Abstract
Cytokinesis is a fundamental cellular process, which ensures equal abscission and fosters diploid progenies. Aberrant cytokinesis may result in genomic instability and cell transformation. However, the underlying regulatory machinery of cytokinesis is largely undefined. Here, we demonstrate that Nlp (Ninein-like protein), a recently identified BRCA1-associated centrosomal protein that is required for centrosomes maturation at interphase and spindle formation in mitosis, also contributes to the accomplishment of cytokinesis. Through immunofluorescent analysis, Nlp is found to localize at midbody during cytokinesis. Depletion of endogenous Nlp triggers aborted division and subsequently leads to multinucleated phenotypes. Nlp can be recruited by Aurora B to the midbody apparatus via their physical association at the late stage of mitosis. Disruption of their interaction induces aborted cytokinesis. Importantly, Nlp is characterized as a novel substrate of Aurora B and can be phosphorylated by Aurora B. The specific phosphorylation sites are mapped at Ser-185, Ser-448, and Ser-585. The phosphorylation at Ser-448 and Ser-585 is likely required for Nlp association with Aurora B and localization at midbody. Meanwhile, the phosphorylation at Ser-185 is vital to Nlp protein stability. Disruptions of these phosphorylation sites abolish cytokinesis and lead to chromosomal instability. Collectively, these observations demonstrate that regulation of Nlp by Aurora B is critical for the completion of cytokinesis, providing novel insights into understanding the machinery of cell cycle progression.
Keywords: Carcinogenesis, Cell Cycle, Cell Division, Centrosome, Oncogene
Introduction
Cytokinesis is the final step of cell division by which the daughter nuclei and cytoplasm are separated completely and produce diploid progenies (1). During this process, a group of anti-parallel microtubules form the midbody apparatus to deliver signals and work as a framework for regulatory protein anchoring (2). Numerous proteins, such as PRC1, MKLP1, and BRUC (3–5), are involved in this process. All of these proteins are recruited to the midbody during cytokinesis, activate downstream signal pathways to lead to actin polymerization and myosin activation, and then create shearing force to separate the cleavage furrow (6, 7). Abnormal cytokinesis can induce defects in cell separation and accumulation of nuclei in polyploidy cells and then promote cellular transformation and tumorigenesis.
Aurora B is an important phosphokinase in mammalian cells. Together with the inner centromere protein (INCENP)2 and survivin, it composes the chromosome passenger complex and has been implicated in multiple mitotic events, especially in the regulation of cell division (8, 9, 29). Aurora B moves from centromeres to the midzone spindle at anaphase and to the midbody at telophase and cytokinesis. As the unique kinase center of chromosome passenger complex, the activity of Aurora B is crucial for the functions of the complex, which depend on Aurora B phosphorylation of its substrates at proper time in corresponding cell cycle phases (8). Several substrates of Aurora B have already been identified, such as MKLP1 and MgcRacGAP (10). In the phase of cytokinesis, Aurora B just locates at midbody and recruits important molecules for cytokinesis signal transmission (11). Furthermore, Aurora B is required for ensuring the correct assembly of the midzone and midbody microtubules (24). In mammalian cells, several cytoskeletal proteins are phosphorylated by Aurora B for cytokinesis. Goto et al. (12) have reported that Aurora B can regulate the cleavage of furrow-specific vimentin phosphorylation and then control vimentin filament segregation during the cytokinetic process.
Nlp is a recently identified BRCA1-associated centrosomal protein (28), which was tethered to the interphasic centrosome by the tumor suppressor BRCA1. Such interaction is required for the maintenance of Nlp centrosomal localization and protein stability, which ensures centrosome maturation and spindle formation. Depletion of endogenous Nlp via siRNA hindered bipolar spindle formation and triggered chromosomal missegregation (28). Significantly, several lines of evidence have linked Nlp to human cancers. Deregulated expression of Nlp is found in diverse human primary cancers and tumor-derived cell lines, typically breast, lung, ovarian, and head and neck cancer, which is in part coupled with NLP gene amplification (31–33). Overexpression of Nlp is able to transform NIH3T3 fibroblasts in vitro and initiate tumor formation in nude mice. Consistently, Nlp transgenic mice display spontaneous tumorigenesis in the breast, ovary, and testicle and showed more rapid onset of radiation-induced lymphoma. Therefore, Nlp may be a potential oncogenic protein in tumorigenesis (31). It has been previously reported by Casenghi et al. (13) that Nlp is a γ-tubulin-binding protein and involves centrosome maturation and microtubule nucleation. During interphase, Nlp is transported to the centrosome by the dynein-dynactin motor complex and then contributes to the attachment and nucleation of microtubules (14). Additionally, the phosphorylation of centrosomal Nlp by Plk1 upon entry into mitosis, followed by displacement from the centrosome, is a prerequisite for correct spindle formation (13, 28). In mammalian cells, Nlp is expressed in a cell cycle-dependent manner with a peak at G2/M transition. Further analysis has indicated that Nlp is a relatively short half-life protein and can be ubiquitinated via the anaphase-promoting cyclosome complex pathway (15). Intriguingly, both yeast two-hybrid and biochemical evidences strongly suggest Nlp as a physiological substrate of several essential mitotic kinases, including Plk1, Nek2, and Cdc2 (13, 14, 25, 34) Coordinated phosphorylation of Nlp by these kinases facilitate its dissociation from the centrosome, which is a critical step for centrosome maturation, spindle assembly, and chromosome segregation (14). Apparently, phosphorylation plays a key role in the regulatory machineries of Nlp for proper execution of sequential fundamental mitotic events and cell cycle progression.
In this study, we demonstrate that Nlp is involved in the final events of cell cleavage. Depletion of Nlp perturbs proper cytokinesis and fosters multinucleation. Interestingly, Nlp is recruited to the midbody and phosphorylated by Aurora B during cytokinesis. Such phosphorylation is not only involved in recruiting Nlp to the midbody but also stabilizes Nlp during cytokinesis. These findings suggest that the modulation of Nlp by Aurora B might be vital to the commitment of cytokinesis and maintenance of genomic stability.
EXPERIMENTAL PROCEDURES
Antibodies and Dyes
Anti-GFP, Aurora B, γ-tubulin, and ubiquitin were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-phosphohistone H3.1 antibody was from Bioworld (GeneLinx International, Inc.). FITC/TRITC-conjugated goat anti-mouse/rabbit antibodies were from Zhongshan (Zhongshan Goldenbridge Biotechnology, Co., Ltd., China). Rabbit polyclonal anti-Nlp antibody was produced by MBL (Beijing B&M Biotech Co., Ltd., China). DAPI was purchased from Sigma and High Technology Corp.
Cell Culture
HeLa cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (Biochrom Ltd.), 100 units/ml penicillin, and 100 μg/ml streptomycin and incubated at 37 °C in a humidified atmosphere of 5% carbon dioxide. HeLa cells overexpressing wild type or mutant GFP-Nlp were grown in selective medium (RPMI 1640 medium with G418 at a concentration of 400 μg/ml). To synchronize cells at G2/M phase, cells were treated with 400 ng/ml nocodazole (Sigma) for 16 h and then changed to fresh RPMI 1640 medium until harvest.
Immunoblotting Analysis
To extract cellular proteins, cells were collected, rinsed with PBS, and lysed in lysis buffer. The Western blotting assays were performed as described before (26).
Plasmid Construction, Mutagenesis, and Cell Transfection
To make Aurora B expression vector, human Aurora B coding regions were amplified using the High Fidelity PCR system. To create Nlp site-directed mutations, Muta-directTM mutagenesis kit (SBS Genetech Co., China) was used with pEGFP-Nlp as template.
For transient transfection, 5 μg of DNA and 10 μl of Lipofectamine 2000 (Invitrogen) were used. After being incubated for 6 h, the medium was changed with fresh RPMI 1640 medium. Cells were harvested or fixed at 24 or 48 h after transfection.
Preparation of GST Proteins, Pulldown, and Immunoprecipitation
Expression and extraction of GST fusion proteins were performed as described previously (26).
For pulldown experiments, the purified GST fusion proteins (conjugated with glutathione-agarose beads) were incubated with 1 mg of cell lysate at 4 °C overnight. After washing the beads with PBS, the pulldown complexes were analyzed by Western blotting analysis.
For immunoprecipitation experiments, 1 mg of cellular proteins was first incubated with 10 μl of special antibodies at 4 °C for 6 h, and then 20 μl of protein A/G-agarose beads was added and incubated at 4 °C for 6 h. After centrifugation, the precipitated complexes were analyzed by Western blotting assay.
Immunofluorescence Microscopy
Fixed cells were incubated with the indicated antibodies and then probed with TRITC-conjugated goat anti-mouse/rabbit IgG. DNA was stained with DAPI. Cells were visualized by an Olympus fluorescent microscope. For observing the co-localization of distinct proteins, a laser-scanning confocal microscope (Leica Microsystems Heidelberg GmbH, Am Friedensplatz 3, Germany) was used to visualize the cells.
Aurora B Kinase Assay
Aurora B kinase assay was performed as follows: 1–5 μg of GST fusion proteins were incubated with 20 ng of active Aurora B kinase (Upstate Biotechnology, Inc.), 2.5 μCi of [γ-32P]ATP (PerkinElmer Life Sciences), and 5 μl of magnesium/ATP mixture (75 mm MgCl2, 500 μm ATP in 20 mm MOPS, pH 7.2, 25 mm β-glycerol phosphate, 5 mm EGTA, 1 mm sodium orthovanadate, 1 mm dithiothreitol). The reaction system was incubated for 30 min at room temperature and stopped the by adding 2× protein loading buffer. The phosphorylated GST fusion proteins were run on a polyacrylamide gel. Polyacrylamide gels were dried using a vacuum dryer and exposed to x-ray films under −70 °C for 2 h.
RNA Interference and RT-PCR Analysis
The Nlp siRNA sequence was designed as 5′-GUGAGUCUUGAGGAAUUCC)d(TT)-3′ and 5′-CAUGUAGAUUUGAGAGAGA)d(TT)-3′. The Aurora B siRNA sequence was designed as 5′-CAGCCACGAUCAUGGAGGATTUCCUCCAUGUCGUGGCUGTT)d(TT)-3′ and 5′-GGUGAUGGAGAAUAGCAGUTTACUGCUAUUCUCCAUCACCTT)d(TT)-3′. The nonspecific siRNA (control) sequence was designed as 5′-AUUGUAUGCGAUCGCAGAC)d(TT)-3′. These sequences do not match any other sequences in the GenBankTM.
RESULTS
Nlp Localizes at Midbody during Cytokinesis
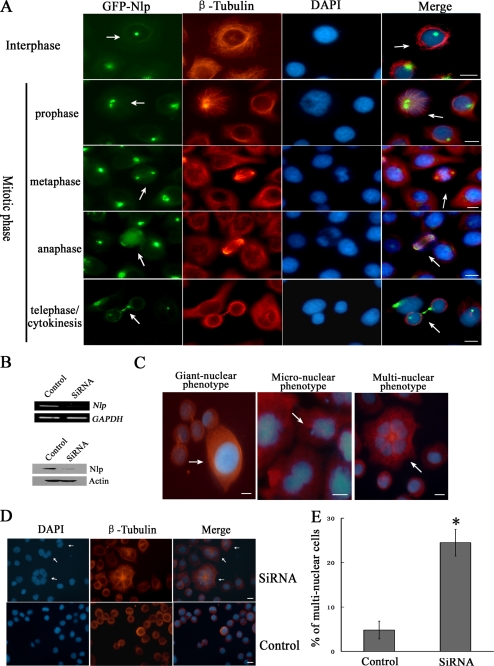
To further explore the functions of Nlp in regulation of cell cycle progression, HeLa cells overexpressing GFP-Nlp were first established. Using an immunofluorescent approach, the localization of GFP-Nlp was found to be dynamic according to cell cycle progression. In this experiment, different phases of the cell cycle were discriminated by special nuclear phenotypes. As shown in Fig. 1A, GFP-Nlp was localized at centrosomes at the beginning of the mitotic phase and then displaced when cells passed into anaphase. Until telophase and cytokinesis when daughter cells were linked only by a narrow cytoplasmic bridge, GFP-Nlp was clearly seen in a double-band region of the midbody.
FIGURE 1.
Nlp is involved in cytokinesis. A, localization of Nlp in different phases of cell cycle. HeLa cells overexpressing GFP-Nlp were immunostained for skeletons (red) with anti-β-tubulin antibody. Different cell cycle phases (interphase, mitotic phase, and cytokinesis) were identified through the special nuclear morphological phenotypes. The arrows show the special location of Nlp during different phases of cell cycle. Scale bars, 10 μm. B, expression of Nlp was inhibited by siRNA. HeLa cells were transfected with Nlp-specific siRNA (nonspecific siRNA was used as control). After 24 h, RT-PCR was performed to examine the mRNA of Nlp; after 48 h, immunoblotting assay was performed to analyze the protein level of Nlp. C, abnormal nuclear phenotypes in HeLa cells silenced for endogenous Nlp. The arrows showed the images of specific phenotypes. D, multinuclear cells in HeLa cells treated with Nlp siRNA. HeLa cells were transfected with Nlp-specific siRNA for 96 h, and cellular nuclei and skeletons were stained as described in A. The arrows showed the images of special multinuclear cells after Nlp depletion. Scale bars, 20 μm. E, quantitative comparison of multinuclear cells between Nlp interference cells and control. The results were obtained from three separate experiments in which more than 500 cells were examined. Asterisk indicates that remarkable discrepancy existed in the statistics between Nlp interference cells and control cells (p < 0.05).
To investigate whether Nlp is involved in cytokinesis, specific Nlp siRNA was used to inhibit the expression of Nlp. The interfering results were examined through RT-PCR and Western blot assays (Fig. 1B). When endogenous Nlp in cells was depressed, several abnormal nuclear phenotypes were observed, including giant nuclear (the nuclear material is much more than normal ones), micronuclear (there is small nuclear material besides the forming nucleus), and multinuclear phenotypes (one cell contains two or more well distributed nuclei) (Fig. 1C). Following the treatment with Nlp siRNA, a number of cells displayed multiple nuclei (Fig. 1D, white arrows). Statistical analysis in three independent experiments showed that the percentage of multinuclear cells in Nlp-depleted teams were about 25%, which was much more than control (Fig. 1E). Because one of the important phenotypes in cytokinesis failure cells is multinucleation (30), we assumed that Nlp may function as an important factor in the process of cytokinesis.
Nlp Physically Interacts with Aurora B in the Phase of Cytokinesis
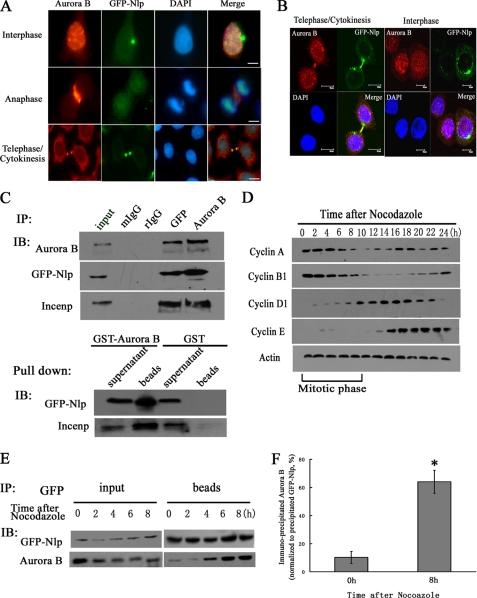
As mentioned above, Aurora B is important for mitosis and cytokinesis, and its subcellular location is cell cycle-dependent. Through immunofluorescent staining, we confirmed that during the progression of mitosis, Aurora B moved from chromatin to the midzone and finally located at the midbody during cytokinesis (Fig. 2A). At the same time, we also found that Nlp was co-localized with Aurora B at the midbody (Fig. 2, A and B, and supplemental Fig. S1A). These observations suggested an interaction between Nlp and Aurora B. Through immunoprecipitation assays with anti-GFP and anti-Aurora B antibodies, we found that Aurora B, GFP-Nlp, and INCENP could be detected in the precipitated complexes (Fig. 2C, top panels). Additionally, the GST-pulldown assay also showed that Nlp was precipitated by GST-Aurora B (Fig. 2C, bottom panels). The physical interaction of endogenous Nlp with Aurora B was further corroborated by antibodies against Nlp and Aurora B through co-immunoprecipitation in HCT116 cells. Apparently, the endogenous Nlp could be immunoprecipitated by Aurora B and vice versa (supplemental Fig. S1B). Collectively, these results indicate that Nlp physically interacts with Aurora B.
FIGURE 2.
Nlp interacts with Aurora B. A, localization of Nlp and Aurora B in different mitotic phases. HeLa cells overexpressing GFP-Nlp were stained with anti-Aurora B antibody (red). DAPI was used to visualize nuclei. Scale bars, 10 μm. B, co-localization of Nlp and Aurora B at midbody during cytokinesis. HeLa cells overexpressing GFP-Nlp were stained as described above. Nlp (green spots), Aurora B (red spots), and nuclei (blue spots) were visualized by the laser-scanning confocal microscope. Scale bars, 10 μm. C, interaction of Nlp and Aurora B. Top panel, immunoprecipitation (IP) was performed with anti-GFP and anti-Aurora B antibodies separately in overexpressing GFP-Nlp cells. Nonspecific rabbit IgG and mouse IgG (2 μg/testing system) were used as negative control. The precipitated complexes were analyzed by immunoblotting (IB) assay with anti-Aurora B, anti-GFP, and anti-INCENP antibodies. Bottom panel, GST-Aurora B fusion protein was incubated with cellular lysate mentioned above, and the precipitated complexes were analyzed by immunoblotting assay with anti-GFP and anti-INCENP antibodies. GST protein was used as negative control. D, synchronization of cell cycle. HeLa cells overexpressing GFP-Nlp were arrested at the mitotic phase by nocodazole treatment. After being released into fresh medium, cells were collected at the indicated time points, and cellular lysates were analyzed through immunoblotting analysis with anti-cyclin A, B1, D1, and E. Mitotic stage of cell cycle is indicated at the bottom panels. E, overexpressing GFP-Nlp cells were synchronized by nocodazole and harvested at the indicated time points. Cellular lysates were immunoprecipitated with anti-GFP antibody, and immunoblotting assay was performed with anti-GFP and anti-Aurora B antibodies. F, quantitative comparison of Nlp-Aurora B interaction at the beginning of mitosis and at the end of cytokinesis. After immunoprecipitating, the precipitated Aurora B protein levels at two different time points were normalized to the corresponding precipitated GFP-Nlp protein. Asterisk indicates that the protein level of the precipitated Aurora B by Nlp at the 8-h time point after release by nocodazole treatment is statistically different from that of 0 h (p value).
However, using immunofluorescent microscopy, we found that co-localization of Nlp and Aurora B was not throughout the whole cell cycle but only in the final stage of cell division (Fig. 2, A and B), suggesting that the interaction of Nlp and Aurora B might be cell cycle-dependent. To confirm this hypothesis, nocodazole (mitotic inhibitor) was employed to synchronize cells at G2/M phase. After releasing into fresh medium, most of cells were synchronized in the same stage of mitosis. According to the protein level of cyclins (cyclin A, B1, D, and E1), the mitotic stages of the cell cycle was well defined (Fig. 2D). Following synchronization, the immunoprecipitation assay with anti-GFP antibody was performed. We found that the level of precipitated Aurora B protein was elevated according to the process of mitosis and cytokinesis (Fig. 2E). Normalizing to the precipitated GFP-Nlp in the same complexes, the precipitated Aurora B in the stage of cytokinesis was much more than that seen at the beginning of mitosis (Fig. 2F). This evidence suggests that interaction of Nlp with Aurora B mostly happened in cytokinesis.
Nlp Is Phosphorylated by Aurora B at Sites of Ser-185, Ser-448, and Ser-585
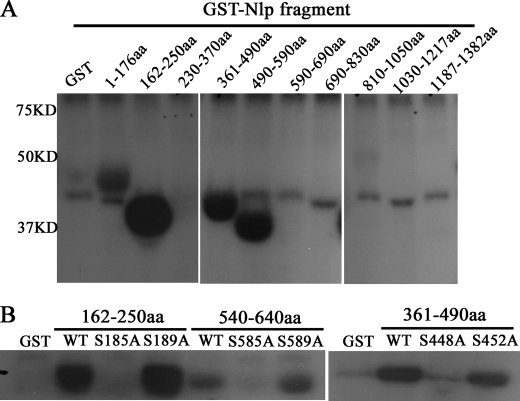
To investigate whether Nlp is phosphorylated by Aurora B, kinase assay was performed. As shown in Fig. 3A, the amino acid fragments 162–250, 361–490, and 490–590 of Nlp exhibited obvious phosphorylation in the presence of Aurora B. Generally, it is believe that Aurora B targets its substrates at special sites ((R/K)1–3X(S/T)) (10). Through sequence analysis, we found three conserved phosphorylating sites (Ser-185, Ser-448, and Ser-585) in these positive fragments. To confirm the phosphorylating sites, Ser-185, Ser-448, and Ser-585 were mutated to nonphosphorylatable alanines by site-directed mutagenesis, respectively. The Aurora B kinase assay showed that compared with wild type fragments, the phosphorylation of those site-mutated fragments was abolished (Fig. 3B). These data indicate that Nlp is phosphorylated by Aurora B at the sites of Ser-185, Ser-448, and Ser-585.
FIGURE 3.
Mapping the phosphorylating sites of Nlp by Aurora B. A, identification of the phosphorylated regions of Nlp. Aurora B kinase (the final concentration was 8 ng/ml) was used in kinase assays, which were performed with a series of GST fusion Nlp fragments covering the full length of Nlp. B, determining the Aurora B phosphorylatable sites of Nlp. In three special fragments (162–250, 361–490, and 540–640 amino acids (aa)) of GST-Nlp, Ser-185, Ser-448, and Ser-585 were mutated to alanines, respectively. These three mutated fragments were employed as substrates in kinase assay. Additionally, three another irrespective sites (Ser-189, Ser-452, and Ser-589) were also mutated and used in kinase assay as controls.
Phosphorylation of Nlp at Ser-448 and Ser-585 Is Necessary for Its Localization at Midbody
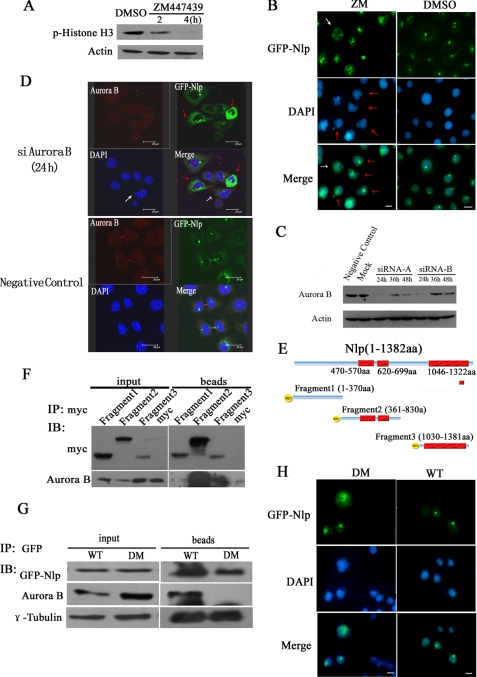
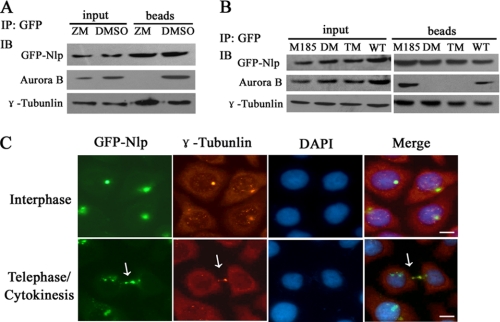
To explore the significance of Nlp phosphorylation by Aurora B in cytokinesis, ZM447439, a specific selective ATP-competitive inhibitor of Aurora B kinase (16), was employed to inhibit the activity of Aurora B in the experiments. First of all, histone H3, an identified substrate of Aurora B, was examined to detect the kinase activity of Aurora B in this assay. Immunoblotting assay with anti-phosphorylated histone H3 antibody showed that after cells were treated by ZM447439 for 4 h, the level of phosphorylated histone H3 was substantially reduced (Fig. 4A), indicating that treatment with ZM447439 resulted in inhibition of Aurora B kinase activity. Next, we performed immunofluorescent assay and found that following inhibition of Aurora B cells displayed abnormal nuclear phenotypes (Fig. 4B). Most of nuclei were not divided completely. There were a number of multinuclear cells that emerged (red arrows shown in Fig. 4B) and multipolar spindles (white arrow showed in Fig. 4B). In addition, after suppression of Aurora B activity, GFP-Nlp diffused widely in the cytoplasm, and none of cells in the cytoplasmic dividing phase were found (red arrows in Fig. 4B). Similar results were also obtained in cells treated with siRNA. After HeLa cells overexpressing GFP-Nlp were treated with Aurora B siRNA (siAurora B), the endogenous Aurora B protein was substantially suppressed (Fig. 4C). Through immunofluorescent staining and laser-scanning confocal examination, we observed that GFP-Nlp aggregated into large assemblies and dispersed into the cytoplasm following suppression of Aurora B by siRNA approach (red arrows showed in Fig. 4D). Additionally, abnormal nuclear types (multiple nuclei, giant nuclei, and micro-nuclei) were also seen in cells treated with Aurora B siRNA (white arrows in Fig. 4D and supplemental Fig. S2). These results indicate that Aurora B is required for the normal localizations of Nlp in cytokinesis.
FIGURE 4.
Aurora B recruits Nlp to the midbody during cytokinesis. A, inhibition of Aurora B activity by ZM447439. HeLa cells were treated with 5 μm ZM447439 for 2 or 4 h and harvested (DMSO was used as control). The cellular lysates were examined through immunoblotting analysis with anti-phosphorylated histone H3. B, inhibition of Aurora B kinase results in Nlp abnormal localization and multiple nuclei formation as well as typical multipolar spindle. HeLa cells overexpressing GFP-Nlp were treated with ZM447439 or DMSO for 48 h and stained with DAPI to display the cellular nuclei. Scale bars, 10 μm. (Red arrows indicate Nlp abnormal localization, and white arrow illustrates a tetrapolar spindle). C, expression of Aurora B was inhibited by siRNA. Cells were transfected with Aurora B-specific siRNA (nonspecific siRNA was used as negative control). siRNA-A and -B means two different targets on Aurora B (for sequence, see “Experimental Procedures”). D, HeLa cells overexpressing GFP-Nlp were treated with siAurora B and nonspecific siRNA (negative control) for 24 h and then stained as in Fig. 2A. Nlp (green spots), Aurora B (red spots), and nuclei (blue spots) were visualized by the laser-scanning confocal microscope. The red arrows show mis-localization of GFP-Nlp; the white arrow shows the abnormal nuclear phenotype. E, schematic representations of Nlp fragments (1–370, 361–830, and 1030–1381 amino acids (aa)) cloned in pCS2+MT expression vector. These vectors could express Myc-fused protein in cells. The red boxes showed the coiled-coil domain in Nlp protein. F, Nlp expression vectors harboring different fragments were transfected into HeLa cells for 48 h, and the cellular lysates were immunoprecipitated (IP) with anti-Myc antibody. The precipitated complexes were analyzed by immunoblotting (IB) assay with anti-Myc and anti-Aurora B antibodies. G, interaction of Nlp with Aurora B. HeLa cells were transfected with GFP-Nlp (WT) or GFP mutant Nlp (DM: double-site mutations in S448A and S585A) for 48 h, and the cellular lysates were immunoprecipitated with anti-GFP antibody. The precipitated complexes were analyzed by immunoblotting assay with anti-GFP, anti-Aurora B, and anti-γ-tubulin antibodies. H, subcellular localization of Nlp mutants and appearance of multiple nuclei. HeLa cells were transfected with GFP-Nlp (WT) or GFP-mutant Nlp (DM) expression vectors for 96 h and then stained with DAPI to display the cellular nuclei. Scale bars, 10 μm.
The analyses of the bioinformatics showed that Nlp contains three coiled-coil motifs (470–570, 620–699, and 1046–1322 amino acids), which are thought of as the functional motifs for protein-protein interactions (27). To determine which part of Nlp was essential for interaction with Aurora B, we constructed three Myc-tagged Nlp deletions (1–370, 361–830, and 1030–1381 amino acids) expressing vectors in which coiled-coil motifs were included (Fig. 4E). Through the immunoprecipitation assay with anti-Myc antibody, we found that the fragments covering 361–830 and 1030–1381 amino acids were able to precipitate Aurora B (Fig. 4F). Similar results were obtained in HEK293T cells expressing these Myc-tagged Nlp deletion mutants (data not shown). Because the phosphorylation sites of Ser-448 and Ser-585 are located only in the interacting positive fragment (361–830 amino acids), we presumed phosphorylations at these two sites might mediate the interaction of Nlp and Aurora B. To prove this hypothesis, Ser-448 and Ser-585 were replaced with nonphosphorylatable alanines. Through immunoprecipitation assays with anti-GFP antibody, we found that the double-site mutant of GFP-Nlp was not able to precipitate Aurora B (Fig. 4G), which indicates that Ser-448 and Ser-585 are required for Nlp interacting with Aurora B.
We also transfected Nlp mutant expression vector with Ser-448 and Ser-585 double-site mutations into cells, and we found that the expression of such a mutant Nlp protein led to multinuclear formation. In parallel, Nlp was not located at midbody but diffused throughout the cytoplasm (Fig. 4H). Thus, it appears that the localization of Nlp at midbody is dependent on its interaction with Aurora B.
Phosphorylation of Nlp by Aurora B Maintains the Stability of Nlp in Cytokinesis
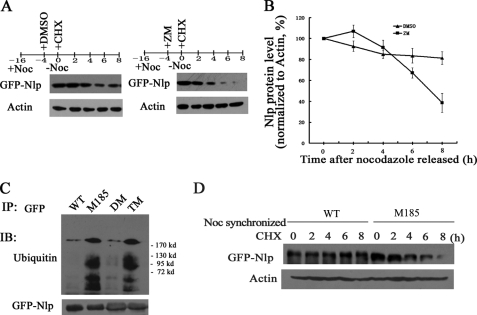
It has been previously reported by Wang et al. (15) that Nlp is a short lived protein, and its degradation is regulated by anaphase-promoting cyclosome complex-mediated ubiquitination. We further examined whether phosphorylation on Nlp could affect its degradation. In this experiment, ZM447439 was employed to inhibit the kinase activity of Aurora B. We synchronized cells with nocodazole and used cycloheximide to block protein synthesis. Immunoblotting assay showed that when cells were treated with ZM447439, Nlp degraded much faster than the cells treated with DMSO (Fig. 5A). Following suppression of Aurora B activity, the protein level of Nlp remained only 40% compared with that seen in control cells (Fig. 5B).
FIGURE 5.
Aurora B activity affects degradation of Nlp. A, HeLa cells overexpressing GFP-Nlp were synchronized with nocodazole (Noc). 4 h before releasing, ZM447439 was added to the medium to inhibit Aurora B kinase activity (DMSO was used as control). At the releasing time, cycloheximide (CHX, 100 μg/ml) was added to medium. The cells were harvested at every 2 h for immunoblotting analysis with anti-GFP or anti-actin antibodies. B, cells were treated as described in A. To quantitatively evaluate the GFP-Nlp protein levels at each time point, the protein levels determined by immunoblotting (IB) were normalized to corresponding actin in three independent experiments. C, HeLa cells were transfected with GFP-Nlp (WT), mutation in Ser-185 (M185), double mutations in Ser-448 and Ser-585 (DM), or triple mutations in Ser-185 and Ser-448 and Ser-585 (TM) vectors, respectively, for 48 h. The cellular lysates were immunoprecipitated with anti-GFP antibody and analyzed by immunoblotting assay with anti-ubiquitin and anti-GFP antibodies. D, HeLa cells overexpressing GFP-Nlp (WT) or GFP-mutant Nlp (M185) were synchronized at G2/M with nocodazole and cycloheximide. The treated cells were harvested at every 2 h for immunoblotting assay with anti-GFP antibody, and the levels of action were used for normalization.
To further investigate the role of phosphorylation in Nlp ubiquitination, we performed immunoprecipitation assay in cells transfected with different expression vectors (WT, GFP-Nlp; M185, mutation in Ser-185; DM, double mutations in Ser-448 and Ser-585; and TM, triple mutations in Ser-185, Ser-448, and Ser-585) and found that the ubiquitination levels of the Ser-185 mutant and the triple-site mutant were much higher than double-site mutant and wild type (Fig. 5C). These findings suggest that phosphorylation at Ser-185 might play a role in the ubiquitination of Nlp.
We also compared the protein degradation of wild type Nlp and mutant Nlp with mutation in Ser-185. HeLa cells overexpressing wild type or Ser-185-mutated GFP-Nlp were synchronized in late mitotic phase by nocodazole, treated with cycloheximide, and followed by immunoblotting assay. Consistently, degradation of Ser-185 mutant Nlp was much faster than wild type Nlp (Fig. 5D), suggesting that phosphorylation at Ser-185 by Aurora B might stabilize Nlp during the phase of cytokinesis.
Phosphorylations of Nlp by Aurora B Have No Effect on the Interaction of Nlp and γ-Tubulin in Cytokinesis
As reported by Casenghi et al. (13), Nlp is a candidate γ-tubulin-binding protein, which interacts with γ-tubulin and stimulates microtubule nucleation. We then examined whether Aurora B regulates the interaction of Nlp with γ-tubulin. Immunoprecipitation assay showed that GFP-Nlp could still precipitate γ-tubulin when the activity of Aurora B was inhibited (Fig. 6A). Furthermore, HeLa cells were transfected with wild type GFP-Nlp, Ser-185 mutant, Ser-448 and Ser-585 double-site mutant, or Ser-185 and Ser-448 and Ser-585 triple-site mutant vectors, respectively, and immunoprecipitation assays were performed with anti-GFP antibody. As showed in Fig. 6B, all the Nlp mutants were still able to precipitate γ-tubulin, regardless of inhibition of Aurora B kinase activity. These results further confirm that phosphorylation of Nlp by Aurora B does not perturb the interaction between Nlp and γ-tubulin at midbody.
FIGURE 6.
Phosphorylation of Nlp has no effect on its interaction with γ-tubulin. A, HeLa cells overexpressing GFP-Nlp were treated with ZM447439 or DMSO for 4 h and then harvested for immunoprecipitation (IP) assay with anti-GFP antibody. The precipitated complexes were analyzed by immunoblotting (IB) assay with anti-GFP, anti-Aurora B, and anti-γ-tubulin antibodies. B, HeLa cells were transfected with GFP-Nlp (WT), GFP-Nlp mutants (M185, DM, and TM), respectively, for 48 h. The cellular lysates were performed through immunoprecipitating assay with anti-GFP antibody and then immunoblotting assay with anti-GFP, anti-Aurora B, and anti-γ-tubulin antibodies. C, HeLa cells overexpressing GFP-Nlp were immunostained with anti-γ-tubulin antibody. Interphase and telophase/cytokinesis were distinguished by the special nuclear phenotypes. The white arrow showed the midbody in cytokinesis. Scale bars, 10 μm.
Interestingly, we found that Nlp and γ-tubulin not only interacted on centrosomes during interphase, but also co-localized at the midbody during cytokinesis (Fig. 6C, white arrow). These observations indicate the necessity for the recruitment of Nlp by Aurora B to the midbody to transiently stimulate microtubule nucleation with γ-tubulin. As a platform, Nlp can coordinately organize γ-tubulin and other unidentified elements to maintain the stability of the midbody apparatus (Fig. 7A).
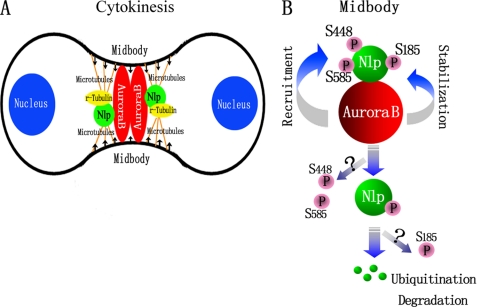
FIGURE 7.
Models for the role of Aurora B regulating Nlp during cytokinesis. A, schematic representative for the role of interaction between Nlp and Aurora B in cytokinesis. Nlp and Aurora B co-localized at midbody and are involved in execution of cytokinesis. In this process, midbody is the major reacting region, and some other proteins are involved, including γ-tubulin and microtubules. B, phosphorylation of Nlp by Aurora B kinesis regulates the function of Nlp. Aurora B phosphorylation at Ser-448 and Ser-585 is required for the localization of Nlp at midbody, and phosphorylation at Ser-185 stabilizes the Nlp protein in cytokinesis.
DISCUSSION
The process of cell division is divided into two consecutive steps as follows: nuclear division and plasmatic division, both of which are critical for chromosome stability of daughter cells. Any aberrations of these procedures will lead to chromosome instability and then result in the formation of heteroploidy cells, which is closely associated with cell transformation and tumorigenesis (17). However, the machinery of controlling cytokinesis is poorly understood. In this study, we demonstrated that Nlp plays an important role in cytokinesis, which is regulated by mitotic kinase Aurora B. RNAi-mediated depletion of Nlp perturbs the process of cytokinesis and leads to heteroploidy. In cells with experimentally exhausting endogenous Nlp, we observed three kinds of aberrant nuclear phenotypes, including giant nuclear, micronuclear, and multinuclear phenotypes (Fig. 1C). It is well accepted that there are several possible mechanisms that contribute to the formation of heteroploidy cells, including dysfunction of mitotic spindles, partial inactivation of mitotic proteins, and failure of cytokinesis (18, 19). For those giant nuclear and micronuclear cells, the daughter nuclei have not divided or separated abnormally, which are caused by defects of spindle formation and unsuccessful chromosomal segregation. For those multinuclear cells, all the nuclei demarcated completely, indicating the process of nuclear division was faultless. However, the phenotype with several nuclei held in one cell indicates that there are faults in the process of cytoplasmic division, in which the complete daughter nuclei were unable to separate equally but gathering together to form polyploidy cells. Therefore, formation of multinuclear cells might be mainly due to aborted cytokinesis.
Cytokinesis is the final stage of cell division in which a critical architecture, the midbody apparatus, must be formed to fulfill this cellular process. Midbody is a narrow tubular intercellular bridge that consists of tightly bundled anti-parallel microtubules, but the origin of these microtubule bundles is largely unknown. According to the observations by Shu et al. (20) and Julian et al. (21), γ-tubulins transiently present at midbody and act as a transient microtubule organizing center for the midbody formation. Piel et al. (22) have also reported that during cytokinesis the centrosomes transiently appear at midbody, which is necessary for completion of cytoplasmic division. All these observations suggest the necessity of a transient microtubule organizing center for the midbody formation. In this study, we found a co-localization of γ-tubulin and Nlp at midbody in the phase of cytokinesis (Fig. 6C). Significantly, phosphorylation of Nlp by Aurora B does not affect its interaction with γ-tubulin at midbody (Fig. 6B). When Nlp expression interfered, midbody formation was disrupted and multinuclear cells substantially increased, indicating that Nlp is one of the key components involved in control of cytokinesis. Thus, we presume that Nlp may work as a platform that coordinately organizes γ-tubulin and other unidentified microtubule-associated proteins to maintain the stability of the midbody apparatus and ensure the commitment of cytokinesis (Fig. 7A).
Aurora B is one of the most notable proteins for regulation of cytokinesis and involved in the formation of midbody (23). Depletion of Aurora B induces failure of cytokinesis and results in polyploidy cells (24). Similarly, we found that when activity of Aurora B was suppressed by ZM447439, or expression of Aurora B protein was directly blocked via siRNA approach, the offspring nuclei did not separate completely (Fig. 4B and supplemental Fig. S2), and even multipolar spindles emerged (Fig. 4B, white arrow). In the cells depleted of Aurora B, GFP-Nlp was unable to localize in the midzone between daughter cells but aggregated into large assemblies and dispersed widely into cytoplasm, and none of the cells could organize the midbody apparatus. All these results suggest that recruitment of Nlp to the midbody by Aurora B is necessary for cytokinesis.
The biological functions of Nlp in centrosome maturation and spindle formation have been reported to be regulated by several important protein kinases such as Plk1, Nek2, and Cdc2 (25, 34). In this study, we have demonstrated that Aurora B is able to phosphorylate Nlp and regulate its function in cytokinesis. Through kinase assays and other biochemical approaches, we have mapped three specific phosphorylation sites at Ser-185, Ser-448, and Ser-585 in Nlp. Significantly, the phosphorylations of Nlp at the three sites by Aurora B may have two distinct functions (Fig. 7B). First, phosphorylations at Ser-448 and Ser-585 might be required for interaction of Nlp with Aurora B. Mutations at these two sites disrupt the localization of Nlp at the midbody and result in impaired cytokinesis (Fig. 4H). Second, phosphorylation at Ser-185 of Nlp is of considerable importance, not only stabilizing the recruitment of Nlp signal by Aurora B but also ensuring no premature destruction of Nlp before completion of cytokinesis. Nlp has recently been characterized as a short life protein, and its degradation is mediated by the ubiquitin-proteasome pathway (15). In this study, we demonstrate that phosphorylation on Ser-185 affects ubiquitination of Nlp and stabilizes the protein and in turn ensures its function in cytokinesis. Thus, Nlp functions as a novel substrate of Aurora B and mediates its role in the control of cytokinesis.
Nlp expression is cell cycle-dependent (15), and its expression is dynamic in mitosis and cytokinesis. A delicate level of Nlp is required for the “healthy” mitotic progression. Deregulated expression of Nlp is closely associated with chromosomal instability and malignant transformation (28). The findings in this study (Fig. 1, C–E) further confirm that depletion of Nlp function results in multiple nuclei and aneuploidy. Recently, we have found that there are abnormal expressions and gene amplifications of NLP (KIAA0980) in clinical human cancers, and overexpression of Nlp induces anchorage-independent growth and tumorigenicity in both cell and animal models. We have also observed centrosome amplification and spontaneous tumors in Nlp transgenic mice (31). Thus, deregulated levels of Nlp would result in genomic instability and tumorigenesis. In conclusion, we have demonstrated that the centrosomal protein Nlp interacts with Aurora B and is phosphorylated by Aurora B, especially during cytokinesis. Prominently, Aurora B regulation of Nlp is required for cytokinesis and plays a role in maintaining chromosome stability. These observations provide novel insights into understanding the molecular mechanism(s) involved in the control of cell cycle progression and cell division.
Supplementary Material
Acknowledgment
We thank Nagase at KAZUSA DNA Research Institute (Japan) for providing KIAA0980/Nlp cDNA clone.
This work was supported by 973 National Key Fundamental Research Program of China Grant 2009CB521801 and National Natural Science Foundation of China Grants 30730046 and 30721001.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- INCENP
- inner centromere protein
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Scholey J. M., Brust-Mascher I., Mogilner A. (2003) Nature 422, 746–752 [DOI] [PubMed] [Google Scholar]
- 2.D'Avino P. P., Savoian M. S., Glover D. M. (2005) J. Cell Sci. 118, 1549–1558 [DOI] [PubMed] [Google Scholar]
- 3.Jiang W., Jimenez G., Wells N. J., Hope T. J., Wahl G. M., Hunter T., Fukunaga R. (1998) Mol. Cell 2, 877–885 [DOI] [PubMed] [Google Scholar]
- 4.Zhu C., Bossy-Wetzel E., Jiang W. (2005) Biochem. J. 389, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pohl C., Jentsch S. (2008) Cell 132, 832–845 [DOI] [PubMed] [Google Scholar]
- 6.Guertin D. A., Trautmann S., McCollum D. (2002) Microbiol. Mol. Biol. Rev. 66, 155–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glotzer M. (2005) Science 307, 1735–1739 [DOI] [PubMed] [Google Scholar]
- 8.Vader G., Medema R. H., Lens S. M. (2006) J. Cell Biol. 173, 833–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vernos I. (2004) Dev. Cell 7, 145–146 [DOI] [PubMed] [Google Scholar]
- 10.Meraldi P., Honda R., Nigg E. A. (2004) Curr. Opin. Genet. Dev. 14, 29–36 [DOI] [PubMed] [Google Scholar]
- 11.Minoshima Y., Kawashima T., Hirose K., Tonozuka Y., Kawajiri A., Bao Y. C., Deng X., Tatsuka M., Narumiya S., May W. S., Jr., Nosaka T., Semba K., Inoue T., Satoh T., Inagaki M., Kitamura T. (2003) Dev. Cell 4, 549–560 [DOI] [PubMed] [Google Scholar]
- 12.Goto H., Yasui Y., Kawajiri A., Nigg E. A., Terada Y., Tatsuka M., Nagata K., Inagaki M. (2003) J. Biol. Chem. 278, 8526–8530 [DOI] [PubMed] [Google Scholar]
- 13.Casenghi M., Meraldi P., Weinhart U., Duncan P. I., Körner R., Nigg E. A. (2003) Dev. Cell 5, 113–125 [DOI] [PubMed] [Google Scholar]
- 14.Casenghi M., Barr F. A., Nigg E. A. (2005) J. Cell Sci. 118, 5101–5108 [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., Zhan Q. (2007) J. Biol. Chem. 282, 17712–17719 [DOI] [PubMed] [Google Scholar]
- 16.Girdler F., Gascoigne K. E., Eyers P. A., Hartmuth S., Crafter C., Foote K. M., Keen N. J., Taylor S. S. (2006) J. Cell Sci. 119, 3664–3675 [DOI] [PubMed] [Google Scholar]
- 17.Jefford C. E., Irminger-Finger I. (2006) Crit. Rev. Oncol. Hematol. 59, 1–14 [DOI] [PubMed] [Google Scholar]
- 18.Storchova Z., Pellman D. (2004) Nat. Rev. Mol. Cell Biol. 5, 45–54 [DOI] [PubMed] [Google Scholar]
- 19.King R. W. (2008) Biochim. Biophys. Acta 1786, 4–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu H. B., Li Z., Palacios M. J., Li Q., Joshi H. C. (1995) J. Cell Sci. 108, 2955–2962 [DOI] [PubMed] [Google Scholar]
- 21.Julian M., Tollon Y., Lajoie-Mazenc I., Moisand A., Mazarguil H., Puget A., Wright M. (1993) J. Cell Sci. 105, 145–156 [DOI] [PubMed] [Google Scholar]
- 22.Piel M., Nordburg J., Euteneuer U., Burnens M. (2001) Science 291, 1550–1553 [DOI] [PubMed] [Google Scholar]
- 23.Giet R., Glover D. M. (2001) J. Cell Biol. 152, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murata-Hori M., Wang Y. L. (2002) J. Cell Biol. 159, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapley J., Baxter J. E., Blot J., Wattam S. L., Casenghi M., Meraldi P., Nigg E. A., Fry A. M. (2005) Mol. Cell. Biol. 25, 1309–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji J., Liu R., Tong T., Song Y., Jin S., Wu M., Zhan Q. (2007) Oncogene 26, 6396–6405 [DOI] [PubMed] [Google Scholar]
- 27.Burkhard P., Stetefeld J., Strelkov S. V. (2001) Trends Cell Biol. 11, 82–88 [DOI] [PubMed] [Google Scholar]
- 28.Jin S., Gao H., Mazzacurati L., Wang Y., Fan W., Chen Q., Yu W., Wang M., Zhu X., Zhang C., Zhan Q. (2009) J. Biol. Chem. 284, 22970–22977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terada Y. (2001) Cell Struct. Funct. 26, 653–657 [DOI] [PubMed] [Google Scholar]
- 30.Eggert U. S., Mitchison T. J., Field C. M. (2006) Annu. Rev. Biochem. 75, 543–566 [DOI] [PubMed] [Google Scholar]
- 31.Shao S., Liu R., Wang Y., Song Y., Zuo L., Xue L., Lu N., Hou N., Wang M., Yang X., Zhan Q. (2010) J. Clin. Invest. 120, 498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu D., Qu H., Fu M., Zhao X., Liu R., Sui L., Zhan Q. (2008) Gynecol. Oncol. 110, 230–236 [DOI] [PubMed] [Google Scholar]
- 33.Yu L., Song Y., Zhang Q., Zhan Q. (2009) Oncol Rep 22, 789–798 [DOI] [PubMed] [Google Scholar]
- 34.Zhao X., Jin S., Song M., Zhan Q. (2010) Cancer Biol. Ther., in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.