Abstract
The recent discovery of induced pluripotent stem cell (iPSC) technology provides an invaluable tool for creating in vitro representations of human genetic conditions. This is particularly relevant for those diseases that lack adequate animal models or where the species comparison is difficult, e.g. imprinting diseases such as the neurogenetic disorder Prader-Willi syndrome (PWS). However, recent reports have unveiled transcriptional and functional differences between iPSCs and embryonic stem cells that in cases are attributable to imprinting errors. This has suggested that human iPSCs may not be useful to model genetic imprinting diseases. Here, we describe the generation of iPSCs from a patient with PWS bearing a partial translocation of the paternally expressed chromosome 15q11-q13 region to chromosome 4. The resulting iPSCs match all standard criteria of bona fide reprogramming and could be readily differentiated into tissues derived from the three germ layers, including neurons. Moreover, these iPSCs retain a high level of DNA methylation in the imprinting center of the maternal allele and show concomitant reduced expression of the disease-associated small nucleolar RNA HBII-85/SNORD116. These results indicate that iPSCs may be a useful tool to study PWS and perhaps other genetic imprinting diseases as well.
Keywords: Embryonic Stem Cell, Epigenetics, Gene Regulation, Genetic Diseases, Neurological Diseases
Introduction
Diploid organisms like mammals have two copies of each chromosome; and therefore, gene expression is the result of two combined alleles. However, it is estimated that at least 1% of human genes are expressed in monoallelic fashion because of repressive inherited modifications in DNA regions near the maternal or parental gene of interest, which is known as imprinting (1, 2). Imprinting modifications frequently consist of changes in the DNA methylation pattern of CpG islands inside the promoter of affected genes, but histone modifications are important as well (3). Among other well known imprinting genes are IGF2, H19, and CDKN1C, and the number of newly identified ones is increasing slowly but steadily (4). A difficulty for the latter is that many imprinting genes are expressed in a tissue-specific manner. Remarkably, gene imprinting is present not only throughout mammals but has also been described in other species such as plants (5), which gives an idea of its evolutionary importance.
During gametogenesis, all imprinting marks are deleted and reset again according to the sex of the donor (6). However, because of further global methylation changes after fertilization, some imprinting marks are only properly established during early embryonic development. As for the physiological significance, the observation that many known imprinting genes are important for proper embryonic growth has suggested that imprinting arose with the objective of controlling the parental influence to promote fetal growth and the maternal tendency to save resources (2). Supporting this idea, parthenogenic embryos of several species, which are produced without need of oocyte fertilization, mostly fail to develop because of lack of imprinting (7). In addition, because of its epigenetic nature and the delicate balance between mother and fetus, imprinting is particularly susceptible to environmental variables during development such as changes in nutrition or oxygenation (8, 9). As a consequence, it is believed that many human diseases have a developmental defect basis determined by accumulated imprinting abnormalities (10). Alterations in imprinting are also observed and thought to play a role during cancer progression (11).
A number of inherited conditions are related to imprinting as well. Among these, Prader-Willi syndrome (PWS)4 and Angelman syndrome were the first identified human imprinting diseases and have ever since been a reference in the field and a paradigm for poorly understood clinical conditions (12, 13). Other imprinting genetic diseases are Beckwith-Wiedemann syndrome and Silver-Russell syndrome, for example (14). PWS is a neurological disorder characterized by neonatal hypotonia, failure to thrive, hypogonadism and short stature, mild-to-moderate mental retardation, and compulsive hyperphagia in early childhood that leads to morbid obesity (12). These symptoms are a consequence of the lack of expression of genes contained in paternal chromosome region 15q11-q13, as the same region in the maternal chromosome is repressed by means of DNA methylation. Conversely, Angelman syndrome is caused by the lack of maternal expression of the UBE3A gene, also located in chromosome 15 (12, 15). Most PWS patients (∼70%) bear large deletions of the paternal 15q11-q13 region that happen during meiosis, but ∼25% have inherited both copies of chromosome 15 from their mother, which is called maternal uniparental disomy (12). The rest (∼5%) are attributed to other errors during spermatogenesis or early development. Notably, several genes contribute to PWS, but the absence of the small nucleolar RNA (snoRNA) HBII-85, also termed SNORD116, may be instrumental (16). However, the role of this snoRNA in regulating cell function is unknown. Although mouse models recapitulate several aspects of the human disease, they fail to reproduce others; and thus, available information on the molecular basis of PWS is limited (12). Besides, the species cross-comparison is always difficult for anything related to alterations in behavior. Another source of study, neurons of affected individuals, can only be accessed post-mortem, as neural stem cells from PWS patients that undergo brain surgery are a rarity (17). In this work, we describe the successful generation of induced pluripotent stem cells (iPSCs) from a patient with PWS and provide evidence that they may represent a valuable alternative model for studying PWS.
EXPERIMENTAL PROCEDURES
Cell Culture and iPSC Generation
H1 and H9 embryonic stem cells (ESCs) were purchased from the Wisconsin International Stem Cell Bank. Fibroblasts from a patient with PWS were obtained from the Coriell Cell Repositories (catalog ID GM21889) and cultured in DMEM (HyClone) containing penicillin/streptomycin, l-glutamine, and 10% (HyClone) before retroviral transduction. 40,000 cells were transduced twice in a 6-well culture dish using pMX-based retroviruses (Addgene) as described previously (18). Transduced cells were maintained in high-glucose DMEM containing 20% defined FBS (HyClone), nonessential amino acids, penicillin/streptomycin, l-glutamine, β-mercaptoethanol, and basic FGF (bFGF; Invitrogen). At day 6 post-infection, they were split in the same medium on 10-cm dishes coated with feeders (mitomycin C-treated mouse fibroblasts). Vitamin C (sodium l-ascorbate, 50 μg/ml; Sigma) (19) and valproic acid (1 mm; Merck) (20) were added from day 7; vitamin C was maintained all the time, but valproic acid was removed at day 14. iPSC clones were initially cultured on feeders using knock-out serum replacement medium (DMEM/F-12 (HyClone), 20% knock-out serum replacement (Invitrogen), nonessential amino acids, penicillin/streptomycin, l-glutamine, β-mercaptoethanol, and bFGF) and later expanded the same way or on Matrigel (BD Biosciences) using mTeSR1 medium (STEMCELL Technologies); the same was applied to human ESCs. During the first passages, iPSC colonies were expanded manually, and those secondary colonies with abnormal morphology were routinely removed by scratching, after which they were routinely passaged with Dispase (Invitrogen). Donor umbilical cord matrix mesenchymal cells (UMCs) and iPSC clone C6 have been reported by us previously (18); normal fibroblasts were extracted from a skin biopsy of a middle-aged Asian individual and reprogrammed similarly to PWS fibroblasts.
Fluorescence in Situ Hybridization
FISH was performed using a PWS region probe (SNRPN) and a chromosome 15 control probe (15qter) purchased from Cytocell according to the manufacturer's instructions. SNRPN hybridization was detected with a red fluorophore (Texas Red), and 15qter hybridization was detected with a green fluorophore (FITC) provided by the manufacturer. Slides were prepared as for normal karyotyping, baked at 56 °C for 2 h, fixed in 10% buffered formalin, and denatured (in a solution containing 7 parts formamide, 1 part standard saline citrate, and 2 parts water, pH 7.0–7.5) for 5 min. After stepwise dehydration with 70, 85, and 100% ethanol, they were hybridized overnight at 37 °C. An Olympus BX51 microscope was used for detection (also for immunofluorescence microscopy; see below).
iPSC Characterization
Alkaline phosphatase staining, immunofluorescence microscopy, transgene integration, karyotyping, and bisulfite sequencing (using nested PCR) were performed as described (18, 19). Nanog antibodies were purchased from R&D Systems, SSEA-3 and SSEA-4 from Abcam, and TRA-1–60 and TRA-1–81 from Millipore. DNA was extracted using the Wizard® genomic DNA purification kit (Promega), and total RNA was extracted using TRIzol (Invitrogen). Quantitative real-time RT-PCR (qPCR) was performed using a Thermal Cycler DiceTM real-time system and SYBR Green Premix EX TaqTM (Takara). β-Actin was used for normalization, and all items were measured in triplicate. qPCR primers for MAGEL2 were 5′-attccaaggtggaggcacag (forward) and 5′-tctccgagcgctggacag (reverse). Primers for SNORD116 were purchased from Guangzhou RiboBio. Primers for bisulfite sequencing of the PWS imprinting center have been reported previously (21): 5′-tccaaaacaaaaaactttaaaacccaaattc (forward 1), 5′-aggtttttttttattgtaatagtgttgtgggg (reverse 1), 5′-tcaatactccaaatcctaaaaacttaaaatatc (forward 2), and 5′-tgtggggttttaggggtttagtagttttttttttttagg (reverse 2). The degree of C-T change after bisulfite conversion was routinely 95%. Other primer sequences for qPCR, semiquantitative PCR primers, and OCT4 and NANOG promoter DNA methylation primers have been described previously (18, 19). Single tandem repeat sequencing was performed using the PowerPlex® 16 system (Promega). Embryoid body (EB) differentiation was performed by detaching iPSCs growing on feeders using Dispase and culturing them in suspension on non-adherent 25-cm2 T-flasks (Corning) in knock-out serum replacement medium without bFGF for several days. EBs were seeded on Matrigel-coated dishes for another few days before RNA was extracted. For teratomas, iPSCs grown on feeders were collected using Dispase, and the cells were resuspended with 30% Matrigel and then injected subcutaneously and intramuscularly into the flanks of SCID mice. Tumors were sectioned after 7 weeks and stained with hematoxylin/eosin.
Neural Differentiation
iPSC colonies were detached from feeder layers and grown in suspension in knock-out serum replacement medium without bFGF for 4 days. Cell aggregates were then treated with neural induction medium consisting of DMEM/F-12, N2 supplement (Invitrogen), heparin (2 mg/ml), and nonessential amino acids for another 2 days. On day 6, iPSC aggregates were adhered to a Matrigel-coated plate. On day 10–14, cells in the center of each colony had differentiated into neuroectodermal cells displaying small columnar morphology, followed by organization of the columnar cells into neural tube-like rosettes. These neuroectodermal cells were isolated from surrounding non-neural cells by manual picking onto a new 24-well plate and cultured for 4 days in N2 medium with bFGF (20 ng/ml) for neuronal induction or for 3 weeks in N2 medium with 0.1% FBS (PAA) for astrocyte differentiation. Polyclonal antibodies against βIII-tubulin were purchased from Covance, MAP2 (microtubule-associated protein 2) from Millipore, and glial fibrillary acidic protein from Sigma.
RESULTS
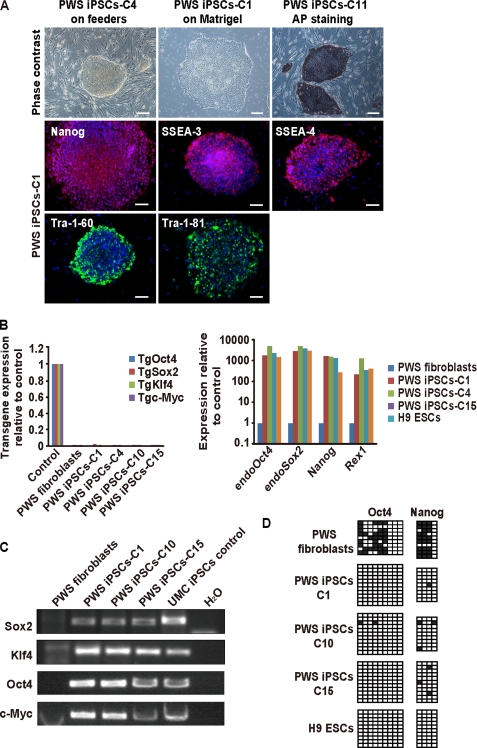
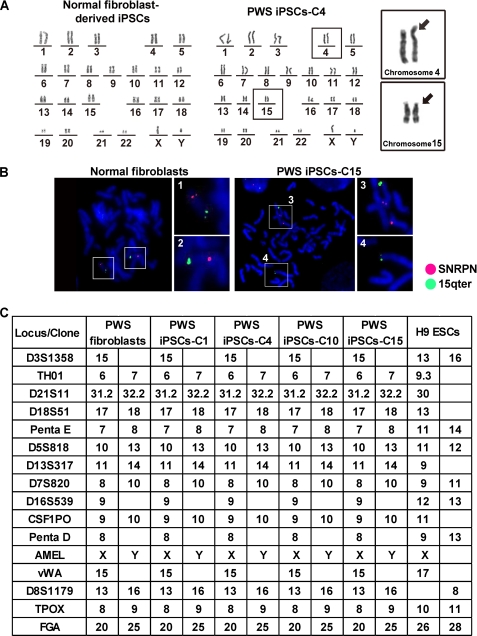
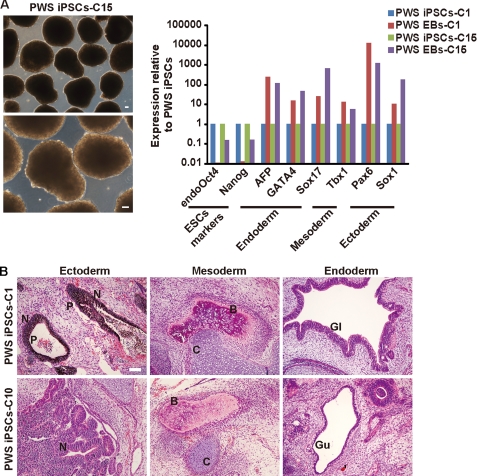
The generation of iPSCs represents a tremendous advance for stem cell biology if not yet practically at least conceptually (22–25). However, there is intense debate regarding, on one side, the feasibility of future clinical application of iPSCs and, on the other, their ability to model human diseases in vitro (26). This debate should help define standards of bona fide reprogramming, map limitations of iPSCs, and find potential solutions to existing problems. In this study, we attempted to answer whether iPSCs from patients with PWS can be produced and are stable after prolonged culture and, more importantly, whether they retain the original pattern of imprinting that would allow modeling this disease in vitro. For this, we obtained fibroblasts from a previously diagnosed PWS patient with de novo balanced translocation from chromosome 15 (break point q11.2) to chromosome 4 (break point q27) and transduced them with retroviruses producing four exogenous factors: Sox2, Klf4, Oct4, and c-Myc. At day 6, cells were split on feeders, and around day 16, small colonies appeared that progressively adopted an ESC-like morphology. After day 25, selected iPSCs were picked manually and expanded, after which they still maintained human ESC-like morphology with prominent nuclei and stained positive for alkaline phosphatase (Fig. 1A). In addition, immunofluorescence microscopy showed positive staining of ESC-like surface antigens SSEA-3, SSEA-4, TRA-1–60, and TRA-1–81 and the ESC transcription factor Nanog (Fig. 1A). In addition, qPCR analysis showed up-regulation of the endogenous ESC transcriptional program (endogenous Oct4, endogenous Sox2, Nanog, and Rex1) comparable with an established embryonic stem cell line (H9) and silencing of the exogenous factors as expected for cell lines that behave as ESCs (Fig. 1B). We then used semiquantitative PCR to demonstrate integration of all four transgenes in the genomic DNA of PWS iPSCs (Fig. 1C) and studied the methylation status of proximal promoters of key ESC transcription factors using bisulfite sequencing. OCT4 and NANOG promoters displayed extensive DNA demethylation in PWS iPSC clones comparable with H9 ESCs and opposed to donor PWS fibroblasts (Fig. 1D). PWS iPSCs also displayed the normal number of chromosomes, but, as expected, translocation from chromosome 15 to chromosome 4 was observed (Fig. 2A). This translocation was verified using FISH and specific probes that bind to the SNURF-SNRPN gene or a control region in chromosome 15 (Fig. 2B). We used single tandem repeat analysis to show that the PWS iPSC colonies originated from the donor PWS fibroblasts and were not mixed with other cells growing in our laboratory (Fig. 2C). In addition, EB and teratoma formation demonstrated that PWS iPSCs are pluripotent. In both cases, PWS iPSCs differentiated into cell lineages corresponding to the three germ layers (Fig. 3, A and B). The teratomas contained complex differentiating structures such as bone, gastrointestinal epithelium, and neural tube-like formations (Fig. 3B). Therefore, iPSCs from PWS patients that match standard characterization criteria for human ESCs can be generated, and these cells could be routinely passaged (up to passage 32 in this study) without obvious alteration in their pluripotent characteristics.
FIGURE 1.
Generation of iPSCs from a patient with PWS. A, upper panels, phase-contrast photographs of representative PWS iPSC clones (C4 P21 and C1 P24, where P indicates passage) grown on feeder layers or Matrigel; alkaline phosphatase (AP) staining of another clone (P17) grown on feeders is also included. Scale bars = 200 μm. Middle and lower panels, immunofluorescence microscopy photographs for the indicated ESC markers of a representative PWS iPSC clone (P22). Nuclei are stained in blue with DAPI. Scale bars = 50 μm. B, left panel, qPCR for the transgenes (Tg) in the indicated PWS iPSC clones. Values refer to transduced donor PWS fibroblasts extracted at day 6; untransduced fibroblasts were included as a control. Right panel, qPCR for selected endogenous ESC transcription factors in representative PWS iPSC clones (C1 P11, C4 P10, and C15 P9). Values refer to donor fibroblasts; H9 ESCs (P36) were used as a positive control. C, semiquantitative PCR shows integration of SOX2, KLF4, OCT4, and c-MYC transgenes in the genomic DNA of three representative PWS iPSC clones. Donor PWS fibroblasts, a previously reported iPSC clone from UMCs, and water were used as controls. D, DNA methylation profile of the proximal OCT4 and NANOG promoters analyzed by bisulfite sequencing. Donor PWS fibroblasts, H9 ESCs (P36), and representative PWS iPSC clones (C1 P11, C10 P10, and C15 P9) were included. Filled squares indicate Cs that failed to be converted to T in the context of CpGs as a result of being methylated; empty squares indicate unmethylated (C-T converted) CpGs.
FIGURE 2.
Additional characterization of PWS iPSCs. A, normal karyotype of an iPSC clone (P20) derived from fibroblasts of a healthy individual (left panel) and a representative PWS iPSC clone (P22; right panel). Insets, chromosomes 4 and 15 in the PWS iPSC clone are magnified to highlight the translocation. B, FISH verified the translocation of the PWS region from chromosome 15 to chromosome 4 in a representative PWS iPSC clone (P23). Normal fibroblasts were used as a control. SNRPN and 15qter are the names of the PWS region and control probes, respectively, which are visualized as red and green dots. C, single tandem repeat analysis verified that the selected PWS iPSC clones originated from the donor PWS fibroblasts. H9 ESCs were included as control.
FIGURE 3.
PWS iPSCs are pluripotent. A, left panel, phase-contrast photographs of EBs from a representative PWS iPSC clone (P22) growing in suspension. Right panel, qPCR analysis of the indicated genes after attaching similar EBs from the indicated PWS iPSC clones (C1 P17 and C15 P17) to gelatin. B, teratomas produced by two representative PWS iPSC clones (C1 P13 and C10 P12) contained tissues corresponding to the three germ layers. N, neural tube-like structures; P, pigmented cells; B, bone; C, cartilage; Gl, ciliated epithelium of a gland-like structure; Gu, gut epithelium. Scale bar = 100 μm.
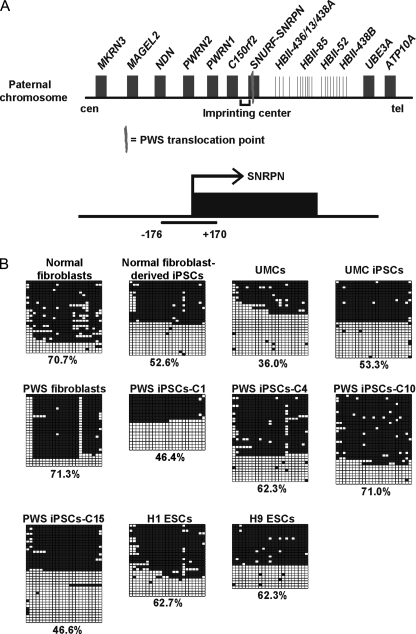
We next sought to study whether a normal pattern of maternal imprinting is conserved in PWS iPSCs, as this is a prerequisite for modeling the disease phenotype in vitro. The PWS imprinting center (IC) consists of a large CpG island that encompasses exon 1 of the SNURF-SNRPN gene (Fig. 4A) (27). This CpG island seems to act as a canonical promoter for SNURF-SNRPN and the following genes, including SNORD116 and other snoRNA clusters. CpGs in the PWS IC of the parental chromosome are normally demethylated, but the maternal chromosome bears heavy DNA methylation that renders transcription inactive. In mice, DNA methylation of the maternal PWS IC is present in the oocyte and remains stable upon fertilization, whereas in humans, this may happen post-fertilization (12). Such consideration is important because it seems plausible that iPSCs and ESCs may have different profiles of imprinting marks that only become reset during early development. To check the status of the PWS IC, we performed PCR on genomic DNA to amplify a fragment containing 346 bp (Fig. 4A) and exposed it to bisulfite treatment and sequencing, as in Fig. 1D. We analyzed donor PWS fibroblasts, four derived PWS iPSCs (clones C1, C4, C10, and C15), fibroblasts from a normal individual, a derived iPSC clone, primary UMCs, a derived iPSC clone (18), and two widely studied human ESCs (H1 and H9). PWS fibroblasts, normal fibroblasts, and donor UMCs displayed higher or lower DNA methylation levels than anticipated (Fig. 4B). One possible way to explain this is that the donor cells represent a heterogeneous population with conserved and altered imprinting marks. In this regard, it is accepted that prolonged in vitro culture can induce epigenetic abnormalities (28). Conversely, embryonic stem cell lines, iPSCs from normal fibroblasts, UMC iPSCs, and PWS iPSC clones C1, C4, and C15 all showed an almost perfect pattern of hemimethylation (Fig. 4B). PWS iPSC clone C10 matched the DNA methylation profile of the donor fibroblasts (∼71%) (Fig. 4B), though this clone did not show any noticeable difference in morphology, growth, or pluripotent characteristics.
FIGURE 4.
Conservation of genomic imprinting in selected PWS iPSCs. A, simplified scheme depicting the genomic organization of the PWS IC and flanking genes in paternal chromosome 15. cen, centromere; tel, telomere. Also shown is a representation of the region amplified by PCR for DNA methylation analysis in B. B, DNA methylation pattern after bisulfite sequencing of a fragment contained in the PWS IC. The methylation percentages for the indicated control and PWS iPSC clones (C1 P11, C4 P12, C10 P10, and C15 P9), UMC iPSCs (P20), normal fibroblast-derived iPSCs (P11), ESCs (H1 P65 and H9 P36), and donor cells (control fibroblasts, UMCs, and PWS fibroblasts) are shown at the bottom of each panel.
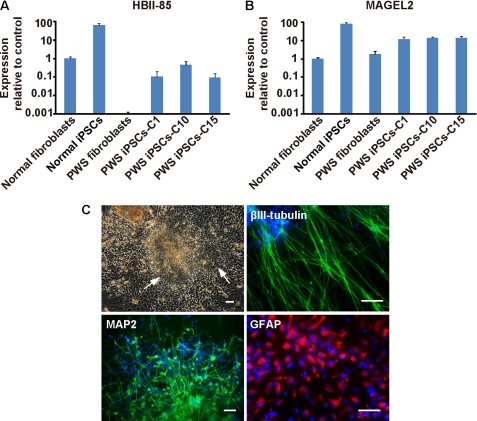
We next analyzed the expression of SNORD116, a snoRNA of the C/D box class contained in the translocated region (16, 29). SNORD116 consists of 29 tandem repeats of the same sequence, followed by 48 copies of a related snoRNA, SNORD115 (30). Compared with other related C/D box snoRNAs, SNORD115 and SNORD116 do not have any significant complementarity to ribosomal RNAs and are thought to regulate protein-coding genes. SNORD115 targets and regulates the splicing of the serotonin 2C receptor (31) but no targets have been identified yet for SNORD116. We performed qPCR for SNORD116 and observed high expression levels in normal fibroblasts and derived iPSCs, whereas donor PWS fibroblasts and PWS iPSC clones, including clone C10, showed low expression (Fig. 5A). The latter is expected, as only decreased DNA methylation at the PWS IC would determine high SNORD116 expression. As a control, we performed qPCR for MAGEL2, a gene located near the PWS IC but outside the translocated region (Fig. 4A). MAGEL2 was similarly expressed in normal fibroblasts, PWS fibroblasts, and iPSCs derived from them (Fig. 5B). These data indicate that our PWS iPSC clones should be valuable for modeling the disease in vitro given the insufficient expression of SNORD116. Because the affected tissue in PWS is the brain, we studied whether PWS iPSCs can be differentiated into neuron-like structures. PWS iPSCs could readily form neurospheres that were further differentiated into neuron-like and astrocyte-like cells, and these cells stained positive for neuronal (βIII-tubulin and MAP2) and astrocyte (glial fibrillary acidic protein) markers (Fig. 5C). Therefore, our PWS iPSCs not only display adequate PWS IC imprinting in most cases but also have the ability to differentiate into the main tissue affected in PWS.
FIGURE 5.
PWS iPSCs express low levels of HBII-85 and can be differentiated into neuron-like cells. A, qPCR for HBII-85 and MAGEL2 in normal fibroblasts, donor PWS fibroblasts, control iPSCs, and PWS iPSCs. Values refer to control fibroblasts and correspond to the mean ± S.D. of three independent experiments (control iPSCs P21, P22, and P23; PWS iPSC clone C1 P18, P21, and P22; PWS iPSC clone C10 P12, P13, and P14; and PWS iPSC clone C15 P19, P22, and P24). B, phase-contrast photograph of EBs from a representative PWS iPSC clone (P14) grown in suspension and attached to a Matrigel-coated surface in neural induction medium. Arrows point to the formation of neural tube-like structures. The immunofluorescence microscopy photographs are of the neuronal markers βIII-tubulin, MAP2, and glial fibrillary acidic protein (GFAP) using neuron-like and astrocyte-like cells produced from picked neural rosettes of a similar representative experiment. Scale bars = 50 μm.
DISCUSSION
Nuclear reprogramming with exogenous factors involves a global erasure of epigenetic marks to acquire pluripotency (32). In particular, changes in DNA methylation are a driving force of the reprogramming process, and for example are needed in the promoters of ESC transcription factors and possibly differentiation related genes as well (33, 34). It is therefore not surprising that subtle epigenetic abnormalities may exist in otherwise bona fide iPSCs that could compromise their ability to model human diseases in vitro or their utility for transplantation purposes. For instance, Benvenisty and co-workers (35) found altered expression of several imprinting genes in a significant number of human iPSCs. However, these abnormalities were not found in all studied iPSC lines, and there did not seem to be a clear tendency to accumulate specific imprinting errors. Because the defect was already observed in early passage iPSCs, the authors argued that it is likely a consequence of the reprogramming procedure rather than colony expansion. Human ESCs contain imprinting abnormalities as well, but their epigenome is considered more stable, and these alterations seem to appear mainly after prolonged cell culture (36). In a different report, Benvenisty and co-workers (37) also found that iPSCs from patients with fragile X syndrome bear DNA methylation and histone modifications of the FMR1 gene promoter similar to donor fragile X fibroblasts and opposed to fragile X ESCs. This suggested that specific sites in the genome that become highly modified upon differentiation might be refractory to reprogramming. In addition, Hochedlinger and co-workers (38) and Daley and co-workers (39) reported epigenetic memory of the donor tissue in mouse iPSCs, and several reports have indicated that this might be the case in human iPSCs as well (40, 41). On the other hand, a number of relevant studies by other groups have supported the idea that iPSCs can reproduce key aspects of given human diseases in vitro, e.g. long QT syndrome, familial dysautonomia, and α-antitrypsin deficiency (42–44).
Here, we have shown that iPSCs could be generated from fibroblasts of a patient with PWS and displayed the normal pattern of maternal imprinting that determines the disease. iPSC clones from two unaffected individuals (corresponding to two unrelated tissues: fibroblasts and UMCs) and human ESCs (45, 46) also showed adequate imprinting of the PWS IC (Fig. 4B), supporting the idea that this genomic site is not only susceptible to faithful reprogramming but also refractory to acquired de novo alterations. Importantly, PWS iPSCs could be readily differentiated into neuron-like cells. Further analysis will be necessary to underscore putative differences between these neurons and those derived from normal iPSCs. Our work is therefore relevant because such an approach may improve our understanding of PWS. For example, genomics or proteomics analysis may allow the identification of targets for SNORD16 or other genes involved in the disease. For this purpose, the generation of iPSCs from different types of PWS patients should provide invaluable phenotype/phenotype correlations, and this could potentially change the way we view PWS. Notably, besides modeling the disease phenotype, neurons from PWS iPSCs may be useful for testing compounds aimed at modifying epigenetic marks at the maternal PWS IC. We cannot exclude that iPSCs, including those presented here, bear epigenetic or transcriptomic abnormalities that render them unable to exert specific functions. Nevertheless, our work supports the idea that iPSCs can be used to increase our understanding of genetic imprinting diseases such as PWS and, given the vicinity of the IC, possibly also Angelman syndrome (12).
Acknowledgments
We thank Dajiang Qin and Xingyan Li for providing normal human fibroblasts and an iPSC clone derived from them.
This work was supported by startup funding from the Guangzhou Institutes of Biomedicine and Health and Grant KSCX2-YW-R244 from the Chinese Academy of Sciences (to M. A. E.); Grant 2008A1-E4011 from the Bureau of Science and Technology of Guangzhou Municipality, Grant 90813033 from the National Natural Science Foundation of China, Grants 2007CB948002 and 2009CB941102 from the 973 Program of the Ministry of Science and Technology of China, and Grant 2010DFB30430 from the Ministry of Science and Technology International Technology Cooperation Program (to D. P.); and Grant 2006AA02A103 from the 863 Program of the Ministry of Science and Technology of China (to L. L.).
- PWS
- Prader-Willi syndrome
- snoRNA
- small nucleolar RNA
- iPSC
- induced pluripotent stem cell
- ESC
- embryonic stem cell
- bFGF
- basic FGF
- UMC
- umbilical cord matrix mesenchymal cell
- qPCR
- quantitative real-time RT-PCR
- EB
- embryoid body
- IC
- imprinting center.
REFERENCES
- 1.Koerner M. V., Barlow D. P. (2010) Curr. Opin. Genet. Dev. 20, 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood A. J., Oakey R. J. (2006) PLoS Genet. 2, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Constância M., Pickard B., Kelsey G., Reik W. (1998) Genome Res. 8, 881–900 [DOI] [PubMed] [Google Scholar]
- 4.Luedi P. P., Hartemink A. J., Jirtle R. L. (2005) Genome Res. 15, 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Law J. A., Jacobsen S. E. (2010) Nat. Rev. Genet. 11, 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reik W., Walter J. (2001) Nat. Rev. Genet. 2, 21–32 [DOI] [PubMed] [Google Scholar]
- 7.Szabó P., Mann J. R. (1994) Development 120, 1651–1660 [DOI] [PubMed] [Google Scholar]
- 8.Jirtle R. L., Skinner M. K. (2007) Nat. Rev. Genet. 8, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerda S., Weitzman S. A. (1997) Mutat. Res. 386, 141–152 [DOI] [PubMed] [Google Scholar]
- 10.Santos-Rebouças C. B., Pimentel M. M. (2007) Eur. J. Hum. Genet. 15, 10–17 [DOI] [PubMed] [Google Scholar]
- 11.Esteller M. (2008) N. Engl. J. Med. 358, 1148–1159 [DOI] [PubMed] [Google Scholar]
- 12.Chamberlain S. J., Lalande M. (2010) Neurobiol. Dis. 39, 13–20 [DOI] [PubMed] [Google Scholar]
- 13.Horsthemke B., Wagstaff J. (2008) Am. J. Med. Genet. A 146A, 2041–2052 [DOI] [PubMed] [Google Scholar]
- 14.Weksberg R., Smith A. C., Squire J., Sadowski P. (2003) Hum. Mol. Genet. 12, Suppl. 1, R61–R68 [DOI] [PubMed] [Google Scholar]
- 15.Kishino T., Lalande M., Wagstaff J. (1997) Nat. Genet. 15, 70–73 [DOI] [PubMed] [Google Scholar]
- 16.Sahoo T., del Gaudio D., German J. R., Shinawi M., Peters S. U., Person R. E., Garnica A., Cheung S. W., Beaudet A. L. (2008) Nat. Genet. 40, 719–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung K. N., Vallero R. O., DuBose A. J., Resnick J. L., LaSalle J. M. (2009) Hum. Mol. Genet. 18, 4227–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai J., Li W., Su H., Qin D., Yang J., Zhu F., Xu J., He W., Guo X., Labuda K., Peterbauer A., Wolbank S., Zhong M., Li Z., Wu W., So K. F., Redl H., Zeng L., Esteban M. A., Pei D. (2010) J. Biol. Chem. 285, 11227–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteban M. A., Wang T., Qin B., Yang J., Qin D., Cai J., Li W., Weng Z., Chen J., Ni S., Chen K., Li Y., Liu X., Xu J., Zhang S., Li F., He W., Labuda K., Song Y., Peterbauer A., Wolbank S., Redl H., Zhong M., Cai D., Zeng L., Pei D. (2010) Cell Stem Cell 6, 71–79 [DOI] [PubMed] [Google Scholar]
- 20.Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D. A. (2008) Nat. Biotechnol. 26, 1269–1275 [DOI] [PubMed] [Google Scholar]
- 21.El-Maarri O., Buiting K., Peery E. G., Kroisel P. M., Balaban B., Wagner K., Urman B., Heyd J., Lich C., Brannan C. I., Walter J., Horsthemke B. (2001) Nat. Genet. 27, 341–344 [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 23.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 24.Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 25.Wilmut I. (2007) Cell Stem Cell 1, 593–594 [DOI] [PubMed] [Google Scholar]
- 26.Lako M., Armstrong L., Stojkovic M. (2010) Stem Cells 28, 845–850 [DOI] [PubMed] [Google Scholar]
- 27.Saitoh S., Buiting K., Rogan P. K., Buxton J. L., Driscoll D. J., Arnemann J., König R., Malcolm S., Horsthemke B., Nicholls R. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7811–7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernández-Gonzalez R., Moreira P., Bilbao A., Jiménez A., Pérez-Crespo M., Ramírez M. A., Rodríguez De Fonseca F., Pintado B., Gutiérrez-Adán A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5880–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallagher R. C., Pils B., Albalwi M., Francke U. (2002) Am. J. Hum. Genet. 71, 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavaillé J., Buiting K., Kiefmann M., Lalande M., Brannan C. I., Horsthemke B., Bachellerie J. P., Brosius J., Hüttenhofer A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14311–14316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kishore S., Stamm S. (2006) Science 311, 230–232 [DOI] [PubMed] [Google Scholar]
- 32.Meissner A. (2010) Nat. Biotechnol. 28, 1079–1088 [DOI] [PubMed] [Google Scholar]
- 33.Li J. Y., Pu M. T., Hirasawa R., Li B. Z., Huang Y. N., Zeng R., Jing N. H., Chen T., Li E., Sasaki H., Xu G. L. (2007) Mol. Cell. Biol. 27, 8748–8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lister R., Pelizzola M., Dowen R. H., Hawkins R. D., Hon G., Tonti-Filippini J., Nery J. R., Lee L., Ye Z., Ngo Q. M., Edsall L., Antosiewicz-Bourget J., Stewart R., Ruotti V., Millar A. H., Thomson J. A., Ren B., Ecker J. R. (2009) Nature 462, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pick M., Stelzer Y., Bar-Nur O., Mayshar Y., Eden A., Benvenisty N. (2009) Stem Cells 27, 2686–2690 [DOI] [PubMed] [Google Scholar]
- 36.Närvä E., Autio R., Rahkonen N., Kong L., Harrison N., Kitsberg D., Borghese L., Itskovitz-Eldor J., Rasool O., Dvorak P., Hovatta O., Otonkoski T., Tuuri T., Cui W., Brüstle O., Baker D., Maltby E., Moore H. D., Benvenisty N., Andrews P. W., Yli-Harja O., Lahesmaa R. (2010) Nat. Biotechnol. 28, 371–377 [DOI] [PubMed] [Google Scholar]
- 37.Urbach A., Bar-Nur O., Daley G. Q., Benvenisty N. (2010) Cell Stem Cell 6, 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polo J. M., Liu S., Figueroa M. E., Kulalert W., Eminli S., Tan K. Y., Apostolou E., Stadtfeld M., Li Y., Shioda T., Natesan S., Wagers A. J., Melnick A., Evans T., Hochedlinger K. (2010) Nat. Biotechnol. 28, 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M. J., Ji H., Ehrlich L. I., Yabuuchi A., Takeuchi A., Cunniff K. C., Hongguang H., McKinney-Freeman S., Naveiras O., Yoon T. J., Irizarry R. A., Jung N., Seita J., Hanna J., Murakami P., Jaenisch R., Weissleder R., Orkin S. H., Weissman I. L., Feinberg A. P., Daley G. Q. (2010) Nature 467, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh Z., Wilson K. D., Wu Y., Hu S., Quertermous T., Wu J. C. (2010) PLoS ONE 5, e8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chin M. H., Pellegrini M., Plath K., Lowry W. E. (2010) Cell Stem Cell 7, 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moretti A., Bellin M., Welling A., Jung C. B., Lam J. T., Bott-Flügel L., Dorn T., Goedel A., Höhnke C., Hofmann F., Seyfarth M., Sinnecker D., Schömig A., Laugwitz K. L. (2010) N. Engl. J. Med. 363, 1397–1409 [DOI] [PubMed] [Google Scholar]
- 43.Lee G., Papapetrou E. P., Kim H., Chambers S. M., Tomishima M. J., Fasano C. A., Ganat Y. M., Menon J., Shimizu F., Viale A., Tabar V., Sadelain M., Studer L. (2009) Nature 461, 402–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rashid S. T., Corbineau S., Hannan N., Marciniak S. J., Miranda E., Alexander G., Huang-Doran I., Griffin J., Ahrlund-Richter L., Skepper J., Semple R., Weber A., Lomas D. A., Vallier L. (2010) J. Clin. Invest. 120, 3127–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rugg-Gunn P. J., Ferguson-Smith A. C., Pedersen R. A. (2007) Hum. Mol. Genet. 16, R243–R251 [DOI] [PubMed] [Google Scholar]
- 46.Kim K. P., Thurston A., Mummery C., Ward-van Oostwaard D., Priddle H., Allegrucci C., Denning C., Young L. (2007) Genome Res. 17, 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]