibition of endogenous Src kinase significantly decreased the phosphorylation of CrkL at the Abl-specific site, Tyr-207 (supplemental Fig. S3). Thus, Abl kinases are activated downstream of the CXCR4 chemokine receptor and are required for SDF1α/CXCR4-mediated invasion and matrix degradation.
FIGURE 3.
Abl kinases are activated downstream of SDF1α and are required for SDF1α-induced invasion and matrix degradation. A, MDA-MB-231 cells were serum-starved overnight and then treated with either vehicle (DMSO) or 10 μm STI571 for 1 h, followed by treatment with 150 ng/ml human recombinant SDF1α for the indicated times. Cells were lysed and analyzed for phosphorylated and total CrkL and Akt proteins. B, MDA-MB-231 breast carcinoma cells expressing either control or Abl/Arg lentiviral miRNAs were serum-starved and then plated in the upper wells of matrigel invasion assay chambers, and allowed to chemotax toward SDF1α. Quantification is shown as fold-difference normalized to controls. C, MDA-MB-231 cells were serum-starved overnight, and then treated with either vehicle (DMSO) or 10 μm STI571 for 1 h. Cells were plated on Oregon Green-conjugated gelatin-coated cover slips and cultured in serum-free medium containing either vehicle alone or 150 ng/ml of SDF1α with or without 10 μm STI571. After 6 h, cells were fixed and stained with Hoechst to stain the nuclei (blue) and analyzed for gelatin degradation (black regions). Experiments are quantified from five randomly selected fields under a 20× objective. Data are quantified as mean ± S.E. and are representative of at least three independent experiments.
Abl Kinases Are Required for Invadopodia Formation
Matrix degradation is mediated by invadopodia in carcinoma cells and activation of Src has been shown to be necessary and sufficient for invadopodia formation (11, 45). Because Abl kinases are required for matrix degradation and invasion in Src-transformed cells (Fig. 2), we asked whether activated Abl kinases were sufficient to promote invadopodia formation. To address this question, untransformed NIH3T3 fibroblasts were transduced with either control vector, constitutively active Src, or constitutively Abl and Arg. Cells expressing constitutively active Abl and Arg were elongated, displayed membrane ruffles, and formed cortactin- and actin-positive podosome-like structures at the cell periphery that were morphologically distinct from the podosome rosettes observed in Src-transformed cells (Fig. 4A). Importantly, the structures at the cell periphery showing colocalization of actin and cortactin overlapped with areas of matrix degradation (supplemental Fig. S2A). Next, we asked whether Abl kinases were localized to invadopodia and were required for their formation. To determine the whether Abl kinases were localized to invadopodia, NIH3T3 fibroblasts expressing constitutively active Src were transduced with either wild type, constitutively active, or kinase-inactive Arg tagged with yellow fluorescent protein (YFP). The Arg-YFP fusion proteins were analyzed for co-localization with cortactin, which marks invadopodia precursors as well as mature invadopodia structures. Wild type and constitutively active Arg co-localized with cortactin at invadopodia assembly sites (Fig. 4B). In contrast, expression of kinase-inactive Arg (ArgKR) disrupted invadopodia formation (Fig. 4B, bottom panels).
FIGURE 4.
Active Abl kinases promote formation of podosome-like structures and localize at invadopodia. A, NIH3T3 fibroblasts were retrovirally transduced with either vector, constitutively active Src (SrcY527F), or simultaneously with constitutively active Abl and Arg (AblPP/ArgPP). Cells were plated on glass cover slips for 6 h, fixed and co-stained for F-actin (green) and cortactin (red). Merged fields demonstrate co-localization between cortactin and F-actin at invadopodia-like sites. Hoechst (blue) stains the nuclei. B, NIH3T3-SrcY527F fibroblasts were transduced with either wild type (WT), constitutively active (PP), or dominant inactive (KR) forms of Arg-YFP. Cells were sorted for YFP expression by flow cytometry, plated on gelatin-coated cover slips for 3 h and stained for cortactin (red), GFP (Arg), and Hoechst (nuclei). Colocalization is indicated by yellow in merged fields.
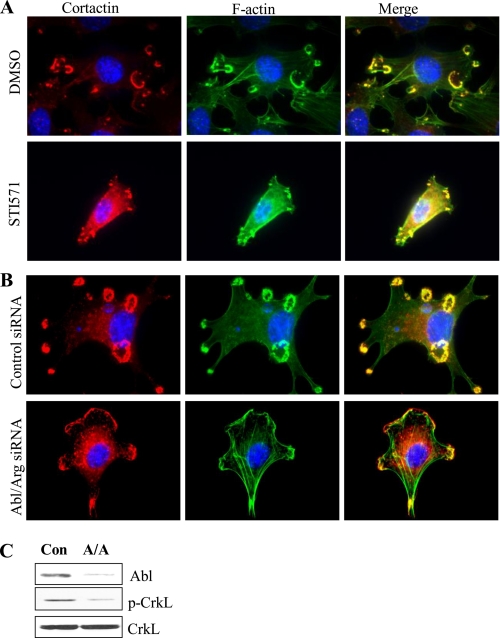
To further examine the requirement for Abl kinases in Src-mediated invadopodia formation, Abl kinases were inhibited by treatment with STI571 or depleted with Abl/Arg siRNAs. Both kinase inhibition and silencing of Abl and Arg abrogated invadopodia formation (Figs. 5, A and B). Taken together, these data reveal that Abl kinases are required for Src-induced invadopodia and that activated Abl kinases promote the accumulation of cortactin-rich structures that resemble invadopodia precursors.
FIGURE 5.
Abl kinases are required for invadopodia formation. A, NIH3T3-SrcY527F cells were treated with DMSO or with 10 μm STI571 for 16 h, fixed and co-stained for cortactin (red, left panels) and F-actin (green, middle panels). Merged fields (right panels) demonstrate co-localization between cortactin and F-actin at invadopodia. B, NIH3T3-SrcY527F cells were transfected with either control or Abl and Arg siRNAs for 48 h, and then fixed and co-stained for cortactin and F-actin. C, lysates from cells transfected with control (Con) or Abl/Arg (A/A) siRNAs were analyzed by Western blotting for the indicated proteins.
Inhibition of Abl Kinases Impairs MT1-MMP and Cortactin Localization
Next, we examined whether Abl kinases modulate invadopodia components responsible for ECM degradation. To this end, we examined the localization of MT1-MMP, which is required for matrix degradation in MDA-MB-231 cells. In control cells, MT1-MMP is localized in a punctate pattern throughout the cell and on the ventral surface. In contrast, cells depleted of Abl kinases with miRNAs or treated with STI571 exhibited perinuclear accumulation of MT1-MMP (Fig. 6, A and B). To determine whether loss of Abl kinases promoted MT1-MMP internalization, cell surface proteins were biotinylated before and after depletion of Abl kinases with miRNAs or after kinase inhibition with STI571. We observed that inhibition of Abl kinases resulted in decreased levels of MT1-MMP at the surface (Fig. 6, C and D). Notably, depletion of Arg appeared to inhibit MT1-MMP cell surface levels to a greater extent than Abl depletion (Fig. 6C). Cortactin has been shown to regulate MT1-MMP recruitment to invadopodia as well as matrix degradation (16, 18). We observed that cortactin also exhibits a perinuclear localization pattern following inhibition of Abl kinases (Fig. 6E). Thus, Abl kinases regulate the accumulation of MT1-MMP and cortactin at invadopodia and the localization of MT1-MMP at the cell surface. These findings suggest that Abl kinases may regulate trafficking and positively regulate the localization of MT1-MMP at the cell surface.
FIGURE 6.
Inhibition of Abl kinases impairs localization of MT1-MMP and cortactin to invadopodia. A, NIH3T3-SrcY527F cells were plated on FITC-gelatin in the presence or absence of STI571 (10 μm) for 3 h. B, MDA-MB-231 cells expressing constitutively active SrcY527F were transduced with lentiviruses encoding either control or Abl/Arg miRNAs, sorted by flow cytometry, and plated on FITC-gelatin-coated cover slips for 6 h. A and B, cells were fixed and stained for MT1-MMP (red). Dark areas of gelatinase activity (black) reveal MMP-mediated degradation. Nuclei were labeled with Hoechst (blue). C, NIH3T3- SrcY527F cells expressing the indicated miRNAs were biotinylated for 30 min, lysed, and incubated with neutravidin beads. Protein complexes bound to beads were analyzed by SDS-PAGE and blotting for cell surface MT1-MMP. Total cell lysates from biotinylated samples were examined for total MT1-MMP and indicated proteins. Densitometry analysis show a ratio of surface MT1-MMP: Total MT1-MMP and were normalized to controls. D, MDA-MB-231 cells were treated with vehicle or 10 μm STI571 for 24 h. Cells were biotinylated for 30 min, lysed, and incubated with neutravidin beads. Protein complexes bound to beads were washed and analyzed by SDS-PAGE for cell surface MT1-MMP and Na,K-ATPase. Results are representative of at least three independent experiments. UB, unbiotinylated control. Densitometry shows a ratio of surface MT1-MMP: surface Na,K-ATPase normalized to controls. E, NIH3T3-SrcY527F cells (left panels) and MDA-MB-231-SrcY527F cells (right panels) were plated on FITC-gelatin-coated cover slips for 6 h, and then fixed and stained for cortactin (red), and nuclei were labeled with Hoechst (blue); dark areas of gelatinase activity (black) reveal MMP-mediated degradation.
Abl Kinases Interact with MT1-MMP
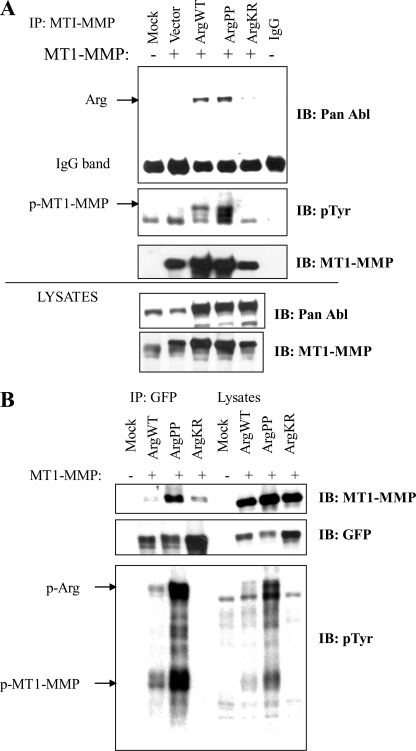
The activity of MT1-MMP is regulated by phosphorylation, degradation, autocatalysis, and endocytosis (46–49). Indeed, phosphorylation of MT1-MMP by Src kinase is important for migration of tumor and endothelial cells (46). We examined whether the Abl kinases interacted with MT1-MMP. We observed that MT1-MMP interacted strongly with exogenous Arg wild type (Arg-WT) and constitutively active Arg (Arg-PP) (Fig. 7, A and B). In contrast, a weak interaction was detected between MT1-MMP and kinase-inactive Arg (Arg-KR) (Fig. 7, A and B). The interaction between Arg and MT1-MMP was detected following immunoprecipitation of either MT1-MMP (Fig. 7A) or Arg (Fig. 7B). Similarly, active Abl but not kinase-inactive Abl interacted with MT1-MMP (supplemental Fig. S4). Expression of wild-type Arg kinase promoted tyrosine phosphorylation of MT1-MMP, which was further augmented by expression of constitutively active Arg (Fig. 7, A and B). The Arg/Abl-induced tyrosine phosphorylation of MT1-MMP is likely indirect because we did not detect tyrosine phosphorylation of purified MT1-MMP with the Abl kinases in an in vitro kinase assay (data not shown). Thus, these data suggest that active Abl kinases may activate endogenous tyrosine kinases to promote MT1-MMP tyrosine phosphorylation and that the interaction of the Abl kinases with MT1-MMP likely requires Abl activation by upstream signals initiated by chemokines, growth factors, or oncogenic tyrosine kinases.
FIGURE 7.
MT1-MMP interacts with Arg kinase. A, active Arg is detected in protein complexes with MT1-MMP and promotes increased tyrosine phosphorylation of MT1-MMP. HEK293T cells were co-transfected with MT1-MMP and either vector or the indicated Arg constructs: WT, constitutively active (PP), or dominant inactive (KR). Lysates were immunoprecipitated with MT1-MMP antibody and blotted for Arg, phosphotyrosine (pTyr), and MT1-MMP. Total lysates are shown in the bottom two panels. B, HEK293T cells were transfected with the indicated Arg-YFP fusion constructs (WT, PP, or KR), and the corresponding lysates were immunoprecipitated with anti-GFP antibodies and analyzed by Western blotting for GFP (Abl), MT1-MMP, and pTyr. Data are representative of at least three independent experiments.
DISCUSSION
In this study, we reveal a functional role for Abl kinases in invadopodia formation, invasion, and matrix degradation. While Abl kinases have long been recognized as oncogenic in leukemias, recent reports also support an oncogenic role for Abl kinases in the progression and metastasis of solid tumors (22–24, 26, 50). Whereas activating mutations of the Abl kinases in solid tumors are rare, endogenous Abl kinases are activated by upstream signals emanating from receptor tyrosine kinases (RTKs), which often are up-regulated in cancers. Our previous work identified Abl kinases as targets of RTKs such as the PDGFR and EGFR, and showed that Abl kinases are also activated downstream of the Src kinase (12, 51). Here we demonstrate that Abl kinases are required for Src-induced invadopodia formation, cell invasion, and matrix degradation. Previous studies have shown that Abl kinases are required for cell proliferation, survival, and migration induced by Src kinase and growth factors (23). Thus, it can be surmised that hyperactive RTK and Src signaling in cancers might lead to the inappropriate activation of Abl kinases. Moreover, pharmacological Src inhibitors such as Dasatinib, also inhibit Abl kinases and are effective in preventing tumor cell growth, invasion and metastasis using in vitro and in vivo models (43, 52). Our findings suggest that the effects of Dasatinib and similar inhibitors may be mediated in part by abrogation of Abl and Arg kinase activities.
The invasive program of metastatic malignancies is promoted by chemokine signaling (53–55). In particular, the chemokine receptor, CXCR4, is highly expressed in melanoma and breast tumor cells while its ligand, SDF1α, is elevated in the microenvironment of metastatic lesions such as lymph nodes, bone marrow, lungs, and liver (56). Activation of CXCR4 is important in stimulating the growth and survival of primary and metastatic tumor cells (53, 57). Moreover, downstream signaling by the Src kinase is required for SDF1α-induced survival and metastasis of tumor cells that overexpress CXCR4 (37, 39). Numerous studies have placed Src kinase downstream of CXCR4 signaling (37, 58–60). Here we show for the first time that Abl kinases are activated downstream of SDF1α/CXCR4 in a Src-dependent manner, and that Abl kinases regulate SDF1α-mediated invasion and matrix degradation. Interestingly, in contrast to Src kinases, which are required for maximal Akt activation downstream of SDF1α/CXCR4 in metastatic breast cancer cells (37), we found that inhibition of Abl kinases did not affect Akt activation. Thus, our findings suggest that Src signaling functions upstream of both Abl kinases and Akt in cells stimulated with SDF1α.
The functions of CXCR4 during invasion and metastasis are mediated in part by regulation of MMPs. In this regard, it has been demonstrated that both CXCR4 and MT1-MMP are required for melanoma cell metastasis to the lungs, with CXCR4 activation required for early stages of metastasis while MT1-MMP function is required for latter stages (55). Moreover, SDF1α stimulates invasion by upregulating MT1-MMP which may be due, in part, to activation of Rac GTPase (61). In this regard, Abl kinases are intimately involved in the regulation of actin cytoskeletal dynamics and modulate the activities of Rho GTPases (34, 62, 63). Specifically, we previously demonstrated that Abl kinases are required for Rac activation (34, 63). Future studies are underway to determine whether Abl kinases regulate the activities of Rho family of GTPases during cancer cell invasion.
Protease-dependent mesenchymal cell motility is mediated by protrusive invadopodia structures (5, 64, 65). Activation of Src kinase is required for induction of invadopodia (11). Here we demonstrate that Abl kinases are required for invadopodia assembly. We show that expression of constitutively active Abl kinases promotes formation of invadopodia-like structures and that loss of Abl kinase activity or down-regulation of Abl and Arg, as well as expression of dominant inactive Arg kinase, abrogates formation of invadopodia. This suggests that phosphorylation of specific targets of Abl kinases are important in the formation of these structures. In this regard, several components of invadopodia such as cortactin and N-WASP have been identified as targets of Abl kinases (20, 28, 66, 67). However, while cortactin is required for assembly of invadopodia precursors, tyrosine phosphorylation of cortactin is not required for invadopodium precursor formation (9). Thus, phosphorylation of other Abl targets may be required for invadopodia assembly. Possible candidates are N-WASP and Abi1. Interestingly, a recent report demonstrated that Abi1, a substrate of Abl kinases, localized at invadopodia, and was required for the formation of these structures (68).
Abl kinases likely regulate distinct stages of invadopodia assembly and function. While dispensable for invadopodia precursor formation, phosphorylation of cortactin is required for invadopodia maturation, recruitment of MT1-MMP- and MMP-mediated matrix degradation (9, 16). Here we demonstrate that inhibition of Abl kinases results in mislocalization of MT1-MMP to the perinuclear region and that Abl kinases regulate the phosphorylation and surface expression of MT1-MMP. In this regard, we have recently shown that Abl kinases regulate lysosomal and protein trafficking as inhibition of Abl kinases induces perinuclear aggregation of lysosomes and impairs the processing of cathepsins and lysosomal hydrolases (35). Taken together, our findings suggest that Abl kinases regulate MT1-MMP trafficking as well as recruitment to invadopodia and matrix degradation activity. Regulation of MT1-MMP by Abl kinases may be mediated in part by cortactin phosphorylation and indirectly through tyrosine phosphorylation of MT1-MMP by yet to be identified protein kinases. Collectively, our findings uncover new roles for the Abl kinases in the regulation of tumor invasion and have implications for the development of therapies in the treatment of metastatic invasive carcinomas.
Supplementary Material
Acknowledgments
We thank Dr. Joan Massague (Memorial Sloan Kettering Cancer Center, NY) for the gift of MDA-MB-231 derivative cell lines. We thank Dr. Sam Johnson and Dr. Yasheng Gao (Duke University Medical Center Light Microscopy Facility) for advice and training. We thank Dr. Elizabeth Harvat (Duke University Flow Cytometry Facility) for dedicated services. We also thank Dr. Alex Strongin (Inflammatory and Infectious Disease Center/Cancer Research Center, Burnham Institute for Medical Research) for providing the MT1-MMP plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants CA 70940 and HL 084102 (to A. M. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- ECM
- extracellular matrix
- Abi-1
- Abelson interactor-1
- Abl
- Abelson
- Arg
- Abelson related gene
- EGFR
- epithelial growth factor receptor
- F-actin
- filamentous actin
- MMP
- matrix metalloproteinase
- MT1-MMP
- membrane type-1 matrix metalloproteinase
- N-WASP
- neural Wiskott-Aldrich syndrome protein
- WAVE
- WASP-family verprolin-homologous protein
- PDGFR
- platelet-derived growth factor
- RTK
- receptor tyrosine kinase
- SDF1α
- stromal-derived factor1-α.
REFERENCES
- 1.Osiak A. E., Zenner G., Linder S. (2005) Exp. Cell Res. 307, 342–353 [DOI] [PubMed] [Google Scholar]
- 2.Carman C. V., Sage P. T., Sciuto T. E., de la Fuente M. A., Geha R. S., Ochs H. D., Dvorak H. F., Dvorak A. M., Springer T. A. (2007) Immunity 26, 784–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linder S., Nelson D., Weiss M., Aepfelbacher M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9648–9653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ory S., Brazier H., Pawlak G., Blangy A. (2008) Eur J. Cell Biol. 87, 469–477 [DOI] [PubMed] [Google Scholar]
- 5.Buccione R., Caldieri G., Ayala I. (2009) Cancer Metastasis Rev. 28, 137–149 [DOI] [PubMed] [Google Scholar]
- 6.David-Pfeuty T., Singer S. J. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 6687–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarone G., Cirillo D., Giancotti F. G., Comoglio P. M., Marchisio P. C. (1985) Exp. Cell Res. 159, 141–157 [DOI] [PubMed] [Google Scholar]
- 8.Gimona M., Buccione R., Courtneidge S. A., Linder S. (2008) Curr. Opin. Cell Biol. 20, 235–241 [DOI] [PubMed] [Google Scholar]
- 9.Oser M., Yamaguchi H., Mader C. C., Bravo-Cordero J. J., Arias M., Chen X., Desmarais V., van Rheenen J., Koleske A. J., Condeelis J. (2009) J. Cell Biol. 186, 571–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W. T., Chen J. M., Parsons S. J., Parsons J. T. (1985) Nature 316, 156–158 [DOI] [PubMed] [Google Scholar]
- 11.Oikawa T., Itoh T., Takenawa T. (2008) J. Cell Biol. 182, 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plattner R., Kadlec L., DeMali K. A., Kazlauskas A., Pendergast A. M. (1999) Genes Dev. 13, 2400–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schade B., Lam S. H., Cernea D., Sanguin-Gendreau V., Cardiff R. D., Jung B. L., Hallett M., Muller W. J. (2007) Cancer Res. 67, 7579–7588 [DOI] [PubMed] [Google Scholar]
- 14.Courtneidge S. A., Azucena E. F., Pass I., Seals D. F., Tesfay L. (2005) Cold Spring Harb. Symp. Quant. Biol. 70, 167–171 [DOI] [PubMed] [Google Scholar]
- 15.Ayala I., Baldassarre M., Giacchetti G., Caldieri G., Tetè S., Luini A., Buccione R. (2008) J. Cell Sci. 121, 369–378 [DOI] [PubMed] [Google Scholar]
- 16.Artym V. V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K. M., Mueller S. C. (2006) Cancer Res. 66, 3034–3043 [DOI] [PubMed] [Google Scholar]
- 17.Clark E. S., Weaver A. M. (2008) Eur. J. Cell Biol. 87, 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark E. S., Whigham A. S., Yarbrough W. G., Weaver A. M. (2007) Cancer Res. 67, 4227–4235 [DOI] [PubMed] [Google Scholar]
- 19.Lapetina S., Mader C. C., Machida K., Mayer B. J., Koleske A. J. (2009) J. Cell Biol. 185, 503–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyle S. N., Michaud G. A., Schweitzer B., Predki P. F., Koleske A. J. (2007) Curr. Biol. 17, 445–451 [DOI] [PubMed] [Google Scholar]
- 21.Pendergast A. M. (2002) Adv. Cancer Res. 85, 51–100 [DOI] [PubMed] [Google Scholar]
- 22.Sirvent A., Boureux A., Simon V., Leroy C., Roche S. (2007) Oncogene 26, 7313–7323 [DOI] [PubMed] [Google Scholar]
- 23.Srinivasan D., Sims J. T., Plattner R. (2008) Oncogene 27, 1095–1105 [DOI] [PubMed] [Google Scholar]
- 24.Chen W. S., Kung H. J., Yang W. K., Lin W. (1999) Int. J. Cancer 83, 579–584 [DOI] [PubMed] [Google Scholar]
- 25.Crnogorac-Jurcevic T., Efthimiou E., Nielsen T., Loader J., Terris B., Stamp G., Baron A., Scarpa A., Lemoine N. R. (2002) Oncogene 21, 4587–4594 [DOI] [PubMed] [Google Scholar]
- 26.Srinivasan D., Plattner R. (2006) Cancer Res. 66, 5648–5655 [DOI] [PubMed] [Google Scholar]
- 27.Ryu J. R., Echarri A., Li R., Pendergast A. M. (2009) Mol. Cell. Biol. 29, 1735–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burton E. A., Oliver T. N., Pendergast A. M. (2005) Mol. Cell. Biol. 25, 8834–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller M. M., Lapetina S., MacGrath S. M., Sfakianos M. K., Pollard T. D., Koleske A. J. (2010) Biochemistry 49, 2227–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai Z., Pendergast A. M. (1995) Genes Dev. 9, 2569–2582 [DOI] [PubMed] [Google Scholar]
- 31.Shi Y., Alin K., Goff S. P. (1995) Genes Dev. 9, 2583–2597 [DOI] [PubMed] [Google Scholar]
- 32.Leng Y., Zhang J., Badour K., Arpaia E., Freeman S., Cheung P., Siu M., Siminovitch K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plattner R., Koleske A. J., Kazlauskas A., Pendergast A. M. (2004) Mol. Cell. Biol. 24, 2573–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zandy N. L., Playford M., Pendergast A. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17686–17691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yogalingam G., Pendergast A. M. (2008) J. Biol. Chem. 283, 35941–35953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLaughlin J., Cheng D., Singer O., Lukacs R. U., Radu C. G., Verma I. M., Witte O. N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20501–20506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X. H., Wang Q., Gerald W., Hudis C. A., Norton L., Smid M., Foekens J. A., Massagué J. (2009) Cancer Cell 16, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minn A. J., Kang Y., Serganova I., Gupta G. P., Giri D. D., Doubrovin M., Ponomarev V., Gerald W. L., Blasberg R., Massagué J. (2005) J. Clin. Invest. 115, 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordón-Cardo C., Guise T. A., Massagué J. (2003) Cancer Cell 3, 537–549 [DOI] [PubMed] [Google Scholar]
- 40.Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massagué J. (2005) Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn R. S. (2008) Ann. Oncol. 19, 1379–1386 [DOI] [PubMed] [Google Scholar]
- 42.Zou D., Yoon H. S., Anjomshoaa A., Perez D., Fukuzawa R., Guilford P., Humar B. (2009) Breast Cancer Res. 11, R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn R. S., Dering J., Ginther C., Wilson C. A., Glaspy P., Tchekmedyian N., Slamon D. J. (2007) Breast Cancer Res. Treat. 105, 319–326 [DOI] [PubMed] [Google Scholar]
- 44.Jallal H., Valentino M. L., Chen G., Boschelli F., Ali S., Rabbani S. A. (2007) Cancer Res. 67, 1580–1588 [DOI] [PubMed] [Google Scholar]
- 45.Bharti S., Inoue H., Bharti K., Hirsch D. S., Nie Z., Yoon H. Y., Artym V., Yamada K. M., Mueller S. C., Barr V. A., Randazzo P. A. (2007) Mol. Cell. Biol. 27, 8271–8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nyalendo C., Michaud M., Beaulieu E., Roghi C., Murphy G., Gingras D., Béliveau R. (2007) J. Biol. Chem. 282, 15690–15699 [DOI] [PubMed] [Google Scholar]
- 47.Cho J. A., Osenkowski P., Zhao H., Kim S., Toth M., Cole K., Aboukameel A., Saliganan A., Schuger L., Bonfil R. D., Fridman R. (2008) J. Biol. Chem. 283, 17391–17405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toth M., Hernandez-Barrantes S., Osenkowski P., Bernardo M. M., Gervasi D. C., Shimura Y., Meroueh O., Kotra L. P., Gálvez B. G., Arroyo A. G., Mobashery S., Fridman R. (2002) J. Biol. Chem. 277, 26340–26350 [DOI] [PubMed] [Google Scholar]
- 49.Wu X., Gan B., Yoo Y., Guan J. L. (2005) Dev. Cell 9, 185–196 [DOI] [PubMed] [Google Scholar]
- 50.Sirvent A., Benistant C., Roche S. (2008) Biol. Cell 100, 617–631 [DOI] [PubMed] [Google Scholar]
- 51.Plattner R., Irvin B. J., Guo S., Blackburn K., Kazlauskas A., Abraham R. T., York J. D., Pendergast A. M. (2003) Nat. Cell Biol. 5, 309–319 [DOI] [PubMed] [Google Scholar]
- 52.Koreckij T., Nguyen H., Brown L. G., Yu E. Y., Vessella R. L., Corey E. (2009) Br. J. Cancer 101, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith M. C., Luker K. E., Garbow J. R., Prior J. L., Jackson E., Piwnica-Worms D., Luker G. D. (2004) Cancer Res. 64, 8604–8612 [DOI] [PubMed] [Google Scholar]
- 54.Müller A., Homey B., Soto H., Ge N., Catron D., Buchanan M. E., McClanahan T., Murphy E., Yuan W., Wagner S. N., Barrera J. L., Mohar A., Verástegui E., Zlotnik A. (2001) Nature 410, 50–56 [DOI] [PubMed] [Google Scholar]
- 55.Bartolomé R. A., Ferreiro S., Miquilena-Colina M. E., Martinez-Prats L., Soto-Montenegro M. L., Garcia-Bernal D., Vaquero J. J., Agami R., Delgado R., Desco M., Sánchez-Mateos P., Teixidó J. (2009) Am. J. Pathol. 174, 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saur D., Seidler B., Schneider G., Algül H., Beck R., Senekowitsch-Schmidtke R., Schwaiger M., Schmid R. M. (2005) Gastroenterology 129, 1237–1250 [DOI] [PubMed] [Google Scholar]
- 57.Dewan M. Z., Ahmed S., Iwasaki Y., Ohba K., Toi M., Yamamoto N. (2006) Biomed. Pharmacother. 60, 273–276 [DOI] [PubMed] [Google Scholar]
- 58.Liu H. Y., Wen G. B., Han J., Hong T., Zhuo D., Liu Z., Cao W. (2008) J. Biol. Chem. 283, 30642–30649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ptasznik A., Urbanowska E., Chinta S., Costa M. A., Katz B. A., Stanislaus M. A., Demir G., Linnekin D., Pan Z. K., Gewirtz A. M. (2002) J. Exp. Med. 196, 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin J., Arlinghaus R. (2008) Oncogene 27, 4385–4391 [DOI] [PubMed] [Google Scholar]
- 61.Bartolomé R. A., Gálvez B. G., Longo N., Baleux F., Van Muijen G. N., Sánchez-Mateos P., Arroyo A. G., Teixidó J. (2004) Cancer Res. 64, 2534–2543 [DOI] [PubMed] [Google Scholar]
- 62.Peacock J. G., Miller A. L., Bradley W. D., Rodriguez O. C., Webb D. J., Koleske A. J. (2007) Mol. Biol. Cell 18, 3860–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zandy N. L., Pendergast A. M. (2008) Cell Cycle 7, 444–448 [DOI] [PubMed] [Google Scholar]
- 64.Yamazaki D., Kurisu S., Takenawa T. (2009) Oncogene 28, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 65.Stylli S. S., Kaye A. H., Lock P. (2008) J. Clin. Neurosci. 15, 725–737 [DOI] [PubMed] [Google Scholar]
- 66.Li Y., Tondravi M., Liu J., Smith E., Haudenschild C. C., Kaczmarek M., Zhan X. (2001) Cancer Res. 61, 6906–6911 [PubMed] [Google Scholar]
- 67.Mizutani K., Miki H., He H., Maruta H., Takenawa T. (2002) Cancer Res. 62, 669–674 [PubMed] [Google Scholar]
- 68.Sun X., Li C., Zhuang C., Gilmore W. C., Cobos E., Tao Y., Dai Z. (2009) Carcinogenesis 30, 2109–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.