Abstract
Following the mutation screening of genes known to cause amyotrophic lateral sclerosis (ALS) in index cases from 107 familial ALS (FALS) kindred, a point mutation was identified in vesicle-associated membrane protein-associated protein B (VAPB), or VAMP-associated protein B, causing an amino acid change from threonine to isoleucine at codon 46 (T46I) in one FALS case but not in 257 controls. This is an important finding because it is only the second mutation identified in this gene that causes ALS. In order to investigate the pathogenic effects of this mutation, we have used a motor neuron cell line and tissue-specific expression of the mutant protein in Drosophila. We provide substantial evidence for the pathogenic effects of this mutation in abolishing the effect of wild type VAPB in the unfolded protein response, promoting ubiquitin aggregate formation, and activating neuronal cell death. We also report that expression of the mutant protein in the Drosophila motor system induces aggregate deposition, endoplasmic reticulum disorganization, and chaperone up-regulation both in neurons and in muscles. Our integrated analysis of the pathogenic effect of the T46I mutation and the previously identified P56S mutation indicate extensive commonalities in the disease mechanism for these two mutations. In summary, we show that this newly identified mutation in human FALS has a pathogenic effect, supporting and reinforcing the role of VAPB as a causative gene of ALS.
Keywords: Amyotropic Lateral Sclerosis (Lou Gehrig's Disease), Apoptosis, Drosophila Genetics, Endoplasmic Reticulum (ER), Ubiquitination
Introduction
Amyotrophic lateral sclerosis (ALS)2 is a common degenerative disease, causing muscle weakness and atrophy, paralysis, and bulbar symptoms, which eventually leads to death due to respiratory failure within 3–5 years of symptom onset. ALS is caused by loss of motor neurons, and a characteristic pathological feature of the disease is the presence of ubiquitinated inclusions containing TDP-43. The mechanism whereby the disease specifically affects motor neurons is unclear, and current therapy is limited in effectiveness. Major advances have occurred recently with the identification of mutations causing the familial form of ALS (FALS), which is clinically indistinguishable from the sporadic form (SALS), accounts for ∼5% of cases, and shares the same pathological features (1). Mutations in seven genes are known to give rise to FALS, superoxide dismutase (SOD1), vesicle-associated membrane protein-associated protein B (VAPB), senataxin, alsin, dynactin, TARDBP, and FUS, which has shed light on the importance of oxidative stress, the unfolded protein response (UPR), protein aggregation, and abnormal RNA processing in the pathogenesis of ALS (2–5). Multiple mutations have been described for all of these genes with the exception of VAPB, where only one missense mutation (P56S) has been reported to date, being present in seven Brazilian kindred exhibiting clinically diverse phenotypes, including ALS and late onset spinal muscular atrophy (6, 7). No other VAPB mutations have been identified in SALS or other ALS populations (8, 9) until now, where we report a second VAPB mutation and further provide evidence for its pathogenic effects.
VAPB is a member of the VAP family, which includes VAPA, an established regulator of vesicle trafficking (10–12). VAPB plays a role in the UPR, which is activated when misfolded proteins accumulate in the endoplasmic reticulum (ER) (13, 14). VAPs are also known to control lipid metabolism and lipid transfer between the ER and other organelles by interacting with proteins such as oxysterol-binding proteins (OSBPs), Nir proteins, ceramide transfer protein (CERT), and Opi1 (15–19). In Drosophila, DVAP-33A (DVAP) controls the number and size of boutons at the neuromuscular junction. Furthermore, loss of DVAP disrupts the synaptic microtubule cytoskeleton (20), causing an increase in miniature junctional potential amplitudes and postsynaptic glutamate receptor clustering (21).
In this study, we have demonstrated pathogenic actions of this novel VAPB mutation, promoting the formation of ubiquitinated aggregates, activating cell death pathways, and leading to the generation of a robust motor phenotype in vivo in larval neurons, adult eye, and neuromuscular junction of Drosophila. We have further developed a model of disease through the integration of our findings with those already reported for the P56S-VAPB mutation.
EXPERIMENTAL PROCEDURES
Sequencing
A cohort of 185 FALS kindred was used for this analysis. Patients and families were referred by general practitioners and physicians, and the diagnosis of ALS was carried out according to the El Escorial World Federation of Neurology criteria for diagnosis of ALS with evidence of both upper and lower motor neuron involvement. The mean age at death of patients lacking SOD1 mutations was 55.6 years (confidence interval (CI), 54.3–56.9 years; n = 389) and mean disease duration was 3.3 years (CI, 2.7–3.9 years; n = 193) (22). In our study, we used 110 FALS cases from this cohort, where DNA was available, in search of additional mutations in VAPB. The sequencing reaction was carried out with a total of 50 ng of DNA and amplified using primers listed in supplemental Table S1. PCR products were purified using a Nucleospin extraction kit (Clontech) and eluted in 20 μl of molecular grade water (Sigma). The purified PCR products were sequenced using a forward or reverse primer and the ABI Prism™ BigDye™ terminator kit. DNA sequencing was carried out using an ABI 377 automatic sequencer.
Molecular Cloning of Wild Type and Mutant VAPBs into GFP and RFP Vectors
To generate the full-length VAPB fragment, the PCR was performed using 5′ and 3′ VAPB primers (GCCAAAGGTGCTCCGCCGC and CTACAAGGCAATCTTCCC) with a VAPB template purchased from OriGene. This fragment was cloned into pcDNA3.1/NT-GFP-TOPO (Invitrogen) via TA cloning, which produced a N-terminal GFP fusion protein. For generating the RFP fusion VAPB, additional restriction enzyme cutting sequences of EcoRI (GAATTC) and BamHI (GGATCC) were placed at the 5′-end of VAPB 5′ and 3′ primers, respectively. The PCR product was cloned into pCR2 (Invitrogen) via TA cloning and then subcloned into pDsRed2-C1 (Clontech) via EcoRI/BamHI. To introduce the mutations, a traditional two-step mutagenesis PCR was performed as described in the supplemental material. The sequences of all PCR products were verified by sequencing.
Transfection and Immunocytochemistry
COS-7, N2a, and NSC-34 cells were cultured in DMEM containing 10% FBS and were seeded the day before transfection. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. To establish VAPB-stable expression cell lines, NSC-34 cells were transfected with either wild type (WT) or T46I-VAPB and were selected in 10% FBS containing DMEM plus 400 μg/ml G418 (Invitrogen).
For western blotting, cells were lysed with radioimmune precipitation assay buffer, except for the solubility analysis, which was carried out as described by Kanekura et al. (13). Protein concentration was determined using the Bio-Rad protein assay kit. Proteins were separated with 10% SDS-PAGE, and blotting was carried out using conditions specified for the antibodies. For immunofluorescence studies, cells were fixed in 4% paraformaldehyde for 30 min at room temperature and permeabilized for 30 min in 0.1% Triton X-100 in PBS. Nuclei were stained with 0.5 μg/ml DAPI in PBS for 10 min. PBS washes were applied between reagent changes. After the final staining and wash, coverslips were mounted onto slides using Fluorsave reagent (Calbiochem) and left to dry at 4 °C overnight.
Antibodies for Immunocytochemistry or Immunofluorescence
Antibodies used were rabbit polyclonal anti-GFP (1:3000; Abcam), rabbit polyclonal anti-ubiquitin (1:200; Dako), rabbit polyclonal anti-phospho-eIF2α (1:1000; Cell Signaling), anti-β-actin (1:25000; Sigma), peroxidase-labeled goat anti-rabbit IgG (1:1000; Vector), and Alexa Fluor 594 goat anti-rabbit IgG (1:2000; Invitrogen).
Cell Death Analysis
NSC-34 cells were trypsinized, washed with ice-cold PBS twice, and stained using the annexin V-phycoerythrin apoptotic detection kit (BD Biosciences) according to the manufacturer's protocol. The cells were then analyzed with a fluorescence-activated cell sorting (FACS) flow cytometer (BD Biosciences).
VAPB SNP Typing
DNA was amplified using 20 ng of genomic DNA, 1.5 mm MgCl2, 0.5 μm forward and reverse primers, 1× PCR buffer, and 1.25 units of Platinum Taq (Invitrogen). PCR was performed using 94 °C for DNA denaturing, 58 °C for annealing, and 72 °C for extension. Restriction enzymes producing different cutting patterns according to SNP status were used to genotype each SNP. Different size fragments were then separated by electrophoresis using a 1.5–2.2% agarose gel. The primer sequences and enzyme used for each SNP are listed in supplemental Table S2. DNA was extracted from the aqueous phase of TRIzol homogenates (23) as described in the supplemental material.
Fly Genetics and Molecular Techniques
Flies were raised on standard medium at 25 °C. To assess the effect of T48I mutation in neurons and muscles, UAST48I-DVAP/UAST48I-DVAP transgenic males were crossed to elav-GAL4 (24) and BG57-GAL4 (25) females, respectively. For temperature pulses, to induce a higher expression of the GAL4 protein, flies of the relevant genotypes were left to mate at room temperature for 24 h, and then the parents were removed, and the progeny was shifted to 30 °C in a water bath. Site-directed mutagenesis on DVAP cDNA was performed using the QuikChange site-directed mutagenesis kit (Stratagene). Transgenic UAS lines were made by cloning the DVAP cDNA carrying the T48I mutation into the PUAST vector (26). Constructs were injected into the embryonic germ cells, and transgenic lines were established following standard procedures.
Cell Transfections and Immunocytochemistry
Full-length T48I-DVAP and DVAP ORF constructs were generated by PCR and cloned into Myc- and FLAG-tagged pCMV expression vectors, respectively. COS-7 cells were cultured in DMEM containing 10% FCS and 1% penicillin/streptomycin. COS-7 cells were seeded on poly-d-lysine-coated glass coverslips (BD BioCoat) at a concentration of 150,000 cells/ml. The day after, cells were transfected with transfection reagent (Roche Applied Science) according to the manufacturer's instructions. 24 h later, cells were fixed in 4% paraformaldehyde for 20 min and blocked in phosphate-buffered saline containing 0.1% Triton-x-100 containing 10% normal goat serum. Staining with the relevant antibodies was performed according to the procedure described under “Immunocytochemistry.” Rabbit anti-Myc (Sigma) and mouse anti-FLAG (Sigma) primary antibodies were used at a concentration of 1:500 and 1:200, respectively. Secondary antibodies (Jackson ImmunoResearch) were all used at a concentration of 1:500.
Scanning Electron Microscopy and Eye Histology
Eyes were fixed in 3% glutaraldehyde in 0.1 m sodium cacodylate buffer, pH 7.3, for 2–3 h and further processed and analyzed as described in the supplemental material.
Microscopy and Morphological Quantification
To acquire the two-dimensional images for processing, the bodies were photographed using a digital camera attached to an Olympus stereomicroscope and processed as described in the supplemental material. Quantification of the eye phenotype was performed using the Oculus version 1.0 software. Oculus version 1.0 is a tool written in MATLAB, which was developed in house to aid measurement of the size of the adult fly eyes. Unlike the shape selection tools provided in image processing software, such as ImageJ (available at the National Institutes of Health Web site; developed by Wayne Rasband), the Oculus version 1.0 software presented each image at a zoomed out level and permitted the border of the eye to be traced pixel-by-pixel; this method encouraged detailed examination of the boundary and therefore increased the precision of the delineation. Subsequent automated validation operations ensured that the highlighted path was closed and skeletonized (27–29), so the outline could be saved to a data base. The area of each eye was then measured using a flood-fill operation in these enclosed regions, and the number of pixels counted was inserted into a spreadsheet for further analysis.
Immunohistochemistry
Body wall muscles from wondering third instar larvae were dissected and stained according to Chai et al. (21). The following primary antibodies were used: rabbit polyclonal anti-HRP at 1:200 (Jackson ImmunoResearch), guinea pig polyclonal anti-DVAP at 1:1000 (20), mouse monoclonal anti-KDEL at 1:50 (Stressgene), mouse monoclonal anti-Hsp70 at 1:200 (Affinity Bioreagents), rabbit polyclonal anti-SERCA antibody at 1:1000, and guinea pig polyclonal anti-Boca antibody at 1:1000. For the co-staining with the anti-Boca antibody, a rat polyclonal DVAP antibody was used at 1:1000 (20). Secondary antibodies (Jackson ImmunoResearch) were all used at a concentration of 1:500. Fixed and labeled larvae were imaged with a Zeiss LSM510 confocal microscope with a 63/1.4 objective. Single high resolution images were acquired as stacks of images with separate channels and processed with the LSM software or Adobe Photoshop and Adobe Illustrator.
Statistics
A one-way ANOVA test was used to compare multiple conditions in an experiment. Bonferroni's multiple comparison test was used as a post-test when significance was found in the ANOVA test. In the fly studies, statistical analysis was performed using the two-tailed Student's t test to compare the test genotype to controls. Error bars represent S.E. Single SNP allele frequency and genotype frequency p values were obtained using the Fisher exact test (see the Quantitative Skills Web site). The EH program was used to test linkage disequilibrium between SNPs and to compare the difference of pairwise haplotype frequency between groups. The six-SNP haplotype was constructed from the population genotyping data by the PHASE program (version 2.1), which also gave a p value of the haplotype frequency difference between groups (30, 31). BIMBAM was used to evaluate the association between the genotypes and the phenotype (32), which, in our case, were SNP haplotype and VAPB expression level. The Mantel-Haenszel common odds ratio was obtained from the SPSS statistics package to evaluate the association of overall major/minor allele with VAPB expression level. The pathogenicity of the T46I mutation was evaluated using the Web program PMut (33), which is based on neural networks that have been trained with a large data base of neutral and pathogenic human mutations, using predictions of secondary structure and conservation of sequence (see the Molecular Modelling and Bioinformatics Group Web site).
RESULTS
Identification of a New ALS-associated Mutation in VAPB, T46I-VAPB
All six coding exons of VAPB plus flanking intronic regions were sequenced in 107 index cases of FALS kindred of United Kingdom origin that lacked mutations in SOD1, TDP-43, and FUS. A point mutation causing an amino acid change from threonine to isoleucine at codon 46 (T46I) was identified in one FALS case presenting with typical ALS but not in 257 healthy controls (Fig. 1A). The index case with the amino acid substitution in VAPB was diagnosed with classical ALS. Signs at onset, revealed on first examination at the age of 73 years, included wasting of the small muscles of the right hand. There were fasciculations in the left vastus medialis and both gastrocnemii. Plantars were flexor. Sensation was normal, whereas speech and swallowing difficulties developed at the age of 75 years. The diagnosis was confirmed by nerve conduction studies. The patient had a brother with ALS who died within 4 months of diagnosis from bronchopneumonia. There were no further DNA samples available from this family and the patient had no children. DNA was not available from any other affected members of this family, and this mutation was not found in any of the other FALS cases screened or in 257 control subjects (Caucasian, which matched the index case).
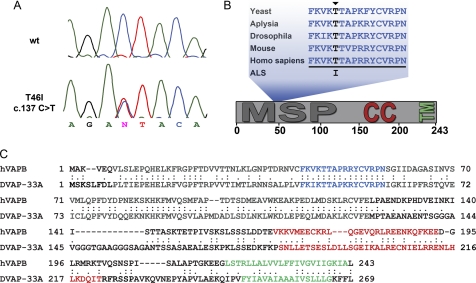
FIGURE 1.
Identification of T46I mutation in a FALS family. A, the gene screening identified a point mutation T46I in VAPB. B, Thr46, which is indicated by an arrowhead, is within the 16-amino acid fragment that is required for VAPB function in yeast, which is highlighted, and is highly conserved in VAP proteins from yeast to humans. C, sequences of human VAPB (hVAPB) protein and its Drosophila ortholog DVAP were aligned by using the ClustalW version 1.82 alignment program available from EMBL-EBI. Colon, an identity match; period, a conserved substitution according to the GONNET 250 matrix. The predicted functional domains in both proteins are preserved: a transmembrane domain (green) at the C terminus, a coil-coiled domain (red) in the middle, and a domain (gray) at the N terminus showing a significant homology to the nematode major sperm protein (MSP).
Although it was not possible to show transmission of the mutation with disease due to the lack of DNA samples from other affected individuals within the family, this mutation is located within a stretch of 16 amino acids that are highly conserved across species (Fig. 1, B and C), which also harbors the P56S mutation that causes ALS8. In support of the importance of this evolutionarily conserved amino acid, the theoretical pathogenicity of the human VAPB sequence variants predicted using the program PMut also suggested that T46I was a pathogenic mutation. The pathogenicity estimate for the T46I coding change is 0.77 (range is from 0 to 1, where values >0.5 indicate pathogenicity), and the confidence index is 5 (where 0 is a low and 9 is a very high level of confidence). By comparison, the theoretical pathogenicity of the P56S sequence variant, found in the Brazilian kindreds, is 0.62 with a confidence index of 2. Furthermore, extensive sequencing of VAPB has failed to reveal this mutation in previously screened control populations (8, 34) or the control subjects that we sequenced in this study. We have therefore carried out a range of functional studies in cell culture and in vivo to elucidate the potential pathological effects of this mutation that it shares with the known ALS-associated mutation P56S and to further elucidate novel aspects of their mechanisms of disease.
Cellular Distribution of T46I and the Formation of Insoluble Aggregates
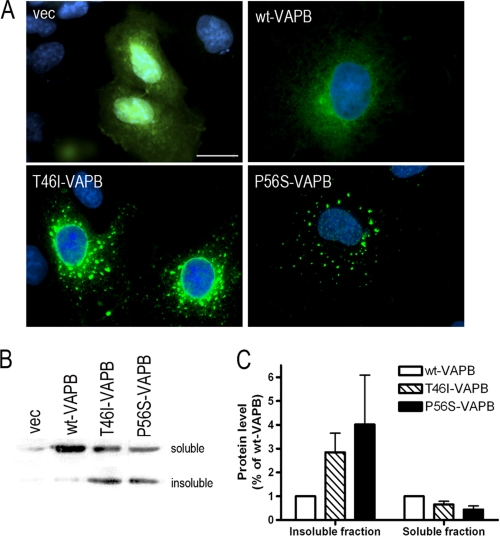
Because studies of the ALS-causing mutation, P56S-VAPB, in cell culture have shown that the mutant protein forms insoluble cytosolic aggregates and is able to interact with wild type VAPB and VAPA (13, 14, 35), affecting the function of the WT protein, we tested whether the T46I mutation affected the subcellular distribution and solubility of the protein. GFP-tagged wild type VAPB (WT-VAPB), T46I-VAPB, and P56S-VAPB constructs were generated and expressed in COS-7 cells. The typical ER localization pattern of a weblike distribution within the cytosol extending from the nuclear surface was shown for the WT-VAPB construct, whereas both T46I-VAPB and P56S-VAPB constructs produced VAPB protein aggregates within cells (Fig. 2A), as reported previously for P56S-VAPB (7, 13, 35). However, there was greater variance in the distribution of T46I-VAPB, which showed different degrees of aggregation ranging from small granules to large coalesced structures with some cells showing a normal ER distribution. The same distribution patterns were found following transfection with these constructs in two neuronal cell lines, the neuroblastoma cell line N2a and the spinal cord motor neuron cell line NSC-34 (supplemental Fig. S1). This marked effect on aggregation was also clearly demonstrated by a shift in distribution from the Triton X-100-soluble fraction of cell lysates to the insoluble fraction (Fig. 2, B and C) compared with WT-VAPB, as previously shown for P56S (13). Although T46I-VAPB showed a different impact on VAPB protein distribution when compared with the P56S mutation, both mutations affected protein solubility to a similar extent (Fig. 2, B and C). This result further supported the observation that both T46I and P56S mutations triggered VAPB protein aggregation.
FIGURE 2.
Protein properties are modified by T46I. A, COS-7 cells were transfected with the indicated plasmids and fixed 48 h after transfection. The distribution of VAPB protein is indicated by GFP (green), and nuclei are shown with DAPI staining (blue). Scale bar, 20 μm. B, NSC-34 cells were harvested 48 h after transfection with empty GFP vector or GFP VAPBs. The separation of Triton X-100-soluble and insoluble fraction was carried out as described by Kanekura et al. (13). The fractions were blotted with anti-GFP antibody. The densitometry measurements of bands for soluble and insoluble VAPB are shown in C. Values are means ± S.E. (error bars) for three separate experiments.
T46I-VAPB Is Unable to Activate the IRE1 Pathway
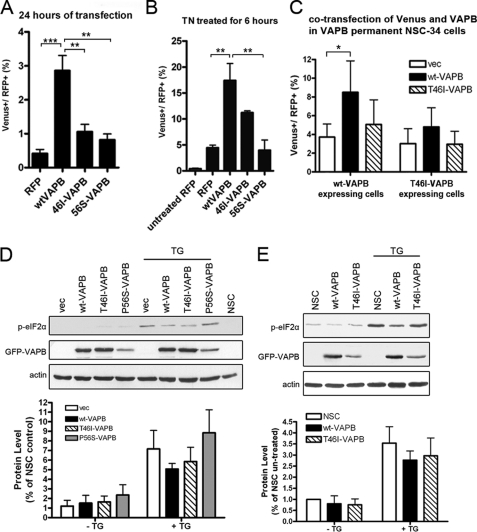
Although VAPB is highly localized to the ER, its actions in ER function are less well characterized. However, a well documented function of VAPB is its ability to potentiate the IRE1 pathway, a component of the UPR, which is disrupted by the P56S-VAPB mutation (13, 14). In order to characterize the role of VAPB in the IRE1/XBP1 pathway, an XBP1-Venus construct was used as a reporter of IRE1/XBP1 activation (36). Under normal conditions, the mRNA for the transcription factor, XBP1, contains a premature stop codon and thus produces an immature protein with a short half-life. When cells are under ER stress, the activated IRE1 causes the splicing of XBP1 mRNA into a mature mRNA, which in turn produces the functional transcription factor. Based on the nature of this alternative RNA splicing, Iwawaki et al. (36) developed an ER stress reporter by fusing a fluorescent protein, Venus, to XBP1, which can only be expressed when the IRE1-mediated splicing takes place. Thus, the presence of the fluorescence indicates the activation of IRE1.
RFP-tagged WT-VAPB, T46I-VAPB, and P56S-VAPB constructs were generated and co-transfected with the XBP1-Venus reporter into NSC-34 cells. The presence of the reporter in RFP-positive cells was quantified for each construct using flow cytometry. Further, the effect of WT and mutant VAPBs on the response to the ER stress inducer, tunicamycin, which is known to inhibit N-linked glycosylation, was also investigated.
WT-VAPB showed a clear induction of IRE1 activity, increasing 9-fold above the vector control after 24-h transfection, an effect that was enhanced in the presence of tunicamycin (ANOVA, p = 0.001 and 0.0035, respectively; Fig. 3, A and B). This property was significantly impaired in both T46I- and P56S-VAPB-expressing cells (p < 0.001; Fig. 3A). The inhibitory effect of T46I was stable, yielding a reduced activation (50%) at 48 h after transfection, compared with WT-VAPB (p < 0.05). Upon tunicamycin treatment, a partial response was seen with T46I-VAPB, but this was not significant.
FIGURE 3.
VAPB participates in the UPR. NSC-34 cells co-transfected with XBP1-Venus and RFP VAPB without (A) or with tunicamycin treatment (2 μg/ml, treated for 6 h before harvested) (B). RFP- and Venus-positive cells were quantified by FACS. The proportions as a percentage of Venus-positive cells in RFP-expressing cells are shown. C, cells stably expressing wild type or T46I-VAPB were transfected with Venus and RFP constructs. 24 h after transfection, cells were harvested and analyzed as in A and B. The p values from one-way ANOVA are 0.001 (A), 0.0035 (B), and 0.0417 (C), respectively. Bonferroni's multiple comparison test was applied as a post-test, and the significance values are shown in the graphs. D and E, the level of phosphorylated eIF2α was examined in transiently transfected cells (D) and in VAPB-stable expression cell lines (E) with or without thapsigargin treatment (500 nm for 30 min before harvest). After correcting with the loading control, actin, the level of phosphorylated eIF2α was quantified. All experiments were repeated at least three times. Values are means ± S.E. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
In a second series of experiments, we used NSC-34 permanent cell lines stably expressing WT-VAPB or T46I-VAPB protein transfected with a RFP-VAPB expression construct and XBP1-Venus to determine whether mutant VAPB interfered with the ability of WT-VAPB to activate this pathway. Although the expression of WT-VAPB in this stable cell line substantially induced IRE1/XPB1 activation, transfection with WT-VAPB significantly increased the magnitude of Venus induction (Fig. 3C). However, in T46I-VAPB-stable cells, transfection with WT-VAPB was not able to induce IRE1/XBP1 activation (Fig. 3C), which indicated that endogenous T46I-VAPB may interact with and prevent exogenously expressed WT-VAPB from promoting IRE1/XBP1 activation. Taken together, these results suggested that T46I-VAPB not only lost the ability to activate IRE1 pathway efficiently but also was able to inhibit the action of the wild type protein.
The activation of another ER stress sensor present in the ER membrane, PERK, was also investigated. This pathway is involved in the regulation of translational suppression and the induction of certain ER chaperone proteins, which are mediated in part through the phosphorylation of eIF2α (37). We monitored the phosphorylation of eIF2α in response to the fast acting ER stress inducer thapsigargin, which acts by increasing cytosolic calcium concentration through inhibiting intracellular calcium pumps (supplemental Fig. S2), and compared the response of VAPB-expressing cells with those of untransfected and vector-transfected cells (Fig. 3, D and E). However, phosphorylation of eIF2α was not significantly modified in the presence of either wild type or mutant VAPBs, using either transiently transfected or permanent cell lines (Fig. 3, D and E). Taken together, these data show that VAPB protein activates the unfolded protein response by acting through the IRE1/XBP1 pathway and that the VAPB protein containing the newly identified T46I mutation lacks this property.
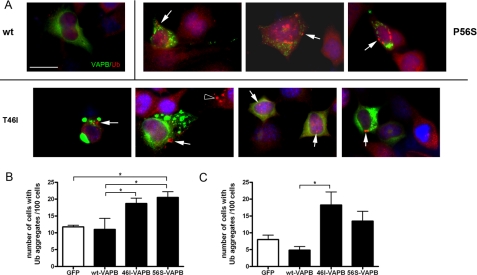
Ubiquitin Aggregation Is Triggered by Mutant VAPB
The presence of ubiquitin-positive aggregates is a widely used marker of the progression of neurodegenerative diseases, which occurs in ALS and the G93A-SOD1 mouse model of ALS. To investigate whether the expression of mutant VAPB triggered the formation of ubiquitin-positive aggregates, NSC-34 cells were transiently transfected with GFP-VAPB constructs and immunostained for endogenous ubiquitin, and cells containing ubiquitin-positive aggregates were quantified for each genotype. Ubiquitinated aggregates were abundant in both T46I- and P56S-VAPB-expressing cells compared with WT-VAPB-expressing cells, whereas GFP-transfected cells contained few or no aggregates (Fig. 4A). A similar increase in the proportion of cells expressing either T46I or P56S-VAPB compared with WT-VAPB was observed (ANOVA, p = 0.0047) (Fig. 4B). The number of ubiquitin-positive cells expressing WT-VAPB was similar to that seen in the vector control (Fig. 4B).
FIGURE 4.
Mutant VAPB induces ubiquitin aggregate formation in both mutant protein-expressing and neighboring cells. A, 3 days after transfection with either vector or GFP-VAPBs constructs, NSC-34 cells were fixed and stained for endogenous ubiquitin. Examples of ubiquitin aggregates are indicated with arrows in each section. Ubiquitin aggregates found in GFP-negative cells are also pointed out with open arrowhead. The number of GFP-positive cells (B) and GFP-negative cells (C) containing ubiquitin aggregates was counted for three independent transfections. The p values from one-way ANOVA test are 0.0047 and 0.022, respectively. Bonferroni's multiple comparison tests were applied as a post-test, and the significance is shown in the graphs. Values are means ± S.E. (error bars). *, p < 0.05; **, p < 0.01.
Previous studies of mutant SOD1 have suggested that a non-cell-autonomous effect occurs, which may be a consequence of secretion of the mutant protein (38, 39), and in order to determine whether this is relevant to VAPB, which is known to be secreted in a truncated form in Drosophila (21, 40, 41), we also analyzed the effect of VAPB-expressing cells on the formation of ubiquitinated aggregates in the neighboring GFP-negative cells. Indeed, we found a significant effect due to T46I-VAPB (ANOVA, p = 0.022) (Fig. 4C), which suggests that mutant VAPB-expressing cells secrete a factor that can influence these neighboring cells. A similar non-significant trend was seen for P56S-VAPB-expressing cells (Fig. 4C). No VAPB protein was detected in the culture medium from the mutant transient transfections (data not shown), in agreement with a previous study (40). Although non-cell-autonomous toxicity appears to be relevant to a number of CNS diseases, whether secreted mutant VAPB is involved in ALS pathogenesis remains to be elucidated.
We further investigated whether the expression of exogenous GFP-VAPBs affected the distribution of endogenous VAPB in untransfected neighboring cells using a VAPB antibody provided by Dr. Lev (15). Expression of endogenous VAPB was detected in NSC-34 cells although present at a much lower level than overexpressed GFP-labeled VAPBs. However, we could detect no evidence that mutant VAPB expressed in the neighboring cells affected the distribution of endogenous VAPB in untransfected cells (supplemental Fig. S3).
In summary, these data show that T46I-VAP induces the formation of ubiquitinated aggregates. Surprisingly, a significant increase in the ubiquitin-positive clusters was observed in untransfected neighboring cells, suggesting a possible non-cell-autonomous effect of the mutant protein-expressing cells on surrounding cells.
In order to investigate whether the accumulation of cellular ubiquitinated aggregates was caused by impaired proteasome activity, we measured the effect of the T46I-VAPB mutation on the activity of the ubiquitin-proteasome system (UPS) in VAPB-expressing cells. Using the UPS reporter GFP-CL1, which leads to an accumulation of GFP in cells with UPS impairment (42), our preliminary results indicate that proteasome activity is impaired in cells expressing T46I-VAPB, whether transiently transfected or permanent cell lines (supplemental Fig. S4).
The Expression of T46I-VAPB Leads to Cell Death
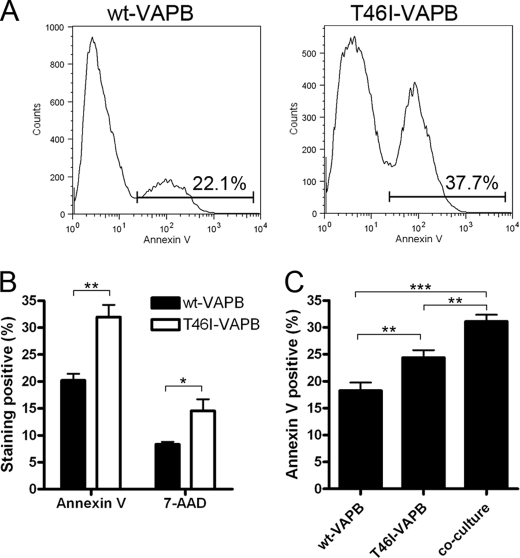
Although the UPR initially plays a cytoprotective role, when the stress is prolonged, a switch from this prosurvival program to a proapoptotic program occurs in which a number of proapoptotic proteins, such as caspase-12 (rodents) and caspase-4 (humans), are activated, leading to cell death (37). We therefore investigated whether the presence of mutant VAPB proteins activated apoptosis by measuring the appearance of both early and late apoptotic markers. We first looked at the presence of annexin V on the cell membrane, an early apoptotic marker, using permanent cell lines. A marked increase in the number of annexin V-positive cells expressing T46I-VAPB compared with cells expressing the WT protein was found, as shown by the flow cytometry profiles (Fig. 5A). When quantified, this increase was highly significant (Fig. 5B). We also examined the appearance of 7-AAD, a late apoptotic marker. In this case as well, we observed that the expression of the mutant protein was accompanied by a significant increase in the number of 7-AAD-positive cells when compared with cells expressing WT-VAPB (Fig. 5B, p = 0.0045). T46I-VAPB-induced apoptosis was further shown to be caspase-dependent because the treatment of benzyloxycarbonyl-VAD efficiently reduced mutant VAPB-induced cell death (supplemental Fig. S5). In contrast, inhibition of caspase-12 has no effect (supplemental Fig. S5). Caspase-12, which is activated downstream of the IRE1/XBP1 pathway, is believed to be involved in ER stress-induced apoptosis (43). Because mutant VAPB is unable to activate IRE1/XBP1, it is not surprising that mutant VAPB-induced apoptosis is caspase-12-independent.
FIGURE 5.
The expression of T46I-VAPB leads to a greater level of cell death. NSC-34 cells that stably express either wild type or T46I-VAPB were harvested for annexin V and 7-AAD staining after 3 days of culture. A, a representative experiment demonstrating the use of FACS to quantify the annexin V-positive cells. The gating of annexin V-positive cells and the percentage the gated cells are displayed. B, the annexin V- and 7-AAD-positive cells from four experiments were quantified. The p values from unpaired t test are 0.0038 and 0.0315 for annexin V and 7-AAD, respectively. C, a total of 3,000 cells were plated in each condition, whereas equal amounts of wild type- and T46I-VAPB-expressing cells were seeded in the co-culture. Cells were harvested and stained for annexin V 3 days after plating. Values are means ± S.E. (error bars) for three separate experiments. The p value from the one-way ANOVA test is 0.0004, and Bonferroni's multiple comparison test was applied as a post-test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
In order to assess the effect of the mutant protein on WT-VAPB, a co-culture was set up, in which the permanent cell line expressing WT-VAPB was co-cultured with the same number of cells expressing T46I-VAPB. The total number of cells in the co-culture was the same as that in the single cultures (total of 3,000 cells in each condition). Although only half of the cells in the co-culture expressed T46I-VAPB, a significantly increased number of annexin V-positive cells were detected (ANOVA; p = 0.0004; Fig. 5C). This suggested that T46I-VAPB interacted with WT-VAPB, promoting apoptosis.
Transgenic Expression of the Mutant Protein in Drosophila Larval Neurons Induces the Formation of Aggregates
In order to gain further insight into the effect of the T46I-VAPB mutation in ALS pathogenesis, we turned to Drosophila, an organism proven to provide a model for VAP-induced ALS (21, 40, 41). The threonine at position 46 in humans corresponds to threonine at position 48 in Drosophila VAP (T48I-DVAP) (Fig. 1C). DVAP cDNA containing the T48I mutation was generated by site-directed mutagenesis and subcloned into Drosophila transformation vectors (UAST48I-DVAP). Several independent transgenic lines were established, and the mutant transgenes were expressed by using the UAS/GAL4 system (26). In Drosophila, the UAS/GAL4 system allows the temporal and tissue-specific expression of a transgene by using a variety of GAL4 drivers (26). We targeted the expression of UAST48I-DVAP in larval neurons by using the pan-neural driver elav-GAL4 (24).
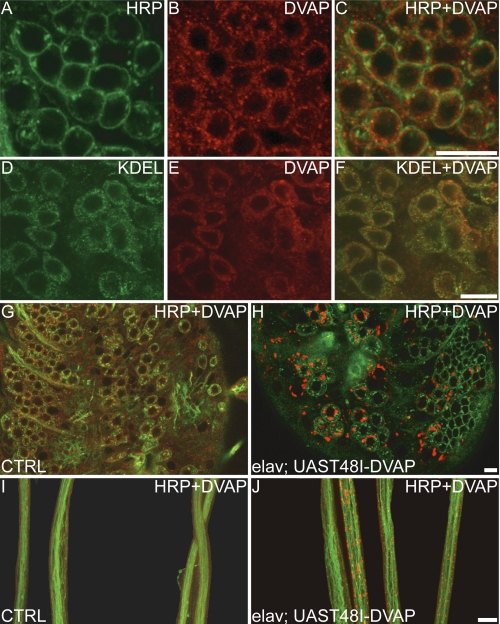
Because VAPB aggregates have been clearly demonstrated for this mutation in cell culture (see above) and for the P56S mutation in flies (21), we assessed whether the accumulation of inclusions containing the protein causing the disease is also common to T48I-DVAP-induced pathology. We performed confocal analysis on larval brains and nerve fibers of T48I-DVAP transgenic larvae stained with antibodies specific for DVAP. In WT flies, DVAP is distributed underneath the cell membrane and in the cytoplasm of neuronal cell bodies (Fig. 6, A–C), where an extensive co-localization with ER-specific markers is observed (Fig. 6, D–F). In contrast, neurons of transgenic larvae expressing T48I-DVAP are filled with brightly fluorescing inclusions of variable sizes (Fig. 6, G and H). In control nerves, VAP immunoreactivity is uniformly distributed, whereas in T48I-DVAP nerves, numerous massive aggregates are visible (Fig. 6, compare I and J). This phenomenon was consistently observed in all transgenic lines examined. Conversely, several lines expressing the WT-DVAP protein at similar or higher levels do not show the formation of aggregates (data not shown) (21). Taken together, these data indicate that transgenic expression of DVAP-T48I in neurons induces the formation of intracellular inclusions brightly immunoreactive for DVAP.
FIGURE 6.
Targeted expression of T48I-DVAP in neurons induces the formation of aggregates immunoreactive for DVAP. A–C, neuronal cell bodies of WT larval brains immunostained with an anti-DVAP antibody (red) and anti-HRP antibody (green), a neuronal cell surface marker. DVAP is located underneath the cell membrane and in the cytoplasm. D–F, immunostaining with an anti-KDEL antibody, an ER-specific marker, shows that the cytoplasmic DVAP immunoreactivity associates with the ER. G–J, brains and nerve fibers of third instar larvae stained with antibodies for DVAP (red) and with antibodies for the neuronal cell surface marker anti-HRP (green). In controls (G), DVAP staining appears dispersed throughout the cytoplasm of neuronal cell bodies, whereas in T48I-DVAP transgenic brains (H), DVAP immunoreactivity is associated with intracellular aggregates of variable size. Nerve fibers of control larvae (I) and transgenic larvae (J) were stained with anti-HRP (green) and anti-DVAP antibodies (red). In control nerves, a faint and uniform staining for DVAP is observed, whereas in transgenic nerves, large aggregates strongly immunoreactive for DVAP accumulate along the nerves. Scale bars, 20 μm (J) and 10 μm (C, F, and H).
Targeted Expression of T48I-DVAP in the Adult Eye Causes Neuronal Degeneration
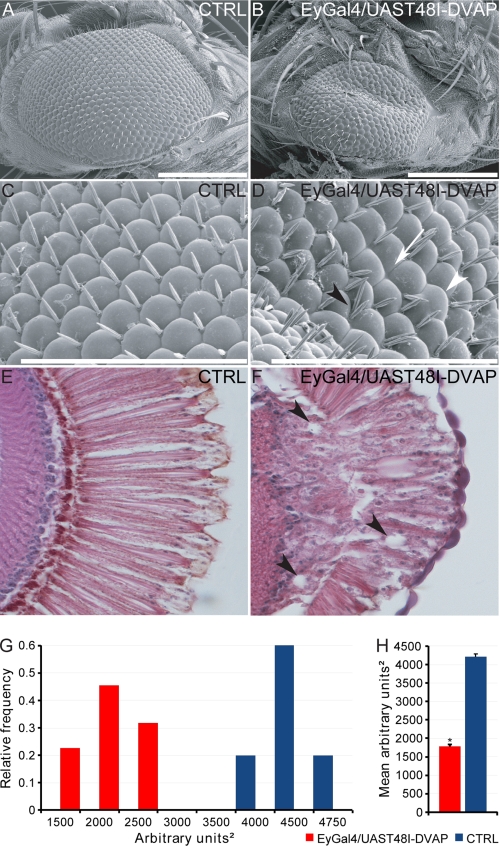
To assess the potential role of T48I-DVAP toxicity in neurons other than motor neurons, we targeted the expression of T48I-DVAP in the nervous system of adult flies. This would allow us to study the progression of the phenotype on the temporal axis because the life span of an adult fly is much longer than that of a larva. When the T48I-DVAP transgene was expressed into the adult brain by using the elav-GAL4 driver, flies of the correct genotype failed to hatch. This early death phenotype is consistent with a severe loss of integrity of the nervous system. We then targeted the expression of T48I-DVAP transgene in the eye using the eyeless-GAL4 driver because adult vision is not required for viability (44, 45). T48I-DVAP transgenic lines exhibited an abnormal eye phenotype upon eclosion. As compared with controls, the eyes expressing the T48I-DVAP transgene were reduced in size and displayed a rough phenotype over the entire surface. Analysis using scanning electron microscopy revealed disorganized and fused ommatidia with missing, irregular, and occasional supernumerary mechano-sensory bristles (Fig. 7, A–D). Paraffin sections of the heads of T48I-DVAP transgenic flies confirmed the loss of cells beneath the external surface of the eyes. Photoreceptor morphology was severely disrupted, and several vacuoles were visible (Fig. 7, E and F). Vacuolization has been observed in several fly models of neurodegenerative diseases, and the appearance of these vacuoles has been correlated to neurodegeneration (46–48).
FIGURE 7.
Transgenic expression of T48I-DVAP in the adult fly eyes causes a severe disruption of eye morphology. A and B, scanning electron micrographs of controls (A) and transgenic adult fly eyes expressing the UAST48I-DVAP transgene under the control of the eye-specific driver, eyeless-GAL4 (B). C and D, higher magnifications of A and B, respectively. Whereas the adult Drosophila eye is composed of an ordered array of ommatidia and interspersed bristles, transgenic eyes appear smaller, with missing or extra bristles and fused ommatidia. Fused ommatidia are indicated by a white arrowhead, whereas a black arrowhead points to ommatidia with extra bristles. The white arrow points to missing bristles. Frontal sections of fly heads of control animals (E) and from flies expressing T48I-DVAP in the eye (F) are shown. Transgenic eyes show a severely disrupted internal morphology, cell degeneration, and numerous vacuoles (see arrowheads, for instance). G, summary of the relative frequency of eye sizes for WT and transgenes. H, quantification of the eye surface area showing that in transgenic flies expressing T48I-DVAP in the eye, the surface area is, on average, 50% smaller than controls. Scale bars, 200 μm (A and B) and 100 μm (C and D). Error bars, S.E.
Another important aspect of the T48I-DVAP-induced eye phenotype seen in adult flies is the fact that the size of the eye exhibits a significant degree of heterogeneity in the severity of the phenotype. We performed a detailed quantification of the eye size in both mutants and controls by using Oculus version 1.0, a MATLAB software recently developed in our laboratory (see “Experimental Procedures” for a detailed description of this software). The distribution of the eye size was measured over a cohort of at least 25 flies carrying the T48I-DVAP transgene. Eye sizes from a similar number of control flies were also measured. The majority of the mutant flies (65%) possessed eye sizes between 1000 and 2000 arbitrary square units, whereas the remaining flies exhibited a larger size ranging between 2000 and 2500 square units. Control flies on the other hand have eye sizes centered between 4000 and 4750 square units (Fig. 7, G and H). Overall, the eye size in T48I-DVAP transgenic flies is less than 50% of the size of controls (Fig. 7H, p < 0.0001).
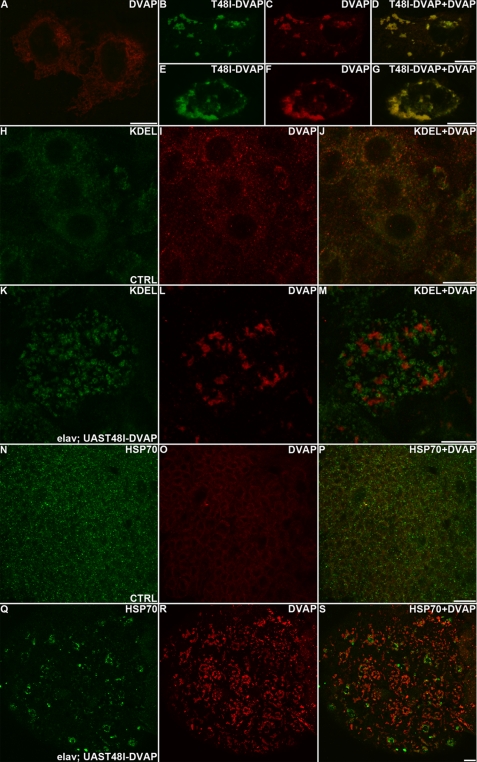
Although the accumulation of aggregates strongly immunoreactive for T48I-DVAP was evident in transgenic animals expressing the mutant protein in neurons (Fig. 6), the pattern of expression of the WT protein was no longer detectable (Fig. 6, compare G with H and compare I with J). To test whether the mutant protein can recruit the WT protein into the aggregates, we co-transfected COS-7 cell lines with plasmids encoding a FLAG-tagged version of DVAP and a MycT48I-DVAP plasmid. The subcellular localization of both proteins was determined by using tag-specific antibodies. In COS7 cells transfected with DVAP, the protein forms a perinuclear reticular staining, as expected for an ER-associated protein (Fig. 8A). When COS-7 cells were cotransfected with both FLAG-DVAP and MycT48I-DVAP, a diffuse punctate staining reminiscent of the aggregates present in cell bodies and motor fibers of T48I-DAVP transgenic larvae, was observed (Fig. 8, B–G). Interestingly, the staining of the WT and the mutant proteins perfectly overlap, indicating that both proteins are components of the intracellular inclusions (Fig. 8, B–G).
FIGURE 8.
Transgenic expression of T48I-DVAP induces formation of aggregates composed of wild type and mutant proteins, ER fragmentation, and Hsp70 up-regulation. A–G, COS7 cells transfected with DVAP-FLAG (A) or FLAG-DVAP and MycT48I-DVAP (B–G) were stained with antibodies specific for the two tags. H–M, brains from control (H–J) and T48I-DVAP transgenic (K–M) larvae were stained for the ER marker, KDEL (green), and DVAP (red). N–S, brains from control (N–P) and T48I-DVAP transgenic (Q–S) larvae were stained for DVAP (red) and Hsp70 (green). Scale bars, 10 μm.
Transgenic Expression of T48I-DVAP in Larval Brains Causes ER Fragmentation and Heat Shock Protein Up-regulation
As reported above for the expression of the mutant protein in motor neuron cell lines, expression of the same protein in Drosophila neurons induces cell degeneration and accumulation of aggregates both in cell bodies and in nerves. We also showed that the accumulation of misfolded proteins in the aggregates triggers a UPR response in cell culture. In Drosophila, the UPR pathway is not well characterized due, at least in part, to the fact that the UPR genes are essential during development (49). We therefore investigated whether other neuropathological features that are usually associated with the UPR response were present in our fly model.
The presence of aggregates in the form of intracellular inclusions containing the disease-causing protein is thought to arise from the unfolding or misfolding of the same protein. Protein folding occurs in the cytoplasm or within the lumen of the ER. In the ER a specialized set of chaperones and components of quality control machinery ensures correct folding. Interestingly, structural abnormalities in the ER characterized by ER fragmentation and Golgi cisternal dilation have been reported in motor neurons of brain stem and spinal cord of patients affected by sporadic ALS (50). To assess whether such a neuropathological feature is also common to T48I-DVAP-induced ALS, we stained T48I-DVAP transgenic brains with an anti-KDEL antibody, a marker specific for ER-resident proteins. In controls, the anti-KDEL staining detects a reticular, perinuclear pattern, as expected for a marker highlighting the ER (Fig. 8, H–J). In transgenic brains, the staining appeared fragmented and dispersed throughout the cell (Fig. 8, K–M). Occasionally, the staining was found to co-localize with the aggregates immunoreactive for DVAP (data not shown). This phenotype was confirmed by using an antibody specific for the Boca protein (51), another independent ER marker (supplemental Fig. S6).
Affected tissues in neurodegenerative diseases often exhibit the induction of a chaperone stress response. Heat shock protein induction has been reported in a mouse model with widespread mutant SOD1 expression (52), and chaperones have been found to be associated with SOD1 inclusions in motor neurons (52–54). Recent data suggest that up-regulation of Hsp70 is beneficial to disease progression in mouse models of ALS (55). Brains of transgenic larvae expressing T48I-DVAP were stained with an anti-Hsp70 antibody. In control brains, Hsp70 staining was faint and spread throughout the cytoplasm of neurons, whereas in the transgenic brains, clusters of specific immunoreactivity were observed. These clusters are closely associated but do not overlap with DVAP aggregates (Fig. 8, N–S). In summary, as already reported for the first P56S-DVAP fly model (21, 40, 41), targeting the expression of T48I-DVAP in neurons induces aggregate formation, cell degeneration, ER fragmentation, and up-regulation of chaperone proteins.
The Skeletal Muscle Is a Direct Target of T48I-DVAP-mediated Toxicity
The advantage of using an intact organism as a model for human diseases is that it allows the study of the effect of a pathogenic mutation in different cell types. Alterations in skeletal muscles during ALS have been considered to result from toxicity of the mutant protein in motor neurons rather than a direct effect of the same protein on muscle cells. In order to explore whether skeletal muscle is a direct target for T48I-DVAP-mediated toxicity, we generated transgenic flies in which the DVAP mutant gene was selectively expressed in skeletal muscles. To this end, the BG57-GAL4 line was used because GAL4 expression in this line is under the control of a muscle-specific enhancer (56). We first assessed the presence of DVAP in larval skeletal muscles and its specific subcellular localization within the muscle. Dissected neuromuscular junctions were stained with a range of muscle-specific markers. As expected from previous studies, we observed DVAP immunoreactivity to be distributed throughout the cytosol of the muscle (35, 57). The staining appears to be concentrated around the nuclei of the muscle syncytium largely co-localizing with SERCA, a calcium ATPase, associated with the sarcoendoplasmic reticulum of the muscle (58) (Fig. 9, A–F).
FIGURE 9.
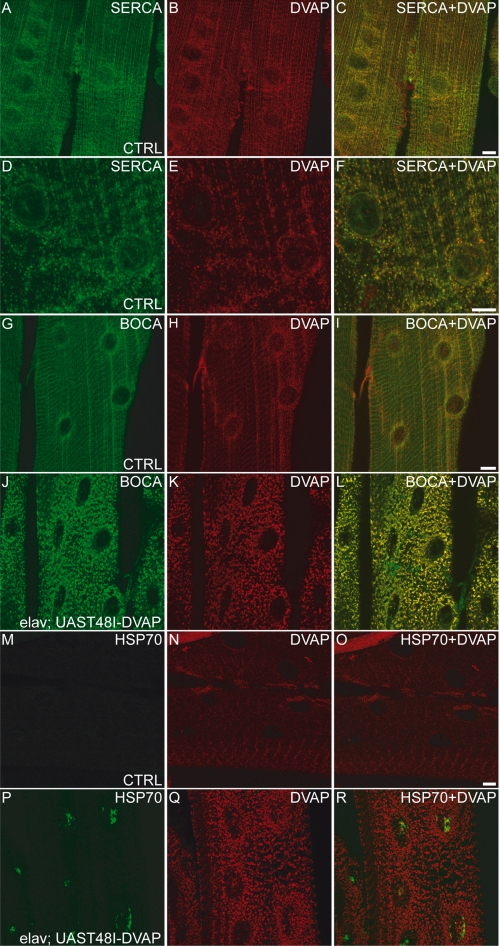
Muscle-specific expression of T48I-DVAP causes cellular stress characterized by aggregate deposition, fragmentation of the sarcotubular system, and Hsp70 up-regulation. A–F, body wall muscle fibers from control larvae were stained for SERCA (green), as ER-sarcoplasmic reticulum marker and DVAP (red). A higher magnification of a smaller area is shown in D–F, demonstrating a high degree of overlap. G–L, control (G–I) and transgenic (J–L) muscles were stained for DVAP (red) and the ER marker, Boca (green). M–R, control (M–O) and transgenic (P–R) muscles were stained for DVAP (red) and the Hsp70 (green). Scale bars, 10 μm.
As already observed for transgenic expression of the mutant allele in neurons, the expression of the same protein in muscles using BG57-GAL4 driver, induced the formation of large aggregates strongly immunoreactive for DVAP (Fig. 9, K and Q). In the same lines, fragmentation of the ER was also observed when the transgenic flies expressing T48I-DVAP specifically in muscle were stained with an antibody specific for Boca, an ER marker in Drosophila (51) (Fig. 9J). In controls, however, anti-Boca staining of muscles exhibited a reticular and perinuclear pattern (Fig. 9G). Interestingly, immunostaining of transgenic muscles with both anti-DVAP and anti-Boca antibodies showed that DVAP-positive aggregates extensively colocalize with the fragmented ER (Fig. 9L). This phenotype was confirmed by using SERCA, another independent ER/sarcoplasmic reticulum marker (data not shown).
In many neurodegenerative diseases, including ALS, aggregates are thought to be formed from the accumulation of misfolded proteins that trigger the up-regulation of chaperone proteins. We assessed whether this was also the case for muscles expressing T48I-DVAP. In controls, Hsp70 protein is very low and almost under the detection limit of our antibody (Fig. 9, M–O). In transgenic animals expressing T48I-DVAP specifically in skeletal muscles, there is an increase in the expression levels of Hsp70 that appears to concentrate in puncta mainly localized to the nucleus (Fig. 9, P–R). Although the phenomenon of relocation of Hsp70s to the nucleus is still poorly understood, it is clear that this relocation is directly linked to cellular stress (59, 60). Similar results were observed when a P58S-DVAP transgene expressing the other DVAP mutation causing ALS8 in humans was targeted to the muscle using the BG57-GAL4 driver (data not shown).
Genotype Correlations with VAPB Expression in SALS
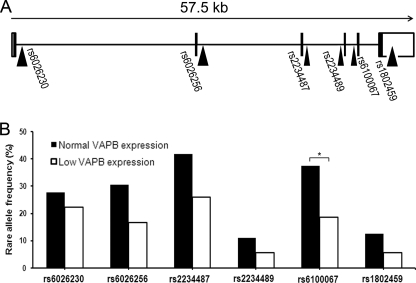
In view of our previous observation that the expression of VAPB is significantly decreased in SALS spinal cord compared with controls (23), we investigated whether VAPB SNP variants were associated with the level of expression of VAPB in spinal cord in this large cohort of cases described previously (23). Because there are no reported functional SNPs in the coding region or splice sites, we chose SNPs that are close to exon boundaries, which might also detect uncharacterized functional variants. The six SNPs selected for this study were rs6026230T/C, rs6026256T/G, rs2234487C/G, rs2234489G/A, rs6100067C/T, and rs1802459A/G, and their locations within the gene are shown in Fig. 10A. A total of 63 samples composed of 21 healthy controls and 42 ALS were genotyped and divided into normal (n = 36) and low (n = 27) expression groups. Normal levels of expression were defined by the 90% CIs of the control group, which contained 19 of 21 of control cases. 60% of the ALS cases were outside this CI. Two groups were formed using this threshold; group 1 contained samples with normal expression (90% CI of control group), and group 2 contained samples with low expression and was composed of 27 samples, including 24 SALS, 1 FALS, and 2 controls.
FIGURE 10.
The rare allele frequency comparison between normal and low VAPB expression groups. A, the positions of the six SNPs within the VAPB gene are shown. The whole VAPB gene, containing six exons (represented with filled boxes), spans to 57.5 kb. Five SNPs closest to each exon and one in the 3′-UTR (represented with an open box) were genotyped. B, the rare allele frequencies of the six SNPs are shown where the normal VAPB expression group is represented as filled bars, and the low VAPB expression group is represented with open bars. The group with normal VAPB expression level is composed of 16 SLAS, 1 FALS, and 19 controls, whereas the group with low VAPB expression level is composed of 24 SALS, 1 FALS, and 2 controls. Fisher exact test was applied for each SNP, and a significant difference of SNP rs6100067 was found (p = 0.029).
A significant difference in allele frequency for SNP rs6100067 was found between normal and low VAPB expression (Fig. 10B) using both the Fisher exact test (p = 0.029) and the BIMBAM program (p = 0.026). The rare allele was more abundant in the group with normal VAPB expression levels compared with the group with low VAPB expression levels, which suggests that this SNP has a beneficial effect preventing decreased VAPB gene expression. Although no significant association was found between expression levels and the other SNPs per se, there was a consistent trend in which the rare allele frequencies of all six SNPs were lower in the low VAPB-expressing samples compared with the normal expressing group (Fig. 10B). Indeed, a significant linkage was found using the Mantel-Haenszel test, which indicates that these six VAPB SNPs show an overall association of major allele with VAPB low expression (p < 0.0001; Mantel-Haenszel common odds ratio = 2.04). The two-SNP haplotype was then constructed and examined by the EH program, and only the rs6026230-rs6100067 haplotype was found to be significantly different between the two expression groups (p = 0.047), where the C-T haplotype was decreased by 14% in the low expression group. The population-attributable risk (61) of disease associated with low expression of VAPB mRNA is 50.79% (p = 5.4 × 10−5), of which the fractional change in VAPB expression attributable to the minor allele rs6100067 is 18.98% (p < 0.02). This equates to an overall protective effect of 9.6% for the minor allele. Despite the fact that this SNP has no known function and is outside regulatory regions involved in RNA splicing, it may be linked to a functional sequence variant that is responsible for the VAPB expression difference.
DISCUSSION
In this study, we report the identification of a novel mutation in VAPB, which is not only the second mutation in VAPB to be found associated with FALS but is the first to be found in a non-Brazilian kindred. Although it was not possible to show transmission of the mutation with disease due to the lack of availability of additional DNA samples within the family and the late onset of disease in the index case, we have obtained extensive evidence for the pathogenicity of the T46I-VAPB mutation based on functional studies in cell culture and in vivo. Calculation of the theoretical pathogenicity of this mutation, which is greater than that of the P56S-VAPB mutation, provides additional support. Furthermore, extensive sequencing of VAPB has failed to reveal this mutation in control populations (8, 34) (this study).
Evidence for the pathogenic effect of this mutation is provided using motor neuron cell lines and motor system-targeted expression in Drosophila. We have shown for the first time that mutant VAPB causes cell death and that this T46I mutant protein in turn affects neighboring cells that do not express the mutant protein. Furthermore, we previously reported a substantial down-regulation of VAPB mRNA in SALS spinal cord (23), and in this study we have shown a SNP association with levels of expression, suggesting that VAPB is widely involved in ALS pathogenesis, which is further borne out by studies in G93A-SOD1 transgenic mice (35).
VAPB and ER Stress
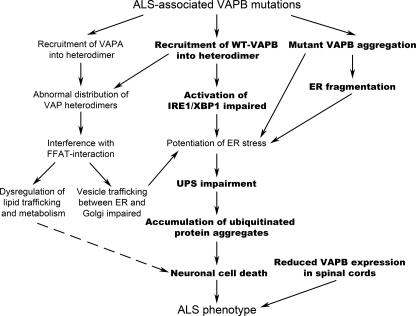
ER stress is a well established feature of ALS (1, 62), and the discovery of ALS-associated mutations in VAPB, a key component of this response, highlights the importance of this essential response in neuronal survival. Early studies showed that the yeast homolog VAP, Scs2, restored inositol auxotrophy IRE1/Hac1 mutant cells (63, 64), which implies a role for VAP in lipid transfer and/or synthesis and sheds light on the cross-talk between VAP and IRE1/XBP1.The latter was also verified in mammalian systems, where VAPB triggers the cleavage of XBP1, an initial signal for IRE1 activation (13, 14). The ALS-associated mutations in VAPB, T46I and P56S, abolish the ability of WT-VAPB to activate IRE1/XBP1, as shown in this study. Furthermore, both P56S-VAPB and T46I-VAPB cause a redistribution of wild type protein, trapping it in VAPB inclusions in mammalian cells and Drosophila (this study) (21, 40), and due to their tendency to form heterodimers with WT-VAPB, they may also prevent WT-VAPB from fulfilling its role during the UPR (13, 14). The results presented here support the hypothesis that affected motor neurons fail to activate a functional UPR to counteract ER stress, which, if prolonged, leads to cell death (Fig. 11).
FIGURE 11.
Cellular disturbances triggered by mutant VAPB. In this study, we demonstrated (highlighted in boldface type) that the expression of ALS-linked mutant VAPB triggers VAPB protein aggregation as well as wild type protein redistribution. As a consequence, structure and function of ER are disrupted, which, together with the inability to activate the unfolded protein-responsive pathway, IRE1/XBP1, leads to potentiation of ER stress, the accumulation of ubiquitinated aggregates, and, eventually, cell death.
The detailed mechanism whereby VAPB participates in UPR is still unclear. There is as yet no evidence showing a direct interaction between VAP and IRE1 or XBP1 (65). However, the key amino acid residues involved in the trafficking of lipid-associated proteins mediated by the FFAT (two phenylalanines in an acidic tract) domain, such as CERT, OSBP, and Nir (15, 17), are contained in a highly conserved 16-amino acid region containing Thr46 (Fig. 1). It has been proposed that this FFAT binding domain coincides with the domain necessary for UPR activation via IRE1/XBP1 (14). Although the P56S mutation would not be expected to affect FFAT binding by itself (66), it still prevents IRE1 activation (13). It was recently demonstrated that P56S obstructs FFAT binding through enhancing VAPB oligomerization and thus decreases accessibility to the VAPB binding domain (67). Therefore, the inability of mutant VAPB to activate IRE1/XBP1 could be a result of the mutations per se or could be the effect of protein conformational changes imparted by the mutation, as is the case for FFAT binding.
This is the first study in which the activation of the PERK pathway has been monitored in VAPB-expressing cells, and no significant effect of VAPB and VAPB mutations was detected on the level of phosphorylated eIF2α. However, we found that high levels of phosphorylated eIF2α could be detected 30 min to 2 h after thapsigargin treatment in NSC-34 cells and thereafter decreased to undetectable levels at 6 h, suggesting that the phosphorylation of eIF2α is a rapid and short term response that is then switched off upon the prolonged stressed condition. Therefore, we cannot exclude the possibility that VAPB mutations may have rapid and transient effects on the PERK pathway.
We have also shown that the expression of T46I-VAPB in NSC-34 cells or T48I-DVAP in flies causes cell death without any further challenge. An earlier report did show that P56S-VAPB reduced cell viability in NSC-34 cells but only after thapsigargin treatment (14). This difference may reflect the fact that we are using permanent expression systems in vitro and in vivo.
Non-cell-autonomous Effect Caused by Mutant VAPB
Emerging evidence indicates that the pathogenesis of many neurodegenerative diseases including ALS, involves a non-cell-autonomous process, but the mechanism mediating the transfer of toxicity between cells is still unclear (1). Interestingly, chromogranin-associated mutant SOD1 is secreted into the extracellular milieu and induces microgliosis and neuronal cell death (38, 39). Here we report that the co-culture of WT- and T46I-VAPB-expressing cells results in much greater cell death compared with T46I alone. This observation, together with the finding that untransfected cells cultured with mutant VAPB transfected cells are more likely to contain ubiquitinated aggregates than controls, suggests that mutant VAPB-expressing cells are able to affect neighboring cells that, although not producing the mutant protein themselves, become stressed and eventually undergo cell death. Recent findings have drawn attention to features of amyloid β and τ pathology in Alzheimer disease that are common to prion disease, whereby an abnormal protein causes propagation of protein misfolding between cells (for a review, see Ref. 68). Such propagation may also occur following transfer from one cell to the next by exocytotic release followed by neuronal uptake, which may also be relevant to non-cell-autonomous processes occurring in ALS.
In Drosophila, the N-terminal MSP domain of DVAP is secreted and may act as a ligand of the Eph receptor, and the P58S mutation in DVAP, equivalent to the human P56S mutation in VAPB, inhibits the ability of the WT protein to be secreted (40, 69). However, because we have been unable to recover any VAPB fragments from the culture medium, it is still undetermined whether the non-cell-autonomous effect is caused by VAPB directly or by the release of other factors triggered by VAPB. It is possible that mutant VAPB is released within exosomes, as has previously been shown for α-synuclein (70), where the authors suggested that this was a potential process involved in Parkinson disease and other neurodegenerative conditions.
Fly Model Expressing Mutant VAPB
The effect of the newly identified mutation on cellular processes known to play a role in neurodegeneration, such as UPR, accumulation of ubiquitinated aggregates, and cell death, were studied in cell lines. In a parallel and complementary approach, we used the Drosophila model system to assess in vivo effects on relevant ALS mechanisms. More specifically, the contributions of different tissues and systems (peripheral motor system, muscle, and central nervous system) to the pathogenic effects induced by the novel mutation were investigated in our Drosophila model.
Previous studies have established Drosophila as a suitable model system to study VAP-induced ALS. In particular, a wealth of experimental evidence indicates that human VAPB and DVAP are functionally interchangeable both in loss-of-function and overexpression paradigms. First, many of the fundamental biochemical properties of human VAPB, including its ability to oligomerize and to interact with lipid-binding proteins, are conserved in DVAP (21, 71). Second, transgenic expression of DVAP carrying the ALS-causing mutation recapitulates many of hallmarks of the human disease (21, 40, 41) (this work). Furthermore, expression of human VAPB in a mutant background for DVAP fully rescues the lethality and the morphological and the electrophysiological phenotypes associated with DVAP loss-of-function mutations (21, 40). Taken together, these results strongly indicate that Drosophila is an excellent model organism to investigate both the WT function of VAP proteins and their role in ALS pathogenesis.
Expression of T48I-DVAP throughout the nervous system in Drosophila is lethal, and it causes premature death. To investigate the pathogenic mechanisms in more detail, we analyzed the effect of transgenic expression of the mutant allele in particular areas of the nervous system. We have shown that expression of T48I-DVP in neurons of the larval motor system leads to accumulation of intracellular aggregates, and targeted expression into the eye causes cell degeneration. Furthermore, expression of the mutant allele depletes the WT protein from its normal localization and recruits it into the aggregates. As already shown for the P56S mutation (e.g. see Ref. 35), this observation is consistent with the hypothesis that the pathogenic allele causes a loss-of-function of WT VAP by a dominant negative mechanism.
Dramatic ER restructuring has been reported in cell culture systems expressing P56S-VAP allele (72). Furthermore structural abnormalities in the ER characterized by ER fragmentation and Golgi cisternal dilation have been reported in motor neurons of patients affected by sporadic ALS (50). To test whether this is also the case for the novel identified mutation, we analyzed the ER structure in larval neurons expressing T48I-DVAP. We found that the ER in these transgenic animals appears fragmented, supporting the hypothesis that structural alterations of the ER are a consistent hallmark of ALS and a possible cause of motor neuron degeneration. Finally, these neuropathological features, together with the up-regulation and accumulation of chaperone proteins in clusters, indicate a gradual overwhelming of the neuronal capacity to properly handle the pathogenic DVAPs and identify cellular processes that are likely to be crucial for ALS pathogenesis.
Heat shock protein induction has been reported in a mouse model with widespread mutant SOD1 expression (52), with chaperones being found co-localized with SOD1 inclusions in motor neurons (52–54). The fact that chaperones do not overlap with DVAP-immunoreactive aggregates is an interesting and distinct feature of fly models for motor neuron diseases when compared with mouse models for the same disease (73). A similar result has been reported for the expression of the pathogenic version of SOD1 in Drosophila, in which a chaperone response was not present in neurons but in a subset of surrounding glial cells (74).
Although the presence of inclusions suggests an overwhelming of the neuronal capacity to properly handle the accumulation of the pathogenic protein, the lack of co-localization of these aggregates with Hsp70 suggests that Hsp70 induction may reflect its cytoprotective effect in regulating other pathways, such as autophagy and apoptosis.
The conventional belief that ALS is exclusively a motor neuron disorder has been recently challenged by the fact that the disease is associated with a broad range of neuropsychological and behavioral deficits (69) and affects non-neuronal tissues, such as muscle, that may exacerbate the disease (1). In support of this concept, we showed that expression of T48I-DVAP in muscle is sufficient to induce the accumulation of large intracellular aggregates of the mutant protein. This would, in turn, trigger a local stress response, as occurs in motor neurons, which is characterized by sarcotubular fragmentation and up-regulation of HSP70. The results of this study show that muscle expression of the mutant protein can induce muscle stress and dysfunction independently from the consequences of motor neuron degeneration.
Effect of T46I-VAP on Lipid Dysfunction in ALS
VAP proteins interact with lipid-binding proteins, such as CERT (17–19, 66), a ceramide transfer protein required for sphingolipid synthesis (75). Interestingly, the Thr46 residue is located in a conserved core of VAP responsible for binding the FFAT motif and is essential for binding CERT (16, 66). Therefore, it is very likely that the T46I mutation affects the ability of VAP to interact with these proteins and as a consequence causes dysfunction in lipid metabolism. Furthermore, flies lacking CERT activity exhibit increased sensitivity to oxidative damage, motor disabilities, and reduced life span (76), and inactivation of CERT in mice causes dilation of the ER, induction of UPR, and impaired mitochondrial activity (77), all of which may play a crucial role in ALS pathogenesis. Indeed, ALS patients carrying the P56S-VAPB mutation exhibit hyperlipidemia and increased cholesterol and triglyceride levels (6), and more recent studies have revealed that ALS patients are more likely to show evidence of hyperlipidemia compared with controls (78). To conclude, data reported here and in earlier studies suggest that a correlation may exist between dysfunction in lipid metabolism and ALS pathogenesis, which warrants further investigation.
In summary, based on the data presented here and from earlier studies, we propose a model for the effects of ALS-associated VAPB mutations and define putative pathogenic pathways (Fig. 11). As we have shown here, mutant VAPB has a profound effect on the IRE1 component of UPR, abolishing the ability of VAPB to activate this pathway and thereby impairing induction of ER chaperones and ERAD activity. This in turn prolongs the effects of ER stress. Accumulation of ubiquitinated protein aggregates reflects this abnormality and promotes cell death. This pathway would be augmented by aggregation of mutant VAPB and ER fragmentation. In addition, recruitment of VAPA and wild type VAPB to mutant VAPB dimers could cause dysregulation of lipid trafficking and further reduce cell viability.
Supplementary Material
Acknowledgments
We are indebted to Ismael Al-Ramahi for advice on the eye section protocol, to Trudy Gillespie for assistance with the confocal images, and to Steven Mitchell for help with scanning electron microscopy analysis. We are also grateful to Joaquim Culi for the gift of the anti-Boca antibody, to Mani Ramaswami for providing the anti-SERCA antibody, and to Professor Miura (University of Tokyo) for kindly providing the XBP1-Venus reporter.
This work was supported by the ALS Association, Motor Neurone Disease Association and Wellcome Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
- ALS
- amyotrophic lateral sclerosis
- FALS
- familial ALS
- SALS
- sporadic ALS
- UPR
- unfolded protein response
- CI
- confidence interval
- ANOVA
- analysis of variance
- UPS
- ubiquitin-proteasome system
- 7-AAD
- 7-aminoactinomycin D
- RFP
- red fluorescent protein.
REFERENCES
- 1.Ilieva H., Polymenidou M., Cleveland D. W. (2009) J. Cell Biol. 187, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dion P. A., Daoud H., Rouleau G. A. (2009) Nat. Rev. Genet. 10, 769–782 [DOI] [PubMed] [Google Scholar]
- 3.Lagier-Tourenne C., Cleveland D. W. (2009) Cell 136, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasinelli P., Brown R. H. (2006) Nat. Rev. Neurosci. 7, 710–723 [DOI] [PubMed] [Google Scholar]
- 5.Maruyama H., Morino H., Ito H., Izumi Y., Kato H., Watanabe Y., Kinoshita Y., Kamada M., Nodera H., Suzuki H., Komure O., Matsuura S., Kobatake K., Morimoto N., Abe K., Suzuki N., Aoki M., Kawata A., Hirai T., Kato T., Ogasawara K., Hirano A., Takumi T., Kusaka H., Hagiwara K., Kaji R., Kawakami H. (2010) Nature 465, 223–226 [DOI] [PubMed] [Google Scholar]
- 6.Marques V. D., Barreira A. A., Davis M. B., Abou-Sleiman P. M., Silva W. A., Jr., Zago M. A., Sobreira C., Fazan V., Marques W., Jr. (2006) Muscle Nerve 34, 731–739 [DOI] [PubMed] [Google Scholar]
- 7.Nishimura A. L., Mitne-Neto M., Silva H. C., Richieri-Costa A., Middleton S., Cascio D., Kok F., Oliveira J. R., Gillingwater T., Webb J., Skehel P., Zatz M. (2004) Am. J. Hum. Genet. 75, 822–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conforti F. L., Sprovieri T., Mazzei R., Ungaro C., Tessitore A., Tedeschi G., Patitucci A., Magariello A., Gabriele A., Labella V., Simone I. L., Majorana G., Monsurrò M. R., Valentino P., Muglia M., Quattrone A. (2006) J. Negat. Results Biomed. 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landers J. E., Leclerc A. L., Shi L., Virkud A., Cho T., Maxwell M. M., Henry A. F., Polak M., Glass J. D., Kwiatkowski T. J., Al-Chalabi A., Shaw C. E., Leigh P. N., Rodriguez-Leyza I., McKenna-Yasek D., Sapp P. C., Brown R. H., Jr. (2008) Neurology 70, 1179–1185 [DOI] [PubMed] [Google Scholar]
- 10.Prosser D. C., Tran D., Gougeon P. Y., Verly C., Ngsee J. K. (2008) J. Cell Sci. 121, 3052–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soussan L., Burakov D., Daniels M. P., Toister-Achituv M., Porat A., Yarden Y., Elazar Z. (1999) J. Cell Biol. 146, 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyles J. P., McMaster C. R., Ridgway N. D. (2002) J. Biol. Chem. 277, 29908–29918 [DOI] [PubMed] [Google Scholar]
- 13.Kanekura K., Nishimoto I., Aiso S., Matsuoka M. (2006) J. Biol. Chem. 281, 30223–30233 [DOI] [PubMed] [Google Scholar]
- 14.Suzuki H., Kanekura K., Levine T. P., Kohno K., Olkkonen V. M., Aiso S., Matsuoka M. (2009) J. Neurochem. 108, 973–985 [DOI] [PubMed] [Google Scholar]
- 15.Amarilio R., Ramachandran S., Sabanay H., Lev S. (2005) J. Biol. Chem. 280, 5934–5944 [DOI] [PubMed] [Google Scholar]
- 16.Kaiser S. E., Brickner J. H., Reilein A. R., Fenn T. D., Walter P., Brunger A. T. (2005) Structure 13, 1035–1045 [DOI] [PubMed] [Google Scholar]
- 17.Kawano M., Kumagai K., Nishijima M., Hanada K. (2006) J. Biol. Chem. 281, 30279–30288 [DOI] [PubMed] [Google Scholar]
- 18.Ngo M., Ridgway N. D. (2009) Mol. Biol. Cell 20, 1388–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., Neefjes J. (2009) J. Cell Biol. 185, 1209–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pennetta G., Hiesinger P. R., Fabian-Fine R., Meinertzhagen I. A., Bellen H. J. (2002) Neuron 35, 291–306 [DOI] [PubMed] [Google Scholar]
- 21.Chai A., Withers J., Koh Y. H., Parry K., Bao H., Zhang B., Budnik V., Pennetta G. (2008) Hum. Mol. Genet. 17, 266–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradley M., Bradley L., de Belleroche J., Orrell R. W. (2005) Neurology 64, 1628–1631 [DOI] [PubMed] [Google Scholar]
- 23.Anagnostou G., Akbar M. T., Paul P., Angelinetta C., Steiner T. J., de Belleroche J. (2010) Neurobiol. Aging 31, 969–985 [DOI] [PubMed] [Google Scholar]
- 24.Yao K. M., White K. (1994) J. Neurochem. 63, 41–51 [DOI] [PubMed] [Google Scholar]
- 25.Budnik V. (1996) Curr. Opin. Neurobiol. 6, 858–867 [DOI] [PubMed] [Google Scholar]
- 26.Brand A. H., Perrimon N. (1993) Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- 27.Haralick R. M., Shapiro L. G. (1992) Computer and Robot Vision, Vol. 1, pp. 168–173, Addison-Wesley, Reading, MA [Google Scholar]
- 28.Kong T. Y., Rosenfeld A. (1996) Topological Algorithms for Digital Image Processing, pp. 99–144, Elsevier, Amsterdam [Google Scholar]
- 29.Pratt W. K. (2007) Digital Image Processing, 4th Ed., pp. 431–441, John Wiley & Sons, Inc., New York [Google Scholar]
- 30.Stephens M., Donnelly P. (2003) Am. J. Hum. Genet. 73, 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens M., Smith N. J., Donnelly P. (2001) Am. J. Hum. Genet. 68, 978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Servin B., Stephens M. (2007) PLoS Genet. 3, e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrer-Costa C., Gelpí J. L., Zamakola L., Parraga I., de la Cruz X., Orozco M. (2005) Bioinformatics 21, 3176–3178 [DOI] [PubMed] [Google Scholar]
- 34.Millecamps S., Salachas F., Cazeneuve C., Gordon P., Bricka B., Camuzat A., Guillot-Noël L., Russaouen O., Bruneteau G., Pradat P. F., Le Forestier N., Vandenberghe N., Danel-Brunaud V., Guy N., Thauvin-Robinet C., Lacomblez L., Couratier P., Hannequin D., Seilhean D., Le Ber I., Corcia P., Camu W., Brice A., Rouleau G., LeGuern E., Meininger V. (2010) J. Med. Genet. 47, 554–560 [DOI] [PubMed] [Google Scholar]
- 35.Teuling E., Ahmed S., Haasdijk E., Demmers J., Steinmetz M. O., Akhmanova A., Jaarsma D., Hoogenraad C. C. (2007) J. Neurosci. 27, 9801–9815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwawaki T., Akai R., Kohno K., Miura M. (2004) Nat. Med. 10, 98–102 [DOI] [PubMed] [Google Scholar]
- 37.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 38.Zhao W., Beers D. R., Henkel J. S., Zhang W., Urushitani M., Julien J. P., Appel S. H. (2010) Glia 58, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urushitani M., Sik A., Sakurai T., Nukina N., Takahashi R., Julien J. P. (2006) Nat. Neurosci. 9, 108–118 [DOI] [PubMed] [Google Scholar]
- 40.Tsuda H., Han S. M., Yang Y., Tong C., Lin Y. Q., Mohan K., Haueter C., Zoghbi A., Harati Y., Kwan J., Miller M. A., Bellen H. J. (2008) Cell 133, 963–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ratnaparkhi A., Lawless G. M., Schweizer F. E., Golshani P., Jackson G. R. (2008) PLoS One 3, e2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bence N. F., Sampat R. M., Kopito R. R. (2001) Science 292, 1552–1555 [DOI] [PubMed] [Google Scholar]
- 43.Bredesen D. E., Rao R. V., Mehlen P. (2006) Nature 443, 796–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halder G., Callaerts P., Gehring W. J. (1995) Science 267, 1788–1792 [DOI] [PubMed] [Google Scholar]
- 45.Hauck B., Gehring W. J., Walldorf U. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 564–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson G. R., Wiedau-Pazos M., Sang T. K., Wagle N., Brown C. A., Massachi S., Geschwind D. H. (2002) Neuron 34, 509–519 [DOI] [PubMed] [Google Scholar]
- 47.Warrick J. M., Paulson H. L., Gray-Board G. L., Bui Q. T., Fischbeck K. H., Pittman R. N., Bonini N. M. (1998) Cell 93, 939–949 [DOI] [PubMed] [Google Scholar]
- 48.Wittmann C. W., Wszolek M. F., Shulman J. M., Salvaterra P. M., Lewis J., Hutton M., Feany M. B. (2001) Science 293, 711–714 [DOI] [PubMed] [Google Scholar]
- 49.Ryoo H. D., Steller H. (2007) Cell Cycle 6, 830–835 [DOI] [PubMed] [Google Scholar]
- 50.Oyanagi K., Yamazaki M., Takahashi H., Watabe K., Wada M., Komori T., Morita T., Mizutani T. (2008) Neuropathol. Appl. Neurobiol. 34, 650–658 [DOI] [PubMed] [Google Scholar]
- 51.Culi J., Mann R. S. (2003) Cell 112, 343–354 [DOI] [PubMed] [Google Scholar]
- 52.Liu J., Shinobu L. A., Ward C. M., Young D., Cleveland D. W. (2005) J. Neurochem. 93, 875–882 [DOI] [PubMed] [Google Scholar]
- 53.Shinder G. A., Lacourse M. C., Minotti S., Durham H. D. (2001) J. Biol. Chem. 276, 12791–12796 [DOI] [PubMed] [Google Scholar]
- 54.Watanabe M., Dykes-Hoberg M., Culotta V. C., Price D. L., Wong P. C., Rothstein J. D. (2001) Neurobiol. Dis. 8, 933–941 [DOI] [PubMed] [Google Scholar]
- 55.Gifondorwa D. J., Robinson M. B., Hayes C. D., Taylor A. R., Prevette D. M., Oppenheim R. W., Caress J., Milligan C. E. (2007) J. Neurosci. 27, 13173–13180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Budnik V., Koh Y. H., Guan B., Hartmann B., Hough C., Woods D., Gorczyca M. (1996) Neuron 17, 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gkogkas C., Middleton S., Kremer A. M., Wardrope C., Hannah M., Gillingwater T. H., Skehel P. (2008) Hum. Mol. Genet. 17, 1517–1526 [DOI] [PubMed] [Google Scholar]
- 58.Sanyal S., Consoulas C., Kuromi H., Basole A., Mukai L., Kidokoro Y., Krishnan K. S., Ramaswami M. (2005) Genetics 169, 737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welch W. J., Feramisco J. R. (1984) J. Biol. Chem. 259, 4501–4513 [PubMed] [Google Scholar]
- 60.Zeng X. C., Bhasin S., Wu X., Lee J. G., Maffi S., Nichols C. J., Lee K. J., Taylor J. P., Greene L. E., Eisenberg E. (2004) J. Cell Sci. 117, 4991–5000 [DOI] [PubMed] [Google Scholar]
- 61.Fleiss J. L., Levin B. A., Paik M. C. (2003) Statistical Methods for Rates and Proportions, 3rd Ed., John Wiley & Sons, Inc., New York [Google Scholar]
- 62.Atkin J. D., Farg M. A., Walker A. K., McLean C., Tomas D., Horne M. K. (2008) Neurobiol. Dis. 30, 400–407 [DOI] [PubMed] [Google Scholar]
- 63.Kagiwada S., Hosaka K., Murata M., Nikawa J., Takatsuki A. (1998) J. Bacteriol. 180, 1700–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nikawa J., Murakami A., Esumi E., Hosaka K. (1995) J. Biochem. 118, 39–45 [DOI] [PubMed] [Google Scholar]
- 65.Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 66.Loewen C. J., Levine T. P. (2005) J. Biol. Chem. 280, 14097–14104 [DOI] [PubMed] [Google Scholar]
- 67.Kim S., Leal S. S., Ben Halevy D., Gomes C. M., Lev S. (2010) J. Biol. Chem. 285, 13839–13849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frost B., Diamond M. I. (2010) Nat. Rev. Neurosci. 11, 155–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strong M. J. (2008) Amyotroph. Lateral Scler. 9, 323–338 [DOI] [PubMed] [Google Scholar]
- 70.Emmanouilidou E., Melachroinou K., Roumeliotis T., Garbis S. D., Ntzouni M., Margaritis L. H., Stefanis L., Vekrellis K. (2010) J. Neurosci. 30, 6838–6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., Vijayadamodar G., Pochart P., Machineni H., Welsh M., Kong Y., Zerhusen B., Malcolm R., Varrone Z., Collis A., Minto M., Burgess S., McDaniel L., Stimpson E., Spriggs F., Williams J., Neurath K., Ioime N., Agee M., Voss E., Furtak K., Renzulli R., Aanensen N., Carrolla S., Bickelhaupt E., Lazovatsky Y., DaSilva A., Zhong J., Stanyon C. A., Finley R. L., Jr., White K. P., Braverman M., Jarvie T., Gold S., Leach M., Knight J., Shimkets R. A., McKenna M. P., Chant J., Rothberg J. M. (2003) Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 72.Fasana E., Fossati M., Ruggiano A., Brambillasca S., Hoogenraad C. C., Navone F., Francolini M., Borgese N. (2010) FASEB J. 24, 1419–1430 [DOI] [PubMed] [Google Scholar]
- 73.Muchowski P. J., Wacker J. L. (2005) Nat. Rev. Neurosci. 6, 11–22 [DOI] [PubMed] [Google Scholar]
- 74.Watson M. R., Lagow R. D., Xu K., Zhang B., Bonini N. M. (2008) J. Biol. Chem. 283, 24972–24981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Acharya U., Acharya J. K. (2005) Cell Mol. Life Sci. 62, 128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rao R. P., Yuan C., Allegood J. C., Rawat S. S., Edwards M. B., Wang X., Merrill A. H., Jr., Acharya U., Acharya J. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X., Rao R. P., Kosakowska-Cholody T., Masood M. A., Southon E., Zhang H., Berthet C., Nagashim K., Veenstra T. K., Tessarollo L., Acharya U., Acharya J. K. (2009) J. Cell Biol. 184, 143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dupuis L., Corcia P., Fergani A., Gonzalez De Aguilar J. L., Bonnefont-Rousselot D., Bittar R., Seilhean D., Hauw J. J., Lacomblez L., Loeffler J. P., Meininger V. (2008) Neurology 70, 1004–1009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.