Abstract
Nicotinamide mononucleotide (NMN) adenylyltransferase 2 (Nmnat2) catalyzes the synthesis of NAD from NMN and ATP. The Nmnat2 transcript is expressed predominately in the brain; we report here that Nmnat2 is a low abundance protein expressed in neurons. Previous studies indicate that Nmnat2 localizes to Golgi. As Nmnat2 is not predicted to contain a signal sequence, lipid-binding domain, or transmembrane domain, we investigated the nature of this interaction. These experiments reveal that Nmnat2 is palmitoylated in vitro, and this modification is required for membrane association. Surprisingly, exogenous Nmnat2 is toxic to neurons, indicating that protein levels must be tightly regulated. To analyze Nmnat2 localization in neurons (previous experiments relied on exogenous expression in HeLa cells), mouse brains were fractionated, showing that Nmnat2 is enriched in numerous membrane compartments including synaptic terminals. In HeLa cells, in addition to Golgi, Nmnat2 localizes to Rab7-containing late endosomes. These studies show that Nmnat2 is a neuronal protein peripherally attached to membranes via palmitoylation and suggest that Nmnat2 is transported to synaptic terminals via an endosomal pathway.
Keywords: Brain, Endosomes, Golgi, Membrane, NAD, Neuron, Synapses, Nmnat2, Palmitoylation
Introduction
Nicotinamide mononucleotide (NMN)2 adenylyltransferases (Nmnat)2 catalyze the synthesis of NAD from NMN and ATP (1–3). Humans and mice express three isoforms, each from a separate gene: Nmnat1, -2, and -3; whereas invertebrates such as Drosophila melanogaster have only one (4). Nmnat2 is unique in that its transcript is expressed predominately in the brain (5–7). In contrast, Nmnat1 and -3 transcripts are widely expressed, although they do not necessarily overlap (2). Furthermore, Nmnat2 localizes to Golgi (8), whereas Nmnat1 is nuclear (8, 9), and Nmnat3 associates with mitochondria (8, 10). This suggests that 1) Nmnat1, -2, and -3 have evolved specialized roles in vertebrate NAD metabolism, and 2) the subcellular location of NAD synthesis is cell type-specific.
In addition to its role as a cofactor, NAD is also a substrate for numerous enzymes including sirtuins, poly(ADP-ribose) polymerases, and ADP-ribosyl cyclases (e.g. CD38) (11). These enzymes regulate diverse cellular processes including transcription, apoptosis, and calcium signaling (12–14). In eukaryotes, NAD is synthesized either de novo from tryptophan or recycled from nicotinic acid, nicotinamide, or nicotinamide riboside (1, 11). Because neurons require an enormous amount of energy to propagate action potentials, it is not surprising that insufficient dietary intake of NAD precursors results in severe neurological dysfunction (15, 16).
It is unclear what specific role Nmnat2 has in the brain that cannot be met by Nmnat1 or -3. To address this question, we have first sought to understand how Nmnat2 interacts with the Golgi, its cellular and developmental protein expression, and its localization in brain cells. We report here that Nmnat2 is a developmentally regulated, low abundance neuronal protein that localizes not only to Golgi but also to vesicles and synaptic compartments. Overexpression of Nmnat2 is toxic to neurons, suggesting that endogenous protein levels must be tightly regulated and providing a rationale for its low abundance. Additional experiments investigate its interaction with Golgi, revealing that Nmnat2 is peripherally attached to the membrane via palmitoylation. Biochemical analysis shows that endogenous Nmnat2 behaves like a membrane protein but is difficult to solubilize suggesting that 1) palmitoylation directs Nmnat2 to detergent resistant membranes or 2) in addition to palmitoylation, Nmnat2 is held at the membrane via protein-protein interaction. Finally, colocalization studies in HeLa cells suggest that Nmnat2 may traffic through an endosomal pathway via Rab7-containing vesicles. The data presented here provide the basis for future investigation of the specific role of Nmnat2 in neuronal NAD metabolism.
EXPERIMENTAL PROCEDURES
Antibodies
Full-length human NMNAT2 (NM_015039) was cloned into pET-28a (Novagen) and expressed in Rosetta (DE3)pLysS Escherichia coli. The resulting His-tag fusion protein was purified using nickel-nitrilotriacetic acid agarose beads (Qiagen) according to the manufacturer's guidelines. Polyclonal antibody was generated by YenZym Antibodies (San Francisco, CA) and purified by affinity chromatography. Antibodies against the following proteins were purchased from the indicated vendors: neurofilament (heavy chain; Affinity BioReagents); valosin-containing protein (VCP) and GM130 (BD Biosciences); Rab7 and EEA1 (Cell Signaling Technology); and EEA1, FLAG, GAPDH, TGN46, and VDAC (Sigma). Antibodies against Rab5, synaptobrevin 2, synaptotagmin I, and α/β-neurexins were kindly donated by Thomas Südhof (Stanford School of Medicine, Palo Alto, CA). Joachim Seemann (University of Texas Southwestern Medical Center, Dallas, TX) kindly donated anti-β-COP antibody.
Mammalian Expression Vectors and Site-directed Mutagenesis
Human NMNAT2 (NM_015039) was cloned into pEGFP-N1 (Clontech) as described previously (10). This construct is referred to here as Nmnat2-EGFP and was used as template for PCR-based mutagenesis to create the C164S/C165S mutant as well as the low homology domain (LHD; see below) deletion mutants. Nmnat2-FLAG and Nmnat2 C164S/C165S-FLAG were generated by subcloning into a C-terminal FLAG vector. All constructs were verified by DNA sequencing.
Lentivirus Production
Lentiviral shRNA vectors (pLKO.1-puro) targeted against mouse Nmnat2 and EGFP were purchased from Sigma. Nmnat2-EGFP and its derivatives were subcloned into a modified version of FUGW, a lentivirus expression vector, which, along with the packaging constructs pVSVG and pCMVΔ8.9, were kindly provided by Thomas Südhof (Stanford School of Medicine). Lentiviruses were produced from HEK293T triple-transfected (FuGENE 6) with pVSVG, pCMVΔ8.9, and one of the lentiviral expression vectors described above. Approximately 48 h after transfection, virus containing medium was collected and filtered (0.45 μm) then stored in small aliquots at −80 °C until needed. Viruses were titered by quantitative PCR using a protocol adapted from Salmon and Trono (17) and Sastry et al. (18).
Cell Culture and Imaging
HEK293T, Neuro-2a, and SH-SY5Y cell lines were purchased from ATCC. U343 and U87 cells were generously donated by Richard Lu (UT Southwestern Medical Center). HeLa cells were a gift from Joachim Seemann (UT Southwestern Medical Center). Cultures were maintained in growth medium containing 10% FBS/DMEM (Invitrogen), except SH-SY5Y, which were maintained in 10% FBS/Advanced minimum essential medium supplemented with nonessential amino acids, 110 mg/ml sodium pyruvate, and 2 mm glutamine (Invitrogen). SH-SY5Y cells were differentiated using a previously established protocol (19). In brief, SH-SY5Y were seeded at low density and sequentially treated with 10 μm retinoic acid (Sigma) in growth medium for 5 days and then 50 ng/ml human BDNF (Invitrogen) in serum-free medium for an additional 7 days. At this point, most cells developed a rounded shape with an extensive network of neurites emanating from clumps of cell bodies.
Primary neuronal cultures were generated using standard procedures from cortex dissected from late embryonic (E18) mouse brains. In brief, tissues were cut into small uniform pieces and treated for ∼5 min with warm papain solution (20 units/ml, Worthington; dissolved in Hanks' balanced salt solution supplemented with 10 mm HEPES-NaOH, pH ∼ 7.2, Invitrogen). To inactivate the papain, tissue fragments were briefly incubated with a solution of 1.25% trypsin inhibitor (Sigma) and then rinsed with cold Hanks' balanced salt solution/HEPES buffer. Neurons were dissociated by gentle trituration using a fire-polished Pasteur pipette in neuronal growth medium consisting of Neurobasal supplemented with B-27 and 2 mm glutamine (Invitrogen). For expression analysis, cells were plated on poly-d-lysine (0.1 mg/ml, Sigma) coated 6-cm dishes at a density of 1 × 106 cells/plate. Neurons were fed every 3 days and collected at various time points, as indicated. Primary glial cultures were also derived from the cortex using a similar procedure with the following exceptions; tissue fragments were incubated with papain solution for 15 min, and cultures were maintained in 10% FBS/DMEM. The absence of neurotrophic factors results in the selective loss of neurons. Glial cultures were fed every 7 days and were maintained for ∼21 days before harvesting.
For immunofluorescence experiments, cortical neurons were seeded at a density of 1 × 105 cells per coverslip (12 mm diameter, Thermo Fisher Scientific) previously treated with Matrigel (1:100 in PBS, BD Biosciences) and 0.1 mg/ml poly-d-lysine (Sigma). HeLa cells were seeded at a density of 2 × 104 cells per coverslip. Cells were fixed with 3.7% paraformaldehyde, permeabilized with 0.1% Triton X-100, washed several times with PBS, and blocked for 30 min with a solution of 4% BSA (Sigma) and 1% normal goat serum (Jackson ImmunoResearch Laboratories). Incubation with primary antibody was done overnight at 4 °C. Coverslips were mounted with ProLong Gold antifade reagent (Invitrogen), and images were collected using a Zeiss LSM 510 confocal microscope.
Analysis of Nmnat2 Expression in Cells and Tissue
Tissues were collected from age-matched mice and washed with PBS and then immediately flash-frozen in liquid nitrogen and ground to a powder using a mortar and pestle. Cells were washed with ice-cold PBS and then collected with a rubber policeman and centrifuged at 400 × g for 5 min. Cell pellets and tissue powder were lysed with cold 50 mm HEPES-NaOH, 1% LDS, 1 mm DTT, and protease inhibitors (0.2 mm PMSF, 0.5 μg/ml leupeptin, and 1 μm pepstatin A) and then briefly sonicated to reduce viscosity. Protein concentration was measured using the DC protein assay according to the manufacturer's instructions (Bio-Rad). Within a given experiment, the same amount of total protein was loaded for each sample.
Palmitoylation Assay
HEK293T cells transiently expressing Nmnat2-FLAG or Nmnat2 C164S/C165S-FLAG were incubated for 6 h in media containing 0.4 mCi/ml [9,10–3H(N)]palmitic acid (PerkinElmer Life Sciences). Cells were washed with ice-cold PBS, collected using a rubber policeman, and centrifuged at 800 × g and then lysed with 1% LDS/4 m urea and diluted 1:20 with buffer B (1% Nonidet P-40, 0.2 mm PMSF, 0.5 μg/ml leupeptin, and 1 μm pepstatin A). Diluted lysate was incubated with anti-FLAG agarose beads overnight at 4 °C. After extensive washing with PBS, bound protein was eluted in a small volume of sample buffer (60 mm Tris-HCl, pH 6.8, 2% LDS, 10% glycerol, and 0.025% bromphenol blue). Eluted protein was separated by SDS-PAGE and exposed to Biomax XAR film (Kodak) at −80 °C for 14 days. To enhance detection of radiolabeled protein, polyacrylamide gels were impregnated with 2,5-diphenyloxazole (Sigma) as described by Bonner and Laskey (20).
Purification and Activity of Nmnat2 and Nmnat2 C164S/C165S
The coding sequence for human NMNAT2 and NMNAT2 C164S/C165S was cloned into pMBP-parallel1 (21). The resulting maltose-binding protein fusion protein was affinity purified using amylose resin (New England BioLabs). After removal of the maltose-binding protein tag by tobacco etch virus protease treatment, Nmnat2 and Nmnat2 C164S/C165S were further purified using a phenyl-Sepharose column and by gel filtration chromatography (GE Healthcare).
Steady-state kinetic parameters were determined using a coupled assay adapted from Kurnasov et al. (22). Briefly, reaction mixtures were prepared in parallel containing 50 mm HEPES-NaOH, pH 7.5, 115 mm ethanol, 40 mm semicarbazide, 6 units/ml of alcohol dehydrogenase, 5% glycerol, and varying amounts of ATP and NMN. Reactions were started by adding purified Nmnat2 or Nmnat2 C164S/C165S to a final concentration of 0.4 nm. The production of NADH was monitored by measuring absorbance at 340 nm. Apparent values of Km and kcat were calculated using SigmaPlot 11 (Systat Software).
Extraction of Postnuclear Membrane Fractions
Whole mouse brains from adult mice were cut into equally sized pieces and homogenized on ice using a potter-elvehjem homogenizer in buffer A supplemented with 320 mm sucrose. Buffer A consists of 10 mm HEPES-NaOH, pH 7.4, 1 mm DTT, and protease inhibitors (0.2 mm PMSF, 0.5 μg/ml leupeptin, and 1 μm pepstatin A). To reduce nuclear and whole cell contamination, homogenate was filtered through four layers of cheesecloth and centrifuged at 900 × g for 10 min. The resulting supernatant was centrifuged at 100,000 × g for 1 h. The supernatant from this spin is designated as the cytosolic (S100) fraction. The pellet (P100), which is enriched for cytoplasmic membranes, was suspended with buffer A/320 mm sucrose. 100 μl aliquots of P100 (1 mg/ml) were centrifuged at 100,000 × g for 1 h. The resulting pellets were suspended in 100 μl of buffer A supplemented with one of the following: 320 mm sucrose, 1 m NaCl, 4 m urea, 1% CHAPS, 1% octyl glucoside (OG), or 1% lithium dodecyl sulphate (LDS). Suspensions were incubated on ice for 30 min and then centrifuged at 100,000 × g for 1 h. To analyze the hydrophobicity of endogenous Nmnat2, cytoplasmic membranes were extracted with 2% Triton X-114 (EMD Chemicals) overnight on ice. Separation of the detergent and aqueous phases was done essentially as described by Brusca and Radolf (23).
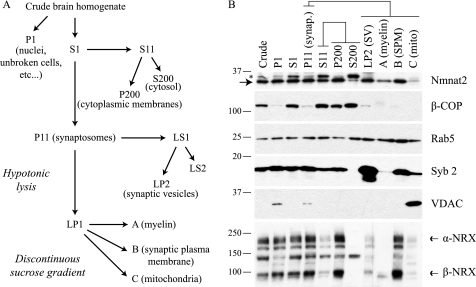
Fractionation of Mouse Brain Synaptosomes
This protocol is based on previously published methods with some modifications (24–27). Whole mouse brains were homogenized in 320 mm sucrose, 10 mm HEPES-NaOH, pH 7.4, 1 mm DTT, and protease inhibitors (0.2 mm PMSF, 0.5 μg/ml leupeptin, and 1 μm pepstatin A) using a potter-elvehjem homogenizer attached to an overhead motor (∼500 rpm). Homogenate was filtered through four layers of cheesecloth and then centrifuged at 900 × g for 10 min to sediment nuclei and unbroken cells (P1). To enrich for synaptosomes and mitochondria, postnuclear supernatant (S1) was centrifuged at 11,000 × g for 15 min. All remaining membranes in the resulting supernatant (S11) were collected by centrifugation at 200,000 × g for 2 h (P200). The synaptosome/mitochondria pellet (P11) was washed once with homogenization buffer and then lysed in hypotonic buffer (5 mm HEPES-NaOH, pH 7.4, supplemented with protease inhibitors). This suspension was then centrifuged at 25,000 × g for 20 min to sediment large membrane fragments (LP1). These membranes (LP1) were further fractionated by flotation through a discontinuous sucrose gradient. The LP1 pellet was suspended with 1.1 m sucrose and formed the bottom layer, with solutions of 0.855 and 0.32 m sucrose layered on top. Gradients were centrifuged at 19,500 rpm for 2.5 h using an SW-41 rotor (Beckman Coulter). Myelin enriched membranes (A) were collected from the 0.32/0.855 m interface and synaptic plasma membranes (B) from the 0.855/1.1 m interface. Mitochondria accumulated at the bottom of the tube in a sticky dark brown pellet (C). Synaptosome supernatant (LS1) was centrifuged at 200,000 × g for 2 h to sediment synaptic and other small vesicles (LP2).
RESULTS
Expression Profile of Nmnat2 Protein
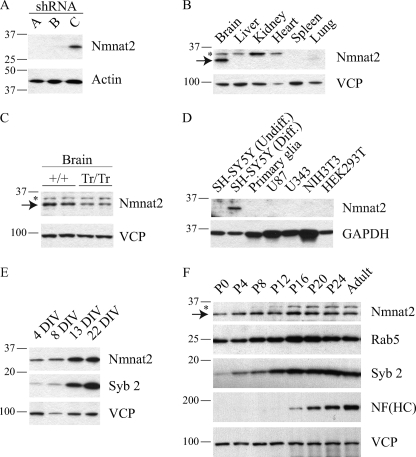
To evaluate the expression of endogenous Nmnat2, we generated several polyclonal antibodies using either peptides or full-length protein. Following an initial assessment, sera against full-length Nmnat2 was chosen for further analysis. To establish its specificity, we looked for cell lines expressing high levels of Nmnat2 transcript (BioGPS). As a result, Neuro-2a cells, which are a mouse neuroblastoma cell line, were infected with three different shRNA-expressing lentiviruses: two targeted against Nmnat2 and one targeted against EGFP (negative control). Total lysate from Neuro-2a was analyzed by Western blot, and a band migrating at the expected molecular mass (34.4 kDa) is observed in cells infected with control virus but is undetectable in cells infected with shRNA targeted against Nmnat2 (Fig. 1A).
FIGURE 1.
Nmnat2 is a neuronal protein and expression is developmentally regulated. A, to assess the specificity of an in-house antibody targeted against full-length Nmnat2, Neuro-2a, a neuroblastoma cell line reported to express Nmnat2 transcript (BioGPS), were infected with lentiviruses (MOI of 1) expressing either shRNA against Nmnat2 (lanes A and B) or EGFP (lane C). Total lysate from each was compared by Western blot. B, to evaluate the tissue expression of Nmnat2, lysate from various tissues were analyzed by Western blot. Note that a slightly slower migrating band is observed in several tissues including brain, marked with an asterisk; this band is nonspecific as shown in C. C, comparison of brain lysate from mice in which both Nmnat2 alleles have been trapped (Tr/Tr) versus wild-type controls (+/+). Each lane represents a single animal. As expected (see text), Nmnat2 expression in gene trap mouse brain is ∼50% of wild-type controls. Note the same nonspecific band observed in B, marked with an asterisk. D, to evaluate the cellular expression of Nmnat2, lysates from various cell types were analyzed by Western blot. Nmnat2 is detected in a human neuroblastoma cell line SH-SY5Y, but not in primary glia, U87 or U343, which are glioblastoma cell lines, or either of the negative controls NIH3T3 or HEK293T. In SH-SY5Y, Nmnat2 is dramatically up-regulated upon differentiation. E, Nmnat2 expression increases during maturation of primary cortical neurons similarly to synaptobrevin 2, a critical synaptic vesicle protein. F, expression of Nmnat2 during mouse development. Mouse brain lysate was obtained from a pair of male and female mice at birth and every 4 days thereafter as indicated. For each panel, an equal amount of protein was loaded in each lane; actin, GAPDH, or VCP are used as loading controls. An asterisk indicates a nonspecific band. NF, neurofilament; Syb 2, synaptobrevin 2; DIV, days in vitro; Diff., differentiated; Undiff., undifferentiated. HC, heavy chain.
Existing studies show that the Nmnat2 transcript is enriched in the brain relative to other tissues (5–7), although the transcript has also been detected in heart and muscle (7). To further test the specificity of this antibody, we compared lysates from various tissues and identified the same ∼34-kDa band detected in Fig. 1A in brain but not heart or any other tissues examined (Fig. 1B), including skeletal muscle (data not shown). Note that a slightly slower migrating band (marked with an asterisk) is observed in several tissues including brain, suggesting that this is a nonspecific band. To determine whether this band is nonspecific, we compared brain lysate derived from mice in which both Nmnat2 alleles have been trapped. The intensity of the same ∼34-kDa band observed in Fig. 1 (A and B) is reduced ∼50% (Fig. 1C), consistent with a 50% reduction in mRNA expression (p = 0.002, n = 3), whereas the intensity of the same slower migrating band observed in Fig. 1B (asterisk) remains constant. This mouse line was created to analyze the physiological function of Nmnat2. A more detailed analysis of these mice will be provided in future studies. Gene traps often produce null mutants (28); however, reduced (or even normal) expression of the underlying gene is not uncommon (29, 30).
Using Western blot analysis, these data show that an antibody targeted against full-length Nmnat2 detects a band migrating at the expected molecular weight in lysate derived from brain and a neuroblastoma cell line. This band is sensitive to targeted knockdown using either shRNA or a gene-trap insertion. The intensity of this band compared with known amounts of purified recombinant Nmnat2 indicates that the abundance of endogenous protein in the brain is very low (∼0.001% of total protein). To the best of our knowledge, this is the first antibody validated to detect endogenous Nmnat2.
Based on its specific expression in the brain, we speculated that Nmnat2 may be further restricted to specific cell types, i.e. neurons or glia. Therefore, we analyzed total lysate from several different cells, including SH-SY5Y, a human neuroblastoma cell line; U87 and U343, which are glioblastoma cell lines; as well as primary cultures. Nmnat2 is expressed in two neuroblastoma cell lines: SH-SY5Y (Fig. 1D), Neuro-2a (Fig. 1A), and primary cortical neurons (Fig. 1E) but is undetectable in primary glia or either of the glioblastoma cell lines tested (Fig. 1D), suggesting that Nmnat2 is more strongly expressed in neurons.
Interestingly, Nmnat2 protein levels increase dramatically upon differentiation of SH-SY5Y (Fig. 1D) and during maturation of primary neuronal cultures (Fig. 1E). This effect was also observed in vivo, as Nmnat2 levels in the brain are relatively low at birth and then peak toward the end of the postnatal period (Fig. 1F). This expression profile is similar to Rab5, which regulates endocytosis and early endosome fusion (31), and synaptobrevin 2, which is a neuronal protein critical for normal synaptic function (32). In contrast, neurofilament (heavy chain) increases dramatically at the end of postnatal development (33), whereas VCP, a chaperone (34), remains steady throughout.
Nmnat2 Is a Palmitoylated Peripheral Membrane Protein
Nmnat2 has previously been reported to localize to the Golgi complex in HeLa cells (8). As it is not predicted to contain a signal sequence or transmembrane domain (or lipid binding domain), we speculated that Nmnat2 localization to this organelle results from peripheral attachment via protein-protein interaction or lipid modification.
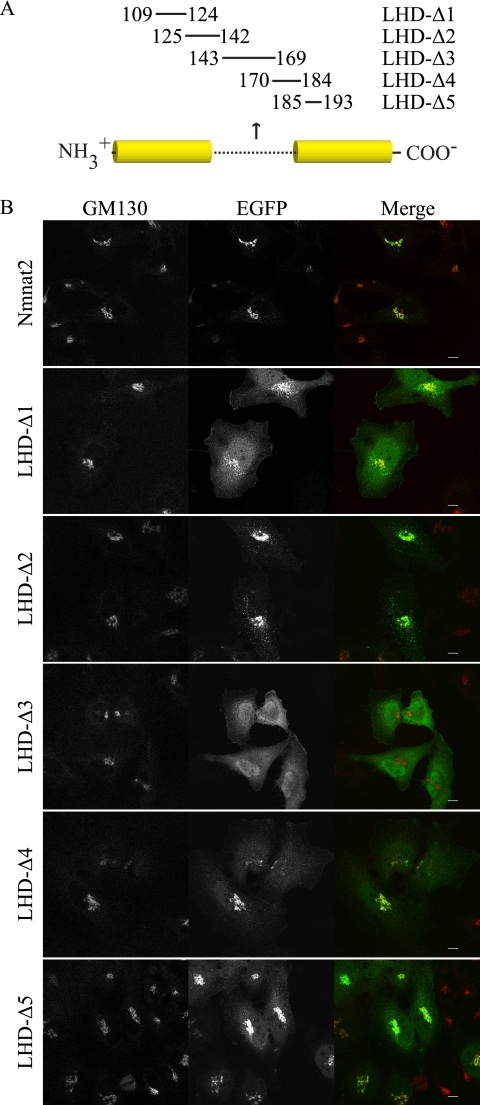
Nmnat1, -2, and -3 consist of a conserved bipartite Rossman fold linked together by a unique stretch of amino acids in the middle of the primary sequence, which is referred to here as the LHD. Given that LHD (Nmnat1) contains the nuclear localization sequence (35), we speculated that LHD (Nmnat2) may contain residues essential for its localization. Therefore, we generated several short deletion mutants (Fig. 2A) and compared their localization relative to GM130, a well characterized Golgi marker (36). We used lentiviruses for gene delivery to reduce potential artifacts due to high level expression resulting from transient transfection. The localization pattern of LHD-Δ2 closely resembles full-length Nmnat2, whereas LHD-Δ1, LHD-Δ4, and LHD-Δ5 retain Golgi localization but have increased cytoplasmic signal (Fig. 2B). This suggests that residues 109–124 (LHD-Δ1) and 170–193 (LHD-Δ4 and -Δ5) may contribute but are not required for Golgi localization. In contrast, LHD-Δ3 does not colocalize with GM130 and shows diffuse cytoplasmic localization.
FIGURE 2.
Residues 143–169 are necessary for Golgi localization and membrane association. A, schematic of human NMNAT2 (NP_055854) indicates residues that have been deleted in a given mutant. A characteristic stretch of residues poorly conserved between Nmnat1, -2, and -3, referred to as the LHD, is illustrated with a dotted line; conserved regions are in solid yellow. B, HeLa cells infected with lentiviruses (MOI ∼ 3) expressing either Nmnat2 or the indicated mutant (with EGFP tag) were fixed and then probed for GM130 to assess Golgi localization. Scale bar, 10 μm.
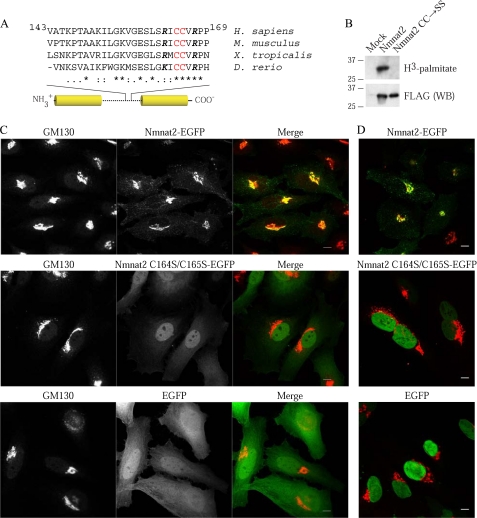
Among the residues deleted in the LHD-Δ3 mutant (amino acids 143–169), we focused on Cys164 and Cys165, given that 1) they are within a highly conserved cluster (Fig. 3A) and 2) cysteines are targets of prenylation and palmitoylation (37). Because prenylation occurs on C-terminal CAAX motifs, we directly investigated the possibility that Nmnat2 is palmitoylated using metabolic labeling. Nmnat2, but not Nmnat2 C164S/C165S, labels with [3H]palmitate, indicating that 1) Nmnat2 is palmitoylated and 2) Cys164 and Cys165 are required for this modification (Fig. 3B).
FIGURE 3.
Nmnat2 is a palmitoylated peripheral membrane protein. A, sequence alignment reveals Cys164 and Cys165 as potential targets of palmitoylation. Palmitoylation lacks a well defined motif but occurs on cysteines frequently with nearby basic residues. Cys164 and Cys165 are highlighted in red, whereas nearby conserved basic residues are in boldface italic. B, HEK293T were transfected and metabolically labeled with [3H]palmitate. Following immunoprecipitation, incorporation of radiolabel was assessed by in-gel fluorography. To assess the efficiency of immunoprecipitation, a 1:10 dilution of eluted protein was analyzed by Western blot (WB). CC, Cys164 and Cys165; SS, Ser164 and Ser165. C, HeLa were infected with lentiviruses (MOI of 3) expressing either Nmnat2-EGFP, Nmnat2 C164S/C165S-EGFP, or EGFP and then fixed and probed for GM130, a well characterized cis-Golgi marker (36). D, infected HeLa cells were permeabilized with digitonin prior to fixation to assess the solubility of wild-type Nmnat2 and C164S/C165S mutant. Note that under these conditions, the nuclear membrane remains intact. Scale bar, 10 μm.
To determine whether palmitoylation is required for Golgi localization, we compared the localization of Nmnat2 C164S/C165S relative to GM130. In contrast to wild-type control, but identical to LHD-Δ3 (see Fig. 2B), Nmnat2 C164S/C165S appears diffuse in the cytoplasm and does not colocalize with GM130 (Fig. 3C). This suggests that Nmnat2 C164S/C165S is soluble. To investigate this possibility, infected cells were treated prior to fixation with digitonin, a mild detergent that selectively permeabilizes the plasma membrane, allowing soluble components of the cytoplasm to rapidly diffuse out of the cell. Under these conditions, cytoplasmic Nmnat2 C164S/C165S and EGFP are washed out, whereas wild-type Nmnat2 is retained in the cell (Fig. 3D).
Although Cys164 and Cys165 are likely targets of palmitoylation, these residues could be necessary for Nmnat2 to acquire and/or maintain its tertiary structure. To test this possibility, we compared the activities of purified recombinant Nmnat2 C164S/C165S to wild-type (in parentheses): kcat = 7.21 ± 0.46 s−1 (0.654 ± 0.50 s−1), KmATP = 112.88 ± 0.14 μm (84.25 ± 0.13 μm) and KmNMN = 2.95 ± 0.40 μm (6.91 ± 1.31 μm). Although it is unclear why Nmnat2 C164S/C165S has a faster turnover rate (kcat), the mutant is fully active, indicating that it is able to maintain its tertiary structure.
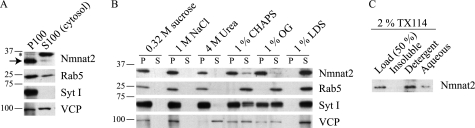
Consistent with membrane localization, most endogenous Nmnat2 sediments with a crude membrane fraction (P100) after subcellular fractionation of mouse brains in the absence of detergent (Fig. 4A). To clarify its interaction with the P100 fraction, we attempted to extract Nmnat2 using a selection of reagents commonly used to solubilize peripheral membrane proteins. VCP is a chaperone that is attached to membranes via protein-protein interaction (34) and is readily soluble in 4 m urea (Fig. 4B). Conversely, Rab5, a small GTPase tethered to membranes via geranylgeranylation (38, 39), and synaptotagmin I, an integral membrane protein (40), and Nmnat2 remain in the pellet fraction. Rab5 is readily soluble in all detergents tested, whereas Nmnat2, VCP, and synaptotagmin I are partially soluble in CHAPS and OG. This is expected for VCP as these detergents lack the necessary polarity to efficiently disrupt protein binding. Similarly, synaptotagmin I also binds numerous proteins at the membrane, which probably accounts for its partial solubility under these conditions. Because endogenous Nmnat2 is resistant to extraction with 4 m urea but slightly soluble in CHAPS and OG, this suggests that 1) palmitoylation recruits Nmnat2 to detergent resistant membranes or 2) Nmnat2 is tightly held at the membrane via protein binding and palmitoylation.
FIGURE 4.
Biochemical characterization of Nmnat2 membrane association. A, mouse brains were homogenized and fractionated in detergent-free buffer as described under “Experimental Procedures.” An asterisk indicates a nonspecific band. B, to characterize the nature of the interaction between Nmnat2 and the membrane, P100 pellets were extracted with either high salt (1 m NaCl), a chemical denaturant (4 m urea), or a selection of detergents: 1% CHAPS (zwitterionic), 1% OG (nonionic), or 1% LDS (anionic). P and S refer to pellet and supernatant collected after a 30-min extraction on ice and a subsequent 100,000 × g spin. C, Nmnat2 is efficiently extracted in 2% Triton X-114 and partitions with detergent following phase separation, as expected for a palmitoylated protein.
Finally, we assessed the behavior of endogenous Nmnat2 in a solution of Triton X-114. Triton X-114 has a similar structure to Triton X-100 but is more hydrophobic and has a lower cloud point. At temperatures above the cloud point, a detergent solution spontaneously separates into aqueous and detergent phases. Because Triton X-114 is a nondenaturing detergent, proteins with hydrophobic surfaces, such as transmembrane and lipid-modified proteins, partition with the detergent phase, whereas hydrophilic proteins remain in the aqueous phase (41). In contrast to its poor solubility in Triton X-100 (data not shown), Nmnat2 is readily soluble in the more hydrophobic Triton X-114, and it is highly enriched in the detergent phase following phase separation (Fig. 4C).
In summary, Nmnat2 localization to the Golgi requires palmitoylation, whereas mutation of Cys164 and Cys165 to serine abolishes this modification and results in a soluble, active protein. Furthermore, despite its lack of a transmembrane domain (or lipid-binding domain) endogenous Nmnat2 has the biochemical characteristics of a membrane protein. Taken together, these data strongly suggest that Nmnat2 is peripherally anchored to membranes via palmitoylation.
Sufficiency of Nmnat2 Low Homology Domain (LHD) for Golgi Localization
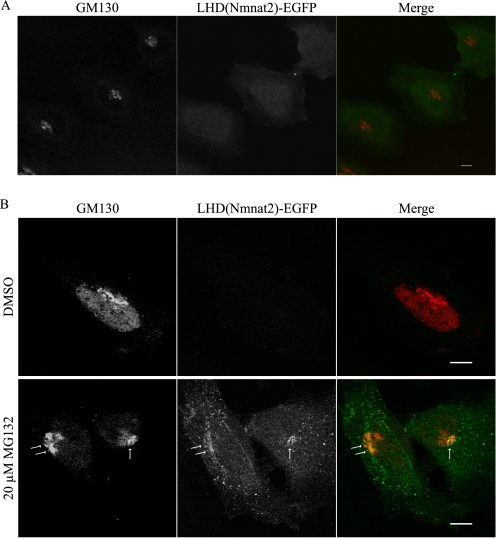
LHD(Nmnat2) was cloned in-frame with EGFP to determine whether this region is sufficient for Golgi localization. Although the fluorescent signal in these cells is low, LHD(Nmnat2)-EGFP shows diffuse cytoplasmic localization (Fig. 5A) similar to Nmnat2 C164S/C165S and LHD-Δ3. Based on this observation, we speculated that LHD(Nmnat2)-EGFP may be unstable. Following treatment with MG132, a proteasome inhibitor, LHD(Nmnat2)-EGFP weakly colocalizes with GM130 (Fig. 5B). This suggests that residues outside the low homology domain are required for stable interaction of Nmnat2 with Golgi.
FIGURE 5.
Nmnat2 low homology domain is insufficient for stable interaction with Golgi. A, HeLa cells infected with lentivirus (MOI ∼ 3) expressing LHD(Nmnat2)-EGFP were fixed and probed for GM130. B, treatment of these cells with a proteasome inhibitor, MG132 (20 μm for 2.5 h), boosts the overall intensity of fluorescent signal revealing weak colocalization between LHD(Nmnat2)-EGFP and GM130 (arrows). Images were taken using the same exposure settings for both dimethyl sulfoxide (DMSO, negative control) and MG132-treated cells. Scale bar, 10 μm.
Overexpression of Nmnat2 Is Toxic to Primary Neurons
Although the antibody described here is useful for detection of endogenous Nmnat2 by immunoblotting, it is unsuitable for immunohistochemistry experiments due to unacceptably high background. All commercially available Nmnat2 antibodies, as well as several other in-house antibodies were also tested but with similar results (data not shown).
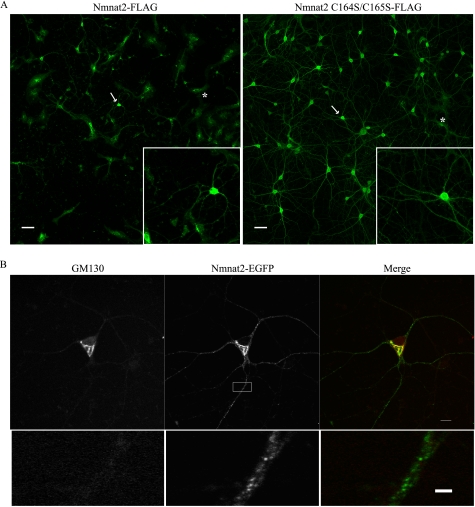
To assess the localization of Nmnat2 in neurons, primary cultures were infected with lentivirus expressing either Nmnat2 or Nmnat2 C164S/C165S. Unexpectedly, overexpression of wild-type Nmnat2 is toxic to neurons, resulting in pervasive cell death (Fig. 6A). In contrast, Nmnat2 C164S/C165S is well tolerated, indicating that increased Nmnat activity alone is not sufficient for toxicity. Furthermore, toxicity is not observed in non-neuronal cells (e.g. HeLa) and is independent of affinity tags (data not shown). This suggests that the levels of endogenous Nmnat2 must be tightly regulated and may account for its low abundance. Nonetheless, prior to the onset of toxicity, exogenous Nmnat2 localizes to Golgi, consistent with its reported localization in HeLa (8), as well as vesicles in dendrites and axons (Fig. 6B).
FIGURE 6.
Exogenous Nmnat2 is toxic to primary neurons. A, primary cortical neurons were infected with lentivirus expressing either Nmnat2-FLAG or Nmnat2 C164S/C165S-FLAG. Approximately 4 days after infection, cultures were fixed and probed with an anti-FLAG antibody. Nmnat2, but not Nmnat2 C164S/C165S, is toxic to neurons, which results in massive neuronal death; arrows indicate neurons shown in the inset. For comparison, a glial cell in each image is marked with an asterisk. Scale bar, 50 μm. B, primary cortical neurons were infected with lentivirus (MOI ∼ 3) expressing Nmnat2-EGFP. Nmnat2 localizes to Golgi (as in HeLa cells) but also dendrites and axons (scale bar, 10 μm). Shown is a magnified view of a dendrite (lower panel; scale bar, 2 μm).
Nmnat2 Localizes to Synaptic Terminals (i.e. synaptosomes)
Palmitoylation is a common mechanism used by neurons to sort peripheral membrane proteins to specific compartments including axons, dendrites, and synapses (42–44). To determine whether endogenous Nmnat2 localizes to any of these structures, mouse brain synaptosomes, which are detached nerve terminals formed during homogenization (45), were purified and fractionated yielding pellets enriched for synaptic vesicles, myelin, synaptic plasma membrane, and mitochondria (Fig. 7A, schematic).
FIGURE 7.
Endogenous Nmnat2 localizes to synaptic terminals. A, schematic of synaptosome purification protocol. Synaptosomes are detached nerve terminals formed during homogenization and contain membranes derived from both pre- and postsynaptic compartments. For P1, S1, P11, S11, P200, and S200, P refers to pellet, and S refers to supernatant, whereas numbers refer to the force of centrifugation for each step (e.g. S200, supernatant resulting from 200,000 × g spin). B, Western blot analysis of fractionated mouse brains; an equal amount of protein was loaded for each sample. An asterisk indicates a nonspecific band. NRX, neurexin; Syb 2, synaptobrevin 2; synap., synaptosome; SV, synaptic vesicles; SPM, synaptic plasma membrane; mito, mitochondria.
Nmnat2 is enriched equally between synaptosome (P11) and cytoplasmic membrane (P200) enriched fractions, unlike β-COP, which in neurons is found exclusively in the cell body (46) and is highly enriched in P200 and cytosol (S200) (Fig. 7B). β-COP is a coatmer protein that continually cycles on/off transport vesicles between the cis-Golgi and endoplasmic reticulum (47–50). In contrast to β-COP, Rab5 localizes to both pre- and postsynaptic terminals (51, 52) and is detected in all fractions. Further purification of synaptosomes (P11) shows that Nmnat2 co-purifies with markers for synaptic vesicles (synaptobrevin 2) (53) and synaptic plasma membrane (α/β-neurexins) (24). Note that α/β-neurexins only minimally contaminate LP2 (synaptic vesicle enriched fraction), suggesting that this fraction is relatively free of plasma membrane. Because Nmnat2 is not detected at the plasma membrane when overexpressed (e.g. Figs. 3C, top panel, and 6B), its co-purification with α/β-neurexins in fraction B may indicate the presence of dendrite-derived endosomes, consistent with the presence of Rab5 in this fraction as well. Taken together with our observation that Nmnat2-EGFP localizes to vesicles in dendrites and axons (Fig. 6B), these data suggest that endogenous Nmnat2 localizes to vesicles in synaptic terminals.
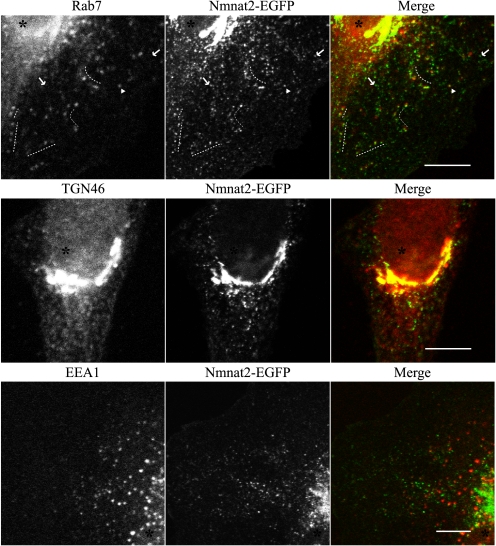
Nmnat2 Localizes to Rab7-containing Late Endosomes and trans-Golgi Network in HeLa Cells
Under normal growth conditions, when overexpressed at low levels via lentiviral infection (MOI of 3), Nmnat2 localizes not only to Golgi but also to cytoplasmic vesicles (e.g. Figs. 3C, top panel, and 6B). Localization is independent of the presence of an affinity tag (data not shown). A subset of these structures colocalize with Rab7 (Fig. 8), a late endosome marker (54). In some instances, Nmnat2 and Rab7 are adjacent or only partially co-localize. Nmnat2 also colocalizes with perinuclear TGN46, a trans-Golgi network marker (55). In contrast, there is no significant overlap with EEA1, an early endosome marker (56), indicating that Nmnat2 localizes to specific compartments in the endosomal pathway. Consistent with its presence at synaptic terminals, these results suggest that Nmnat2 is transported to these compartments via an endosomal pathway.
FIGURE 8.
Nmnat2 localizes to Rab7 late endosomes and trans-Golgi network in HeLa cells. HeLa cells were infected with lentiviruses (MOI of 3) expressing Nmnat2-EGFP and then fixed and probed for Rab7, a late endosome marker (67) (top panel); TGN46, a trans-Golgi network marker (55) (middle panel); or EEA1, an early endosome marker (56) (bottom panel). Dotted lines indicate regions in the cytoplasm where Rab7 and Nmnat2 are closely associated. Arrows indicate where Nmnat2 does not co-localize with Rab7. Arrowhead indicates specific vesicle where Nmnat2 colocalizes with Rab7. An asterisk marks the nucleus in each cell. Scale bar, 10 μm.
DISCUSSION
In this report, we have analyzed the expression and localization of Nmnat2 and characterized its interaction with cellular membranes. To analyze Nmnat2 protein expression, we generated and validated a polyclonal antibody against full-length protein. Of all antibodies tested, including in-house and all commercially available antibodies, the antibody described here had the best specificity and sensitivity. Nmnat2 is detected exclusively in neurons with expression peaking shortly after birth suggesting that these cells require additional NAD synthesis during early postnatal development. Although this coincides with an intense period of synaptogenesis (57), the reason for an increase in expression during this time remains to be determined.
Overexpression of Nmnat1, -2, or -3 in neurons has been demonstrated to slow the rate of axon degeneration in vivo and in vitro (58–62). Although the underlying mechanism remains to be elucidated, axon degeneration precedes neuronal cell body death in many neurodegenerative diseases including Alzheimer and Parkinson diseases, suggesting that manipulation of NAD synthesis pathways may provide new therapeutic targets (63).
Paradoxically, overexpression of Nmnat2 is toxic to primary neurons (but not other cell types such as HeLa), whereas Nmnat2 C164S/C165S is well tolerated, indicating that excess Nmnat activity alone is insufficient to cause toxicity. One possible explanation for this phenotype is that exogenous Nmnat2, but not Nmnat2 C164S/C165S, forms aggregates that are toxic to neurons (but not other cell types). However, this scenario is unlikely, given the highly reducing environment of the cytoplasm. Alternatively, exogenous Nmnat2 may form a spurious interaction with one or more proteins at the membrane. It has recently been suggested that such interactions are a common cause of toxicity resulting from protein overexpression (64). The lack of gross toxicity in non-neuronal cells may therefore be due to the absence (or low abundance) of a specific interacting protein. Another possibility is that exogenous Nmnat2 in neurons binds to a bona fide protein target, but toxicity results from overstimulation of an existing metabolic or signaling pathway.
Using superior cervical ganglia cultures, Gilley and Coleman (65) have recently shown that endogenous Nmnat2 localizes to axons and that Nmnat2-EGFP particles are transported via fast axonal transport. Here, we show prominent localization of Nmnat2-EGFP to dendrites and axons and that endogenous Nmnat2 localizes to synaptic terminals. Consistent with our observation, Zhai et al. (4) report that Drosophila Nmnat localizes to synaptic puncta, suggesting a conserved function for Nmnat at synapses.
While this paper was being reviewed, Lau et al. (66) published an analysis of the low homology domain (LHD) of Nmnat1, -2, and -3 (which they refer to as isoform-specific targeting and interaction domains, i.e. ISTID1, ISTID2, and ISTID3). These authors report that Nmnat2 is palmitoylated on Cys164 and Cys165. We also show that Nmnat2 is palmitoylated but conclude only that Cys164 and Cys165 are required for this modification. Furthermore, we report that endogenous Nmnat2 has the biochemical characteristics of a membrane protein despite lacking a transmembrane domain or lipid binding domain; and Nmnat2 is difficult to solubilize, suggesting that it is either 1) recruited to detergent-resistant membranes or 2) tightly held at the membrane via protein binding and palmitoylation. Additionally, Lau et al. (66) report that expression of FLAG-ISTID2-EGFP, consisting of residues 108–190, is sufficient for Golgi localization. In contrast, we find that LHD(Nmnat2), defined as residues 109–193, is insufficient to stably interact with Golgi membranes. Future experiments will investigate the effect of the low homology domain on overall stability of Nmnat2.
It remains to be determined what specific role Nmnat2 plays in neuronal NAD metabolism, and specifically, what advantages its localization and membrane association provide. However, the data presented here provide a solid foundation from which these questions can better be addressed.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AG029547. This work was also supported by the Ruth K. Broad Biomedical Research Foundation and the Ted Nash Long Life Foundation.
- NMN
- nicotinamide mononucleotide
- Nmnat
- NMN adenylyltransferase
- VCP
- valosin-containing protein
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- MOI
- multiplicity of infection
- LDS
- lithium dodecyl sulfate
- OG
- octyl glucoside
- LHD
- low homology domain.
REFERENCES
- 1.Magni G., Amici A., Emanuelli M., Raffaelli N., Ruggieri S. (1999) Adv. Enzymol. Relat. Areas Mol. Biol. 73, 135–182 [DOI] [PubMed] [Google Scholar]
- 2.Magni G., Orsomando G., Raffelli N., Ruggieri S. (2008) Front. Biosci. 13, 6135–6154 [DOI] [PubMed] [Google Scholar]
- 3.Lau C., Niere M., Ziegler M. (2009) Front. Biosci. 14, 410–431 [DOI] [PubMed] [Google Scholar]
- 4.Zhai R. G., Cao Y., Hiesinger P. R., Zhou Y., Mehta S. Q., Schulze K. L., Verstreken P., Bellen H. J. (2006) PLoS Biol. 4, e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sood R., Bonner T. I., Makalowska I., Stephan D. A., Robbins C. M., Connors T. D., Morgenbesser S. D., Su K., Faruque M. U., Pinkett H., Graham C., Baxevanis A. D., Klinger K. W., Landes G. M., Trent J. M., Carpten J. D. (2001) Genomics 73, 211–222 [DOI] [PubMed] [Google Scholar]
- 6.Raffaelli N., Sorci L., Amici A., Emanuelli M., Mazzola F., Magni G. (2002) Biochem. Biophys. Res. Commun. 297, 835–840 [DOI] [PubMed] [Google Scholar]
- 7.Yalowitz J. A., Xiao S., Biju M. P., Antony A. C., Cummings O. W., Deeg M. A., Jayaram H. N. (2004) Biochem. J. 377, 317–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger F., Lau C., Dahlmann M., Ziegler M. (2005) J. Biol. Chem. 280, 36334–36341 [DOI] [PubMed] [Google Scholar]
- 9.Hogeboom G. H., Schneider W. C. (1952) J. Biol. Chem. 197, 611–620 [PubMed] [Google Scholar]
- 10.Zhang X., Kurnasov O. V., Karthikeyan S., Grishin N. V., Osterman A. L., Zhang H. (2003) J. Biol. Chem. 278, 13503–13511 [DOI] [PubMed] [Google Scholar]
- 11.Belenky P., Bogan K. L., Brenner C. (2007) Trends Biochem. Sci. 32, 12–19 [DOI] [PubMed] [Google Scholar]
- 12.Finkel T., Deng C. X., Mostoslavsky R. (2009) Nature 460, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraus W. L. (2008) Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graeff R., Liu Q., Kriksunov I. A., Kotaka M., Oppenheimer N., Hao Q., Lee H. C. (2009) J. Biol. Chem. 284, 27629–27636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spies T. D., Bean W. B., Ashe W. F. (1939) Ann. Int. Med. 12, 1830–1844 [Google Scholar]
- 16.Hegyi J., Schwartz R. A., Hegyi V. (2004) Int. J. Dermatol. 43, 1–5 [DOI] [PubMed] [Google Scholar]
- 17.Salmon P., Trono D. (2006) Curr. Protoc. Neurosci. 37, 4.21.1–4.21.24 [DOI] [PubMed] [Google Scholar]
- 18.Sastry L., Johnson T., Hobson M. J., Smucker B., Cornetta K. (2002) Gene Ther. 9, 1155–1162 [DOI] [PubMed] [Google Scholar]
- 19.Encinas M., Iglesias M., Liu Y., Wang H., Muhaisen A., Ceña V., Gallego C., Comella J. X. (2000) J. Neurochem. 75, 991–1003 [DOI] [PubMed] [Google Scholar]
- 20.Bonner W. M., Laskey R. A. (1974) Eur. J. Biochem. 46, 83–88 [DOI] [PubMed] [Google Scholar]
- 21.Sheffield P., Garrard S., Derewenda Z. (1999) Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 22.Kurnasov O. V., Polanuyer B. M., Ananta S., Sloutsky R., Tam A., Gerdes S. Y., Osterman A. L. (2002) J. Bacteriol. 184, 6906–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brusca J. S., Radolf J. D. (1994) Methods Enzymol. 228, 182–193 [DOI] [PubMed] [Google Scholar]
- 24.Butz S., Fernandez-Chacon R., Schmitz F., Jahn R., Südhof T. C. (1999) J. Biol. Chem. 274, 18290–18296 [DOI] [PubMed] [Google Scholar]
- 25.Maximov A., Shin O. H., Liu X., Südhof T. C. (2007) J. Cell Biol. 176, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittaker V. P., Michaelson I. A., Kirkland R. J. (1964) Biochem. J. 90, 293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones D. H., Matus A. I. (1974) Biochim. Biophys. Acta 356, 276–287 [DOI] [PubMed] [Google Scholar]
- 28.Abuin A., Hansen G. M., Zambrowicz B. (2007) Handb. Exp. Pharmacol. 178, 129–147 [DOI] [PubMed] [Google Scholar]
- 29.Voss A. K., Thomas T., Gruss P. (1998) Dev. Dyn. 212, 258–266 [DOI] [PubMed] [Google Scholar]
- 30.Voss A. K., Thomas T., Gruss P. (1998) Dev. Dyn. 212, 171–180 [DOI] [PubMed] [Google Scholar]
- 31.Stenmark H. (2009) Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 32.Schoch S., Deák F., Königstorfer A., Mozhayeva M., Sara Y., Südhof T. C., Kavalali E. T. (2001) Science 294, 1117–1122 [DOI] [PubMed] [Google Scholar]
- 33.Shaw G., Weber K. (1982) Nature 298, 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q., Song C., Li C. C. (2004) J. Struct. Biol. 146, 44–57 [DOI] [PubMed] [Google Scholar]
- 35.Schweiger M., Hennig K., Lerner F., Niere M., Hirsch-Kauffmann M., Specht T., Weise C., Oei S. L., Ziegler M. (2001) FEBS Lett. 492, 95–100 [DOI] [PubMed] [Google Scholar]
- 36.Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. (1995) J. Cell Biol. 131, 1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Resh M. D. (2006) Nat. Chem. Biol. 2, 584–590 [DOI] [PubMed] [Google Scholar]
- 38.Kinsella B. T., Maltese W. A. (1991) J. Biol. Chem. 266, 8540–8544 [PubMed] [Google Scholar]
- 39.Kinsella B. T., Maltese W. A. (1992) J. Biol. Chem. 267, 3940–3945 [PubMed] [Google Scholar]
- 40.Perin M. S., Fried V. A., Mignery G. A., Jahn R., Südhof T. C. (1990) Nature 345, 260–263 [DOI] [PubMed] [Google Scholar]
- 41.Bordier C. (1981) J. Biol. Chem. 256, 1604–1607 [PubMed] [Google Scholar]
- 42.Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R., 3rd, Davis N. G., El-Husseini A. (2008) Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang K., El-Husseini A. (2005) Curr. Opin. Neurobiol. 15, 527–535 [DOI] [PubMed] [Google Scholar]
- 44.El-Husseini Ael-D., Craven S. E., Brock S. C., Bredt D. S. (2001) J. Biol. Chem. 276, 44984–44992 [DOI] [PubMed] [Google Scholar]
- 45.Whittaker V. P. (1993) J. Neurocytol. 22, 735–742 [DOI] [PubMed] [Google Scholar]
- 46.Sannerud R., Marie M., Nizak C., Dale H. A., Pernet-Gallay K., Perez F., Goud B., Saraste J. (2006) Mol. Biol. Cell 17, 1514–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffiths G., Pepperkok R., Locker J. K., Kreis T. E. (1995) J. Cell Sci. 108, 2839–2856 [DOI] [PubMed] [Google Scholar]
- 48.Oprins A., Duden R., Kreis T. E., Geuze H. J., Slot J. W. (1993) J. Cell Biol. 121, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duden R., Griffiths G., Frank R., Argos P., Kreis T. E. (1991) Cell 64, 649–665 [DOI] [PubMed] [Google Scholar]
- 50.Duden R. (2003) Mol. Membr. Biol. 20, 197–207 [DOI] [PubMed] [Google Scholar]
- 51.de Hoop M. J., Huber L. A., Stenmark H., Williamson E., Zerial M., Parton R. G., Dotti C. G. (1994) Neuron 13, 11–22 [DOI] [PubMed] [Google Scholar]
- 52.Fischer von Mollard G., Stahl B., Walch-Solimena C., Takei K., Daniels L., Khoklatchev A., De Camilli P., Südhof T. C., Jahn R. (1994) Eur. J. Cell Biol. 65, 319–326 [PubMed] [Google Scholar]
- 53.Takamori S., Holt M., Stenius K., Lemke E. A., Grønborg M., Riedel D., Urlaub H., Schenck S., Brügger B., Ringler P., Müller S. A., Rammner B., Gräter F., Hub J. S., De Groot B. L., Mieskes G., Moriyama Y., Klingauf J., Grubmüller H., Heuser J., Wieland F., Jahn R. (2006) Cell 127, 831–846 [DOI] [PubMed] [Google Scholar]
- 54.Feng Y., Press B., Wandinger-Ness A. (1995) J. Cell Biol. 131, 1435–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ponnambalam S., Girotti M., Yaspo M. L., Owen C. E., Perry A. C., Suganuma T., Nilsson T., Fried M., Banting G., Warren G. (1996) J. Cell Sci. 109, 675–685 [DOI] [PubMed] [Google Scholar]
- 56.Mu F. T., Callaghan J. M., Steele-Mortimer O., Stenmark H., Parton R. G., Campbell P. L., McCluskey J., Yeo J. P., Tock E. P., Toh B. H. (1995) J. Biol. Chem. 270, 13503–13511 [DOI] [PubMed] [Google Scholar]
- 57.Knaus P., Betz H., Rehm H. (1986) J. Neurochem. 47, 1302–1304 [DOI] [PubMed] [Google Scholar]
- 58.Conforti L., Wilbrey A., Morreale G., Janeckova L., Beirowski B., Adalbert R., Mazzola F., Di Stefano M., Hartley R., Babetto E., Smith T., Gilley J., Billington R. A., Genazzani A. A., Ribchester R. R., Magni G., Coleman M. (2009) J. Cell Biol. 184, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beirowski B., Babetto E., Gilley J., Mazzola F., Conforti L., Janeckova L., Magni G., Ribchester R. R., Coleman M. P. (2009) J. Neurosci. 29, 653–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sasaki Y., Araki T., Milbrandt J. (2006) J. Neurosci. 26, 8484–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan T., Feng Y., Zheng J., Ge X., Zhang Y., Wu D., Zhao J., Zhai Q. (2010) Neurochem. Int. 56, 101–106 [DOI] [PubMed] [Google Scholar]
- 62.Press C., Milbrandt J. (2008) J. Neurosci. 28, 4861–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raff M. C., Whitmore A. V., Finn J. T. (2002) Science 296, 868–871 [DOI] [PubMed] [Google Scholar]
- 64.Vavouri T., Semple J. I., Garcia-Verdugo R., Lehner B. (2009) Cell 138, 198–208 [DOI] [PubMed] [Google Scholar]
- 65.Gilley J., Coleman M. P. (2010) PLoS Biol. 8, e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lau C., Dölle C., Gossmann T. I., Agledal L., Niere M., Ziegler M. (2010) J. Biol. Chem. 285, 18868–18876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng G., Mellor R. H., Bernstein M., Keller-Peck C., Nguyen Q. T., Wallace M., Nerbonne J. M., Lichtman J. W., Sanes J. R. (2000) Neuron 28, 41–51 [DOI] [PubMed] [Google Scholar]